Abstract
Although small deletions, splice site abnormalities, missense, and nonsense mutations have been identified in patients with factor VII deficiency, there have been no reports of mutations in the factor VII promoter. We investigated a girl with factor VII levels that were less than 1% of normal in association with a severe bleeding diathesis. The patient is homozygous for a T to G transversion that occurs 61 bp before the translation start site. This nucleotide is in a sequence that is an hepatocyte nuclear factor 4 (HNF-4) binding site within the factor VII promoter (ACTTTG Æ → ACGTTG). Using gel mobility shift assays, we show that the mutation disrupts the binding of HNF-4 to its cognate binding site. In growth hormone reporter gene assays, the activity of a plasmid containing the mutant promoter was 6.7% of the wild-type promoter plasmid. Although HNF-4 was able to transactivate the wild-type factor VII promoter 5.4-fold in HeLa cells, no transactivation could be shown with the mutant promoter. These findings indicate that HNF-4 exerts a major positive regulatory effect on factor VII expression and provides in vivo evidence that binding of this transcription factor is critical for normal factor VII expression.
HUMAN FACTOR VII is a vitamin K-dependent glycoprotein that circulates in blood as an inactive zymogen. It plays a critical role in the initiation of coagulation by binding to tissue factor, which is exposed on vascular injury. This interaction enables factor VII to be rapidly activated to factor VIIa; the factor VIIa-tissue factor complex then initiates blood coagulation by activating factor IX as well as factor X.1,2
The human factor VII gene is located on chromosome 13q34,3 2.8 kb upstream from the coagulation factor X gene.4 It contains 9 exons separated by 8 introns and the entire sequence spans about 12.8 kb.5 During the last 5 years, more than 20 different mutations have been described in patients with factor VII deficiency.6 Most have been missense mutations, but nonsense mutations, small deletions, and splice site abnormalities have also been identified.7
Several groups have recently identified and characterized essential regulatory elements in the 5′-flanking region of the factor VII gene.8-10 Unlike many eukaryotic promoters, it lacks TATA and CAAT boxes. The absence of a CAAT box contrasts with the promoter structure of factor IX and factor X, two closely related vitamin K-dependent coagulation proteins in which a CAAT box is critical for promoter activity.11,12 The major transcription start site for factor VII is located approximately 50 bp upstream from the first base of the codon for the initiation methionine (+1) and is in close proximity to the binding sites of transcription factors Sp1 and hepatocyte nuclear factor 4 (HNF-4), which bind at sites that include nucleotides −100 to −94 and −63 to −58 (ACTTTG), respectively.9 10
We have investigated the molecular mechanism underlying factor VII deficiency in a young girl with a severe bleeding diathesis who is homozygous for a T to G transversion at nucleotide −61 (ACGTTG) in the factor VII promoter. Our data show that this mutation disrupts HNF-4 binding and results in a significant reduction in factor VII promoter activity.
MATERIALS AND METHODS
Clinical history.The proband is a 6-year-old American girl with a severe bleeding diathesis caused by factor VII deficiency. Her parents are of French Canadian ancestry, and there is no familial history of bleeding or known consanguinity. She was noted to have blood in her stools at about 2 months of age and bruising with minimal trauma at 4 months. At 8 months, she presented with a spontaneous cerebral hemorrhage in the right frontal lobe which required neurosurgical decompression after factor VII replacement was administered. She was placed on chronic treatment with factor VII concentrate (Immuno AG, Vienna, Austria) three times per week in the postoperative period and recovered without neurologic sequelae. An attempt to discontinue factor VII concentrate prophylaxis at age 10 months resulted in the development of ecchymoses and a traumatic mouth hemorrhage. She developed spontaneous bleeding in the vitreous of her right eye at 2 years of age. Six months later she presented with a seizure, but a CT scan of the brain was normal. She was placed on carbamazepine, which was effective in controlling seizure activity. Because of the severity of the bleeding, the patient resumed factor VII concentrate treatment at approximately 2 years of age and she continues to receive infusions thrice weekly.
Factor VII assays.Plasma factor VII coagulant activity (VII:C) levels were measured on citrated plasma by one-stage clotting assay using Automated Simplastin (Organon Teknika Corp, Durham, NC). The levels of plasma factor VII antigen (VII:Ag) were determined with an enzyme-linked immunoabsorbent assay (American Bioproducts Co, Parsippany, NJ). A normal pool was constructed by mixing equal volumes of plasma from greater than 30 healthy control subjects.
DNA isolation, in vitro amplification using polymerase chain reaction (PCR), cloning, and sequencing.DNA was purified from leukocyte nuclei obtained from whole blood by standard techniques.13 Two PCR fragments encoding the 5′-flanking region were generated, a 514 bp fragment from nucleotides −416 to +98 and a 247 bp fragment from nucleotides −149 to +98.5 The methods used for amplifying the exons of the factor VII gene, and for subcloning PCR fragment inserts have been described in detail previously.14 The inserts were sequenced by the dideoxy chain termination method15 on an Applied Biosystems 373A DNA Sequencer (Foster City, CA). Four to six independent clones containing the sequence of each exon were analyzed.
Allele specific PCR (ASPCR).Primer-directed allele specific amplification16 with minor modifications14 was used to detect a single base substitution in the 5′-flanking region of the factor VII gene. To this end, sense oligonucleotides of 14 bp containing the mutant (5′-GGAGGCAGAGAACG) or normal base (5′-GGAGGCAGAGAACT) were used with a common antisense primer (5′-ACCAAGTTTATGGAGAAAAC) to amplify a 170 bp fragment from the mutant or normal allele, respectively. PCR reactions were performed in a total volume of 20 μL containing 0.3 μmol/L of each oligonucleotide, 0.01 pmol of template (a PCR fragment spanning from base pair −416 to +98), 0.4 U of Taq DNA polymerase, 70 μmol/L of each dNTP, and 1.0 mmol/L of MgCl2 . Optimal annealing temperatures for each set of primers were previously determined and amplifications were performed for 28 cycles. After completion of the ASPCR, 5 μL of the reaction mixture were subjected to electrophoresis in a 1.5% (wt/vol) agarose gel containing 0.5 μg/mL ethidium bromide and photographed under UV transillumination.
Detection of polymorphic alleles containing the decanucleotide insert at −323.Approximately 200 ng of a 514 bp PCR fragment containing a portion of the 5′-flanking region of the factor VII gene were digested with 10 U of EcoRI (New England Biolabs, Beverly, MA). Electrophoresis was performed in a nondenaturing polyacrylamide gel (8% wt/vol). The gel was stained in 0.5 μg/mL ethidium bromide for 5 minutes and photographed under UV transillumination. Cleavage at an EcoRI restriction site at position −313 in the PCR fragment results in products of 421 and 93 bp, or 421 and 103 bp if the decanucleotide insert is present.
Detection of polymorphic alleles containing Arg353Gln.Msp I restriction enzyme analysis was performed to show the presence of Arg353Gln.14
Construction of growth hormone reporter gene plasmids.The promoterless plasmid pOGH (Nichols Diagnostics Institute, San Juan Capistrano, CA) containing the structural human growth hormone gene was used for reporter gene plasmid constructs.17 The construction of the plasmid p186GH with nucleotides −185 to +1 of the human factor VII gene inserted into plasmid pOGH has been described in detail.10 To generate a plasmid containing the T to G substitution at position −61 of the factor VII promoter,5 we amplified a 214 bp fragment spanning the mutation site by PCR using genomic DNA from the proband as template. HindIII and BamHI restriction sites were artificially introduced at the 5′ and 3′ ends of the fragment, respectively. Restriction digests with these enzymes yielded a 186 bp fragment, which was gel purified and substituted for the homologous wild-type fragment in p186GH. The new plasmid containing the mutation, p186GHm, was sequenced to exclude the presence of other base substitutions that might have been introduced during PCR amplification. All plasmids were purified using DNA purification columns (Qiagen Inc, Chatsworth, CA), resuspended in 10 mmol/L Tris pH 8.5 (25°C), and stored at −80°C until use.
Cell culture and transient transfection assays.Human hepatoma cells HepG2 (ATCC HB 8065) and human cervical adenocarcinoma HeLa cells were grown in 5% CO2 at 37°C in minimum essential medium supplemented with 10% fetal bovine serum, Earle's salts, nonessential amino acids, 1 mmol/L sodium pyruvate, 10 mmol/L HEPES pH 7.2, 100 U/mL of penicillin G, and 100 mg/mL streptomycin. LipofectAmine reagent (GIBCO-BRL, Gaithersburg, MD) was used for all transient transfection assays according to the instructions supplied by the manufacturer. Twenty hours before transfection, HepG2 cells were plated on 60-mm culture dishes at a density of 1 × 106 cells/plate. Three μg of the GH reporter gene construct along with 1.5 μg of plasmid pSV-β-galactosidase (Promega Corp, Madison, WI) used as an internal control for transfection efficiency, were introduced into cells by LipofectAmine transfection. After 12 hours, medium was changed, and 48 hours later, the supernatants and cell lysates were procured and assayed for growth hormone and β-galactosidase activity, respectively. Results of transient assays performed in HepG2 cells are expressed in ng/mL of growth hormone and represent the average ± 1 SD obtained in six independent transfections. Transactivation experiments in HeLa cells were performed with 5 μg of the GH reporter gene construct, 10 μg of the HNF-4 expression plasmid pLEN4S18 or pLEN basic vector, and 2 μg of the internal control plasmid pSV-β-galactosidase. The plasmid pLEN4S (a gift from Dr James E. Darnell Jr, Rockefeller University, New York, NY) contains the entire 3-kb HNF-4 cDNA cloned into pLEN, a 5-kb expression plasmid containing the SV40 enhancer, metallothionein promoter, and the human growth hormone gene 3′-untranslated region. Results are expressed in ng/mL of growth hormone and represent the average ± 1 SD obtained in three independent transfections.
Human growth hormone and β-galactosidase assays.Aliquots of medium from transfected HepG2 or HeLa cells were assayed for growth hormone using a radioimmunoassay (Nichols Diagnostics Institute). Growth hormone levels were calculated from a standard curve that was fitted using a four parameter logistic model. β-Galactosidase activity in cell lysates was assayed colorimetrically at 419 nm at 37°C using a commercially available kit (Promega Corp). The samples were placed in microtiter plates and read by a plate reader (Molecular Devices, Menlo Park, CA) in the kinetic mode over a 1 hour period. β-Galactosidase values were obtained from a standard curve that had been fitted by linear regression using the least-squares method. Growth hormone concentrations were normalized with respect to β-galactosidase activity to correct for differences in transfection efficiency between plates. To account for background growth hormone activity, parallel transfections with the promoterless vector pOGH were performed. Growth hormone levels were then normalized to β-galactosidase activity as described above, and were subtracted from the values obtained for each individual transfection experiment.
Preparation of human liver nuclear extracts.Human liver was obtained from discarded liver transplant specimens under a protocol approved by the Institutional Review Board at the Children's Hospital of Philadelphia. The method described by Gorski et al19 was used to prepare human liver nuclear extracts. Aliquots of 200 μL were frozen in liquid nitrogen at −196°C until use. Protein concentration was determined using the method described by Bradford et al.20
Gel mobility shift assays.The gel mobility shift assay was performed using the method described by Chodosh et al21 with minor modifications. Complementary oligonucleotides of 30 bp (from nucleotide −76 to −47, 5′ to the factor VII translation start site) spanning the HNF-4 binding site in the human factor VII promoter were annealed and end-labeled using [γ−32P]ATP and 10 U T4 polynucleotide kinase at 37°C for 30 minutes. The radiolabeled oligonucleotide was purified with a Sephadex G-50 quick spin column (Boehringer Mannheim, Indianapolis, IN). Approximately 100,000 cpm of probe (0.5 ng) were incubated on ice in 20 μL reactions with 12 μg of human liver extract in the presence or absence of unlabeled competitor oligonucleotides in HEPES binding buffer [25 mmol/L HEPES (pH 7.6), 14 mmol/L KCl, 10% glycerol, 0.1 nmol/L EDTA, 0.75 mmol/L dithiothreitol, 5 mmol/L MgCl2] containing 4 μg salmon sperm DNA (Sigma, St Louis, MO), 1 μg bovine serum albumin, and 5 mg poly(dI⋅dC) (Pharmacia, Piscataway, NJ). Binding reactions were allowed to proceed during 30 minutes. Thereafter, DNA-protein mixtures were electrophoresed and autoradiographed as described.10
Informed consent.Informed consent was obtained from the patient's parents. The study was approved by the Human Studies Committee of Brockton-West Roxbury Department of Veterans Affairs Medical Center and Eastern Maine Medical Center (Bangor, ME).
RESULTS
Coagulation studies.The patient is a 6-year-old girl with a severe bleeding diathesis due to factor VII deficiency. She had a prothrombin time that was markedly prolonged at 28.6 seconds (normal range 10.0 to 13.0 seconds) and a normal activated partial thromboplastin time. Her levels of VII:C and VII:Ag were both less than 1% of normal, while those in her father and mother were 33% and 30% and 44% and 38%, respectively (Table 1).
ASPCR analysis showing the mutation at nucleotide −61 in the proband and her parents. PCR fragments spanning nucleotides −416 to +98 were first generated from genomic DNA, and then used as templates in ASPCR. A sense primer with the normal or mutant sequence at its 3′ end was used with a common antisense primer to generate a fragment of 170 bp. Using primers for the normal allele, fragments of the correct size were amplified in both parents, but not in the patient (left). Using primers for the mutant allele, fragments of the correct size were amplified in all family members (right). The result with a normal control (N) are shown in the first lane of each gel. The DNA mol wt markers are a HaeIII digest of φX174.
ASPCR analysis showing the mutation at nucleotide −61 in the proband and her parents. PCR fragments spanning nucleotides −416 to +98 were first generated from genomic DNA, and then used as templates in ASPCR. A sense primer with the normal or mutant sequence at its 3′ end was used with a common antisense primer to generate a fragment of 170 bp. Using primers for the normal allele, fragments of the correct size were amplified in both parents, but not in the patient (left). Using primers for the mutant allele, fragments of the correct size were amplified in all family members (right). The result with a normal control (N) are shown in the first lane of each gel. The DNA mol wt markers are a HaeIII digest of φX174.
Genomic studies.Sequencing of subcloned PCR fragments containing 149 bp of the 5′-flanking region demonstrated a T to G substitution at nucleotide −61 of the factor VII gene (data not shown). This abnormality was identified in each of six independent clones, and no sequence alterations were identified in any of the exons of the factor VII gene.
To demonstrate homozygosity definitively, we tested for the presence of the T to G substitution at nucleotide −61 in the patient and her parents using ASPCR because the abnormality could not be detected using restriction enzymes. Using primers designed to amplify the normal allele, a fragment of 170 bp was amplified only in the parents (Fig 1, left panel). Using primers for the mutant allele, a fragment of the same size was obtained in the patient, her mother, and her father (Fig 1, right panel). This indicates that the patient is homozygous for the sequence alteration, while both parents are heterozygous.
Polymorphisms in the factor VII gene also significantly influence factor VII levels. Therefore we assayed for the presence of two polymorphisms in the factor VII gene that are associated with modest reductions in factor VII levels, Arg353Gln in exon 822 and a decanucleotide insert at position −323 in the 5′-flanking region of the factor VII gene.23 Neither the patient nor her mother had these polymorphisms. The patient's father, who had a VII:C level that was 33% lower than his wife, was heterozygous for both polymorphisms (data not shown). These two polymorphisms must therefore be on the allele that was not transmitted to the proband (Table 1). Studies performed on healthy individuals in Italy24 and the UK25 show strong allelic association between Arg353Gln and the decanucleotide insert.
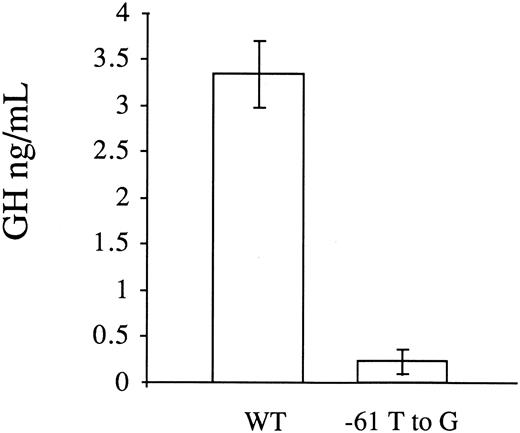
Transient transfection assays in HepG2 cells.To investigate whether the T to G substitution at position −61 influenced factor VII promoter activity, we analyzed the activity of the wild-type (p186GH) and mutant promoters (p186GHm) in transiently transfected HepG2 cells. To control for transfection efficiency, a β-galactosidase plasmid was cotransfected along with the growth hormone reporter gene constructs and the results were normalized with respect to β-galactosidase expression. In six independent transfections with each construct, the mean activity of the mutant plasmid in directing growth hormone transcription was 6.7 ± 3.8 (1 SD) percent of the wild-type plasmid (0.22 ± 0.13 ng/mL v 3.34 ± 0.36 ng/mL, Fig 2). Transfection with the promoterless vector pOGH yielded a growth hormone level in the media of 0.20 ± 0.06 ng/mL.
Functional analysis of the wild-type (WT) and mutant (−61 T to G) factor VII promoters by transient transfections with reporter gene constructs in HepG2 cells. A PCR fragment with the mutation was cloned into a reporter plasmid containing the human growth hormone structural gene. A β-galactosidase plasmid was cotransfected with the growth hormone reporter gene constructs to correct for variations in transfection efficiency. The results are expressed in ng/mL of secreted growth hormone and represent the mean ± 1 SD of six independent transfections. The promoter activity of the mutant plasmid (p186GHm) was 6.7% of the wild-type plasmid (p186GH).
Functional analysis of the wild-type (WT) and mutant (−61 T to G) factor VII promoters by transient transfections with reporter gene constructs in HepG2 cells. A PCR fragment with the mutation was cloned into a reporter plasmid containing the human growth hormone structural gene. A β-galactosidase plasmid was cotransfected with the growth hormone reporter gene constructs to correct for variations in transfection efficiency. The results are expressed in ng/mL of secreted growth hormone and represent the mean ± 1 SD of six independent transfections. The promoter activity of the mutant plasmid (p186GHm) was 6.7% of the wild-type plasmid (p186GH).
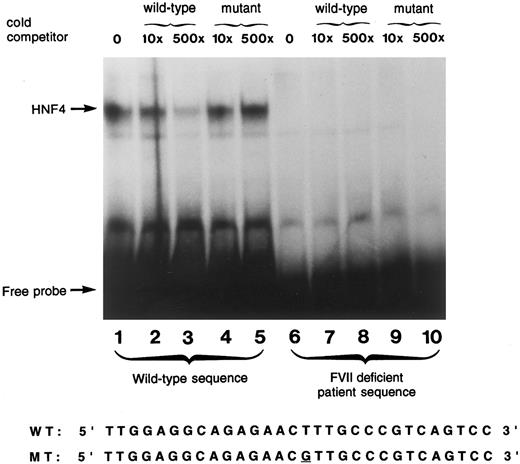
Gel mobility shift assay.A gel mobility shift assay using a 30 bp oligonucleotide spanning a previously shown HNF-4 binding site in the human factor VII promoter10 was used to show that the −61 T to G mutation prevented HNF-4 binding to the human factor VII promoter. To accomplish this, the gel was exposed to film for a time interval that was approximately three times as long as usual. The wild-type oligonucleotide bound a protein present in human liver nuclear extract (Fig 3, lane 1). Unlabeled wild-type sequence efficiently competed for binding at this site (lanes 2, 3) demonstrating the specificity of the probe for this protein, previously identified to be HNF-4.8-10 The faint signal in lane 3, which contains a 500-fold excess of unlabeled competitor, results from the long exposure of the gel and was not seen with more typical exposure times.10 The mutant factor VII promoter sequence, containing the transversion at −61 present in the propositus, did not compete away HNF-4 binding (lanes 4 and 5). The mutant sequence did not bind any liver nuclear extract protein (lanes 6 through 10); no signal was detected even after prolonged exposure of the gel.
Gel mobility shift assay using wild-type and mutant oligonucleotide sequences. A radiolabeled wild-type oligonucleotide encompassing the HNF-4 binding site (−76 to −47) in the factor VII gene shows specific binding to a protein present in human liver nuclear extracts (lane 1). Reactions performed with incubation of unlabeled competitor oligonucleotides at 10× and 500× concentrations of wild-type (lanes 2 and 3, respectively) and the mutant sequence (lanes 4 and 5, respectively) are shown. Lanes 6 through 10 show the absence of binding using a 30 bp radiolabeled oligonucleotide containing the T to G mutation at position −61 in the absence of cold competitor (lane 6), and in the presence of 10× and 500× concentration of cold competitor with either the wild-type sequence (lanes 7 and 8) or the mutant sequence (lanes 9 and 10). The nucleotide sequences of the wild-type and mutant probes are shown below.
Gel mobility shift assay using wild-type and mutant oligonucleotide sequences. A radiolabeled wild-type oligonucleotide encompassing the HNF-4 binding site (−76 to −47) in the factor VII gene shows specific binding to a protein present in human liver nuclear extracts (lane 1). Reactions performed with incubation of unlabeled competitor oligonucleotides at 10× and 500× concentrations of wild-type (lanes 2 and 3, respectively) and the mutant sequence (lanes 4 and 5, respectively) are shown. Lanes 6 through 10 show the absence of binding using a 30 bp radiolabeled oligonucleotide containing the T to G mutation at position −61 in the absence of cold competitor (lane 6), and in the presence of 10× and 500× concentration of cold competitor with either the wild-type sequence (lanes 7 and 8) or the mutant sequence (lanes 9 and 10). The nucleotide sequences of the wild-type and mutant probes are shown below.
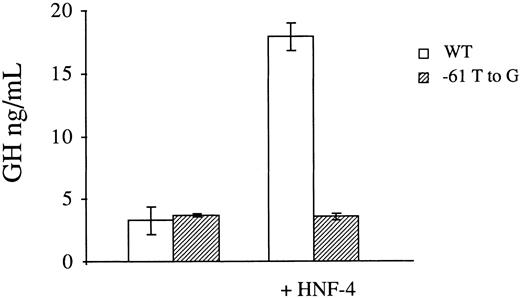
Transactivation studies in HeLa cells.To provide further evidence that defective binding of HNF-4 to the mutant factor VII promoter is responsible for its reduced transcriptional activity, we performed transactivation studies in a nonhepatocyte cell line which does not constitutively support factor VII promoter activity.10 To this end, the expression vector pLEN4S that codes for HNF-4 or the empty pLEN basic vector were cotransfected in HeLa cells with the normal (p186GH) or mutant (p186GHm) growth hormone reporter plasmids. In three independent transfections with each construct, the presence of HNF-4 in trans resulted in a 5.4-fold increase in the promoter activity of p186GH (3.1 ± 0.65 ng/mL versus 17.90 ± 1.07 ng/mL, Fig 4). Conversely, no increase was observed with p186GHm, which contains the −61 T to G transversion at the HNF-4 binding site of the factor VII promoter.
Transactivation of the wild-type (WT) and mutant (−61 T to G) factor VII promoters by HNF-4 in HeLa cells. Growth hormone reporter plasmids containing the wild-type or mutant promoter element were transfected with the expression vector pLEN4S that codes for HNF-4 or the empty pLEN basic vector. A β-galactosidase plasmid was also cotransfected to correct for variations in transfection efficiency. The results are expressed in ng/mL of secreted growth hormone and represent the mean ± 1 SD of three independent transfections. HNF-4 transactivated the wild-type factor VII promoter 5.4-fold, while no effect was seen with the mutant promoter.
Transactivation of the wild-type (WT) and mutant (−61 T to G) factor VII promoters by HNF-4 in HeLa cells. Growth hormone reporter plasmids containing the wild-type or mutant promoter element were transfected with the expression vector pLEN4S that codes for HNF-4 or the empty pLEN basic vector. A β-galactosidase plasmid was also cotransfected to correct for variations in transfection efficiency. The results are expressed in ng/mL of secreted growth hormone and represent the mean ± 1 SD of three independent transfections. HNF-4 transactivated the wild-type factor VII promoter 5.4-fold, while no effect was seen with the mutant promoter.
The higher growth hormone levels in HeLa cells (Fig 4) as compared to HepG2 cells (Fig 2) are attributable to differences in transfection efficiencies and experimental conditions. Before normalizing data with respect to β-galactosidase activity, the activities of the wild-type and mutant promoters in directing growth hormone transcription in HepG2 cells (2.21 ± 0.30 ng/mL and 0.39 ± 0.05 ng/mL, respectively) were actually similar to those in HeLa cells that had been cotransfected with the HNF-4 expression vector (2.63 ± 0.23 ng/mL and 0.58 ± 0.04 ng/mL, respectively).
DISCUSSION
Factor VII deficiency, an autosomal recessive disorder, occurs with an estimated incidence of 1 per 500,000 in the general population. Afflicted patients exhibit a highly variable hemorrhagic predisposition, and the correlation between bleeding tendency and plasma level of factor VII activity is often discordant.26 27 Patients with levels of less than 1% can experience severe bleeding episodes including hemarthroses and intracranial hemorrhage. The propositus of this study was severely affected and sustained life-threatening bleeds that eventually required maintenance therapy with factor VII concentrate. We found that the propositus was homozygous for a T to G transversion at nucleotide −61, which is the first instance of factor VII deficiency caused by a promoter mutation. It is likely that her parents share a common ancestor who carried this genetic defect.
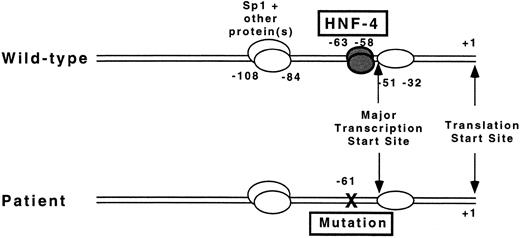
The factor VII promoter has only recently been analyzed in detail.8-10 The major transcription start site has been reported within a strong initiator element (−57 CCCGTCAGTCCC −46)28 at position −5110 and −509 upstream from the start site of translation (+1), in close proximity to the mutation reported here. Analysis of promoter activity using varying lengths of 5′-flanking sequence adjacent to the translation start site in reporter gene assays demonstrated that the first 185 bp are sufficient to provide maximal promoter activity.10 DNase I footprint analysis and gel mobility shift assays identified protein binding sites at positions −51 to −32, −63 to −58, −108 to −84, and −233 to −215 (see Fig 5).10 Erdmann et al8 have also shown an additional site at −13 to +6. The sequence from −100 to −94 was shown to be a binding site for the ubiquitous transcription factor Sp1.10 The identity of the liver-enriched transcription factor HNF-4 as the protein that binds at nucleotides −63 to −58 was established by “supershift” experiments with a specific antibody to HNF-4,8-10 by competition experiments with the oligonucleotide from the apolipoprotein CIII promoter that was originally used as an affinity ligand to purify HNF-4 from nuclear extracts,9,10 and by experiments using in vitro transcribed and translated HNF-4.29 The functional significance of the Sp1 and HNF-4 binding sites was shown in mutagenesis experiments, which showed dramatically reduced promoter activity when the nucleotide sequence of the binding sites was disrupted.8 10
Proposed mechanism by which a T to G mutation at −61 results in the absence of factor VII protein. The diagrams depict the proximal promoter region for the factor VII gene of the wild-type sequence (top) and mutant sequence (bottom) in a patient with severe factor VII deficiency. A mutation at nucleotide −61 disrupts binding of the transcription factor HNF-4 to its cognate binding site located directly 5′ to the major transcription start site at −51 before the translation start site (+1).
Proposed mechanism by which a T to G mutation at −61 results in the absence of factor VII protein. The diagrams depict the proximal promoter region for the factor VII gene of the wild-type sequence (top) and mutant sequence (bottom) in a patient with severe factor VII deficiency. A mutation at nucleotide −61 disrupts binding of the transcription factor HNF-4 to its cognate binding site located directly 5′ to the major transcription start site at −51 before the translation start site (+1).
In this patient with severe factor VII deficiency, the mutation at position −61 occurs within the previously shown HNF-4 binding site in the factor VII promoter (ACTTTG Æ → ACGTTG). Using gel mobility shift assays, we show that the mutation disrupts binding of HNF-4 to its cognate binding site. Using reporter gene assays, we show that the mutation dramatically reduces factor VII promoter activity in HepG2 cells compared to wild-type. Furthermore, while HNF-4 transactivates the wild-type factor VII promoter 5.4-fold in a nonhepatic environment, no transactivation can be shown with the mutated promoter. Taken together, these findings suggest that disruption of the HNF-4 binding site is sufficient to cause the patient's severe factor VII deficiency. The patient's severe clinical phenotype supports the in vitro finding that HNF-4 exerts a major positive regulatory effect on factor VII expression and provides in vivo evidence that binding of this transcription factor is critical for normal factor VII expression.
It is of interest that Greenberg et al9 mutated position −60 (ACTTTG Æ → ACTCTG) and position −59 (ACTTTG Æ → ACTTCG) in the factor VII promoter and observed decreases in promoter activity to 17% and 22% of wild-type, respectively. Our in vitro data showing that a mutation at −61 causes a greater than 93% reduction in factor VII promoter activity might suggest that this nucleotide is more critical than −60 or −59 in HNF-4 binding. Definitive conclusions regarding the relative importance of these nucleotides in HNF-4 binding however requires the evaluation of nucleotide substitutions at −60 and −59 in the same construct that was used to test the −61 mutation, which has not been done here.
Current models of transcription suggest that the complete set of general transcription factors (RNA polymerase II and TFIIA-TFIIH) are sufficient to direct basal transcription from strong promoters such as those containing TATA boxes.28 However, studies involving transcriptional activation in TATA-less promoters suggest the importance of direct interactions between general transcription factors and site-specific accessory transcription factors that serve as activators. In the context of the apoAI promoter, HNF-4 was recently shown to facilitate the formation of the preinitiation complex of transcription through a direct contact with TFIIB,30 a general transcription factor which, in association with RNA polymerase II, facilitates promoter recognition and accurate transcription initiation.28 In the factor VII promoter, the presence of an HNF-4 binding site several bases upstream of the transcription start site coupled with the evidence from this study that severe factor VII deficiency results from a mutation that disrupts HNF-4 binding, suggest that HNF-4 does act as an activator in the TATA-less factor VII promoter. Furthermore, an ACTTTG motif, conserved within the factor VII, IX, and X promoters (at −63 to −58, −56 to −51, and −53 to −48 before the translation start sites, respectively) is present within an HNF-4 binding site in each of these TATA-less vitamin K-dependent coagulation protein promoters and contributes to liver-specific expression of all three of these genes.11,12,31 Improvement in the stability of the transcriptional initiation complex has previously been hypothesized to allow increased expression of factor IX through enhanced binding at the HNF-4 site in an analogous position in the closely related factor IX promoter,32 and may be operative here as well.
The mutation that we have identified in the HNF-4 binding site of the factor VII promoter is also of particular interest because it occurs at a site that, in the factor IX promoter, has been associated with a clinical phenotype that has been termed hemophilia B Leyden.11 It is characterized by very low plasma levels of factor IX during childhood, which gradually rise following puberty.33 The molecular mechanisms underlying this disorder are complex and incompletely understood. Clearly, disruption of the HNF-4 site can account for the low levels of factor IX during childhood, but the mechanism by which levels undergo a steady gradual increase during adolescence and young adulthood is unknown. Several groups have proposed that the effect is androgen-mediated.34 In support of this notion is the fact that treatment of children with hemophilia B Leyden with androgenic or anabolic steroids results in a modest rise in factor IX levels.35 Moreover, a sequence with (imperfect) homology to an androgen-response element (ARE) exists in the 5′-flanking sequence of factor IX, in a region that partially overlaps the HNF-4 binding site.34 Thus, it is tempting to speculate that loss of the high-affinity HNF-4 binding site may permit binding to the low affinity ARE. However, despite attempts by several groups, no one has shown binding to this site by DNase I footprint analysis and results with gel mobility shift assays and cotransfection of an androgen receptor expression plasmid in transient transfections are conflicting.36 37
Because it is unclear why male children with hemophilia B Leyden exhibit an increase in clotting factor expression following puberty, it is impossible to predict whether our female patient with factor VII deficiency, who also has a mutation at the HNF-4 binding site, will have an increase in factor VII expression after puberty. The site containing the mutation (AGAACGTTGCCCGTC) exhibits strong homology to the ARE (AGNACANNNTGTNCT) at its 5′ end (8 of 9 nucleotides match), but little homology at its 3′ end (1 of 9 nucleotides match). The −61 T to G mutation changes a pyrimidine to a purine, which results in a closer match with the ARE than the wild-type sequence. Interestingly, carriers of mutations that result in Hemophilia B Leyden have not been shown to exhibit the same postpubertal increase in factor IX levels exhibited by affected patients.38 Thus the fact that the parents of the propositus in this report have subnormal levels of factor VII does not necessarily imply that their child will not experience a developmental rise in her factor VII levels.
ACKNOWLEDGMENT
The authors thank Fernanda Frank for her invaluable technical assistance and Hsiao-Ling Hung for providing human liver nuclear extracts.
Supported by the Medical Research Service of the Department of Veterans Affairs (K.A.B.) and National Institute of Health Grant No. HL48322 (K.A.H.). A.A.A. is a recipient of a Research Award from the A. Bianchi Bonomi Foundation and the Maggiore Hospital, University of Milan (Milan, Italy).
Address reprint requests to Kenneth A. Bauer, MD, Department of Veterans Affairs Medical Center, 1400 VFW Parkway, W Roxbury, MA 02132.