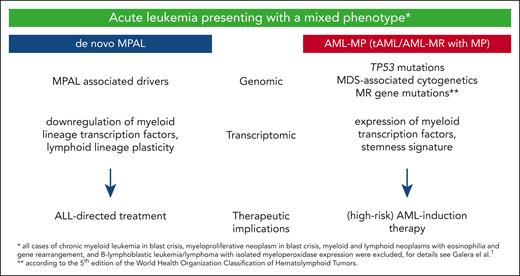
In this issue of Blood, Galera and colleagues demonstrate that de novo mixed phenotype acute leukemia (MPAL) is biologically distinct from acute myeloid leukemia with a mixed phenotype (AML-MP) and outlined the potential clinical implications of this distinction.1 By comprehensive clinical, immunophenotypic, genomic, and transcriptomic characterization of a large cohort of 100 acute leukemia cases presenting with a mixed phenotype (MP) as defined by the revised 4th edition of the World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues,2 they devised a novel genomic classification separating MPAL from therapy-related AML (tAML) and myelodysplasia-related AML (AML-MR) presenting as AML-MP. Although the genomic differences in leukemia biology were also nicely reflected on the transcriptomic level, the authors’ study showed AML-MP has a high-risk AML phenotype, whereas MPAL was characterized by downregulation of critical myeloid lineage transcription factors, lymphoid lineage plasticity, and good response to acute lymphoblastic leukemia (ALL)-directed treatment, which was not negatively impacted by the presence of RUNX1 mutations or a complex karyotype (see figure).
Differences of MPAL and AML-MP according to Galera et al. MDS, myelodysplasia.
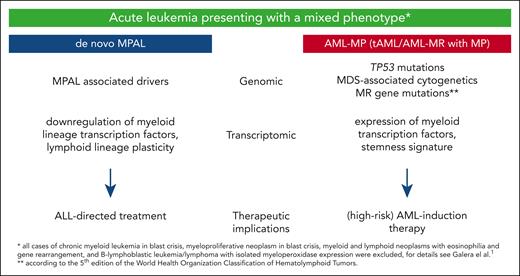
Differences of MPAL and AML-MP according to Galera et al. MDS, myelodysplasia.
As treatment regimens for AML and ALL are markedly different, a more accurate prediction whether an acute leukemia presenting with an MP is more likely to behave like an AML or ALL is of the utmost importance. However, in 2025, even with the availability of the revised WHO classification3 and the International Consensus Classification (ICC),4 an acute leukemia with ambiguous lineage still poses a major diagnostic and therapeutic challenge, especially with regard to the subclassification of MPAL.5 Thus, a refined classification based on driver mutations and accounting for distinct mutations mainly associated with a myeloid phenotype might help to further disentangle the leukemic strands. Ultimately, this should help to better explain the outcome differences reported for MPAL that range from 36% to 80%.5
Recently, the poor prognostic outcome of tAML was clearly linked to the underlying genomic aberrations found and not the tAML per se.6 Analysis of a large tAML cohort showed that tAML is not an independent risk factor for overall survival and concluded that therapeutic algorithms in tAML should be guided by European LeukemiaNet genetic risk7 rather than classification as tAML alone.6 This supports the revised WHO and ICC 2022 classifications,3,4 which include tAML as diagnostic qualifier rather than a separate subcategory. In the same manner, the work by Galera et al supports that MP acute leukemia associated with myeloid drivers should rather be treated as an AML, and, conversely, MPAL cases with a more lymphoid determined phenotype should receive ALL therapy.
These data do support the findings of an earlier comprehensive omics study that used integrative genomic analysis of 31 MPAL samples and compared the molecular profiles from the samples to other defined hematologic malignancies.8 Of the 31 cases, 18 MPAL cases received a therapy that matched with their methylation-defined phenotype (ie, an AML-directed therapy for an AML-like MPAL and an ALL-directed therapy for an ALL-like disease, respectively), 9 patients received unmatched therapy, and the remainder received other combination therapy. Despite the small sample numbers, it could be demonstrated that patients who received matched therapies were more likely to achieve a complete remission than patients receiving unmatched therapy.8
Accordingly, more refined analyses including not only comprehensive genomic and transcriptomic profiling but also epigenomic information and more refined single-cell analyses might help to better guide MPAL therapy. However, considering the work by Galera and colleagues, single-cell profiling studies have to be carefully evaluated, and results cautiously interpreted, especially as the number of samples analyzed is usually still small. For example, a recent retrospective study analyzing 14 MPAL cases showed many mutational signatures that might actually reflect AML-MP cases being driven by TP53 and MR gene mutations.9 Thus, it is not surprising that the respective expression profiles were not really associated with the immunophenotype, but rather the underlying genomic aberrations. Similarly, the comparative mutational analysis of MPAL in recently published series, which were nicely put together by Weinberg,10 also have to be revisited and interpreted in the light of the current study, as AML-MP cases might also be contained in these cohorts.
In summary, the study by Galera and colleagues provides a straightforward algorithm that advances the current understanding of acute leukemia with MP and can help to more easily direct the best treatment decisions in the real-world setting. However, for the MPAL cases without diagnostic gene markers, in the future epigenomic or transcriptomic profiling (eg, through novel long-read sequencing approaches) might help to further distinguish ALL from AML-like MPALs. Finally, in addition to more retrospective studies confirming the current observations, trials prospectively evaluating omics-driven diagnostic approaches are warranted to ultimately “unmix” MPAL and improve treatment outcome by using strategies that target the underlying leukemia biology (see figure).
Conflict-of-interest disclosure: L.B. received honoraria from AbbVie, Amgen, Astellas, Bristol Myers Squibb, Celgene, Daiichi Sankyo, Gilead, Hexal, Janssen, Jazz Pharmaceuticals, Menarini, Novartis, Otsuka, Pfizer, Roche, and Sanofi and research funding from Bayer and Jazz Pharmaceuticals.