In this issue of Blood, Mayer et al1 analyze hematopoietic stem cells (HSCs) isolated from mice with mutations in the cell-cycle kinase cyclin dependent-kinase 6 (CDK6) to study how the kinase-inactivated form of CDK6 regulates HSC self-renewal, differentiation, and quiescence. By carefully quantifying HSC function and gene expression in CDK6 wild-type, knockout (CDK6−/−), and kinase-inactivated (CDK6KM/KM) cells, they discovered that HSCs with the kinase-inactivated CDK6 retain stem cell properties, and loss of CDK6 impairs HSC self-renewal. CDK6, besides its canonical role in cell cycle regulation, thus also regulates gene expression programs required for HSC maintenance and activation.
HSCs regenerate the blood system for the entire life of an organism. Their capacity to self-renew and differentiate long term is protected by their ability to enter a reversible state of quiescence that stops cell cycle progression and prevents proliferative stress. Quiescent HSCs reside in the G0 phase of the cell cycle and reenter a cycling state to replenish the bone marrow when instructed by environmental signals. Although the cell cycle was identified decades ago, a detailed understanding of how HSCs coordinate cell cycle progression and gene expression to regulate self-renewal, differentiation, and quiescence to ensure lifelong hematopoiesis is lacking.
CDK6 regulates HSC cell cycle entry and progression and is activated by binding of cyclin D. Active CDK6 then phosphorylates retinoblastoma, which induces E2F transcription and mediates cell cycle progression.2 Besides its role in cell cycle regulation, CDK6 also interacts with transcription factors and works as a transcriptional cofactor. In HSCs, previous reports showed that CDK6 overexpression increases long-term function, and its loss inhibits cell cycle entry and activation.3
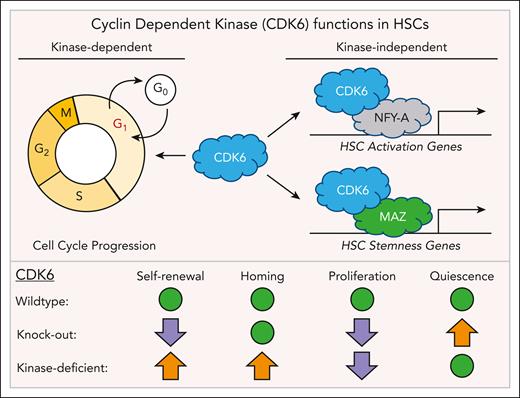
Using in vitro colony forming assays and in vivo serial transplants to assess HSC function, single-cell RNA sequencing (scRNAseq), and transcription factor motif analysis, Mayer et al show that the kinase-independent function of CDK6 is critical to maintain the gene expression programs for HSC quiescence and stemness. They identified CDK6-MAZ and CDK6-NFY-A interactions as critical regulators of gene expression regulating HSC fate, and they provide insights into how HSCs coordinate cell cycle progression and fate decisions to balance self-renewal and differentiation (see figure).
Kinase-deficient CDK6 promotes HSC self-renewal and homing. Besides its canonical role in the regulation of cell cycle progression, CDK6 also acts as a transcriptional cofactor and interacts with the transcription factors NFY-A and MAZ to regulate genes required for HSC activation and stem cell maintenance. HSCs expressing the kinase-deficient form of CDK6 have increased self-renewal and homing potential (orange arrows) compared with wild-type and CDK6 knockout HSCs. CDK6 knockout HSCs have increased quiescence and reduced self-renewal potential (purple arrows) compared with wild-type HSCs. Green dots indicate which stem cell properties of CDK6 kinase-deficient and knockout HSCs have not changed compared with wild-type cells.
Kinase-deficient CDK6 promotes HSC self-renewal and homing. Besides its canonical role in the regulation of cell cycle progression, CDK6 also acts as a transcriptional cofactor and interacts with the transcription factors NFY-A and MAZ to regulate genes required for HSC activation and stem cell maintenance. HSCs expressing the kinase-deficient form of CDK6 have increased self-renewal and homing potential (orange arrows) compared with wild-type and CDK6 knockout HSCs. CDK6 knockout HSCs have increased quiescence and reduced self-renewal potential (purple arrows) compared with wild-type HSCs. Green dots indicate which stem cell properties of CDK6 kinase-deficient and knockout HSCs have not changed compared with wild-type cells.
Using flow cytometry and scRNAseq of wild-type CDK6−/− and CDK6KM/KM bone marrow, they first show that CDK6−/− and CDK6KM/KM mice have more HSCs. Despite similar HSC numbers, the transcriptomes of CDK6KM/KM and CDK6−/− differ; CDK6KM/KM HSCs have a lower proliferation gene expression signature, and CDK6−/− HSCs express more genes associated with quiescence, showing that CDK6 can regulate HSC gene expression independently of its kinase. To obtain further insights into how CDK6 regulates HSC fates, they then stressed wild-type, CDK6−/− and CDK6KM/KM mice by injecting polyinosinic:polycytidylic acid to induce inflammation and HSC cell cycle entry. Activated HSCs were then isolated and serially replated in vitro to assess cell cycle progression, colony-forming potential, and their ability to differentiate. Although both CDK6−/− and CDK6KM/KM HSCs enter the cell cycle slower than wild-type HSCs, only CDK6KM/KM, but not CDK6−/− HSCs, could form colonies with myeloid and lymphoid cells. CDK6 kinase-deficient HSCs are thus functional, which the authors further validated in vivo by serially transplanting CDK6−/− and CDK6KM/KM HSCs into recipient mice. These data show that CDK6KM/KM HSCs have enhanced repopulation and homing potential compared with wild-type and CDK6−/− HSCs. Next, they analyzed the gene expression profile of serially transplanted HSCs and discovered that CDK6’s kinase-independent functions are critical in maintaining quiescent gene expression signatures. Transcription factor motif analysis validated by coimmunoprecipitation and proximity ligation assays further show that CDK6KM/KM interacts with the transcription factors NFY-A and MAZ, to regulate the expression of HSC activation and stemness genes, respectively. Finally, using scRNAseq, serial transplants, and replating assays, the authors show that CDK6 inhibition using the kinase inhibitor palbociclib improves HSC function in normal mouse and human hematopoietic stem and progenitor cells. Taken together, CDK6 regulates HSC fates independently of its kinase domain and its canonical role in cell cycle regulation. These findings provide new insights into how HSCs coordinate cell cycle progression and fate decisions and suggest that CDK6’s kinase-independent functions are at least partially mediated by interactions with the transcription factors NFY-A and MAZ.
Although this study provides novel insights into how HSCs coordinate cell cycle progression and fate decisions, many questions about the precise molecular mechanism remain. For instance, how exactly do CDK6-NFY-A and CDK6-MAZ regulate HSC self-renewal and differentiation? It was previously shown that human HSCs express little or no CDK6 and enter the cell cycle slower than CDK6-positive cells after mitogenic stimulation.3 If these studies are correct and CDK6 is absent in human HSCs, how can CDK6 drive gene expression programs required for stem cell maintenance? Although the data of both studies are compelling, it is not intuitively clear how to reconcile these observations.
Here Mayer et al provide gene expression data from wild-type CDK6−/− and CDK6KM/KM HSCs with lists of differentially expressed genes, including Runx1, Stat3, and Mlec3. How these changes translate into HSC fate decisions is not addressed. Although scRNAseq has become an invaluable tool for understanding biological systems, it is important to remember that it provides only snapshots. As HSCs are heterogeneous, cycle asynchronously, and can acquire different fates,4 important information about the molecular dynamics in single cells is lost.5 scRNAseq alone is thus not sufficient to understand what molecular mechanisms lead to changes in cell fates as “only” changes in gene expression are detected.6 However, scRNAseq is useful for generating hypotheses. Mayer et al thus provide important clues for future mechanistic studies by linking NFY-A7 and MAZ,8 known regulators of HSCs function, to the cell cycle regulator CDK6.
More work, however, is required to better understand how HSCs integrate cell cycle control and fate decisions. In general, the field needs to move beyond reporting changes in gene expression, and alternative and/or complementary approaches are needed to fully recapitulate the molecular dynamics regulating cell behavior and fates in single cells over time. Advanced long-term live-cell imaging,9,10 and quantifying the response of individual cells to precise spatiotemporal manipulation, can provide new insights into how HSCs coordinate cell cycle progression and fate decisions.
Conflict-of-interest disclosure: D.L. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal