Abstract
Acute leukemia has a high concordance rate in young identical twins and in infants this is known, from molecular analysis, to reflect an in utero origin in one twin followed by prenatal metastasis to the other twin via intraplacental anastomoses. The situation in older twins with leukemia has been less clear. We describe a pair of identical twins who were diagnosed with a T-cell malignancy at 9 and 11 years of age, one with T-cell non-Hodgkin's lymphoma and the other with T-cell acute lymphoblastic leukemia. Leukemic cells from the twins shared the same TCRβ gene rearrangement with an identical 11 bp N region. The most plausible interpretation of this result is that these malignancies were initiated in one twin fetus in utero, in a single T-lineage cell that had stable bi-allelic TCRβ rearrangements. Progeny of this cell then spread to the other twin before birth via shared placental vasculature. This was then followed by a 9- and 11-year preleukemic latent period before clinical disease manifestation as leukemia or lymphoma. This result has considerable implications for the etiology and natural history of pediatric leukemia.
THE ETIOLOGY of acute leukemia in young children is likely to involve critical events that occur in infancy, perinatally, or prenatally.1 Backtracking individual leukemias to these periods is difficult experimentally and definitive evidence for such early events has been, until recently, lacking. In rare cases of pediatric leukemias, inherited genetic abnormalities are involved2,3 but in the majority of cases it is assumed that constitutive mutations are absent and that initiating mutations occur postfertilization.1 For those relatively infrequent acute leukemias that present at birth or neonatally,4-9 an origin during fetal hematopoiesis is obvious. Many of these newborn or congenital cases have chromosome alterations involving 11q237 and rearrangement of the MLL gene.8,9 Other evidence has also been very suggestive of prenatal origins of some pediatric leukemias. In particular, prenatal or pregnancy associated risk factors, eg, diagnostic X-irradiation, alcohol or other exposures of the pregnant mother10-13 have been linked in epidemiological studies to increased risk of leukemia in offspring. The lack of N region nucleotides in rearranged IGH genes has been taken as an indication of a prenatal origin in B-cell precursor acute lymphoblastic leukemia (ALL) in young (<3 year) patients on the grounds that TdT activity is low in B-cell lymphopoiesis before birth.14,15 Definitive evidence of an in utero origin of some pediatric leukemias has been provided by studies of clonal markers in concordant leukemias of identical twins. Infant identical twins have a high (∼25%) concordance rate of acute leukemia16 and long-standing evidence has suggested that this probably reflects a prenatal clonal origin in one twin followed by metastatic spread to the other twin via intraplacental anastomoses.1,17 Convincing evidence for this interpretation of concordance was provided by the observation that each of four pairs of infant twins with leukemia shared the same unique MLL gene rearrangement.18-20 In two of these cases,18,20 plus another infant (Siamese) twin pair21 that we reported, there was also a common IGH gene rearrangement as judged by restriction fragment size in Southern blots. An in utero origin of infant acute leukemia is therefore likely in most if not all cases, but no such unambiguous data has existed for leukemia in older children. We report here a unique pair of identical twins with T-cell malignancy that provides such evidence.
MATERIALS AND METHODS
Preparation of DNA
DNA was prepared essentially as described in Ford et al22 either from fresh leukemic cells isolated from bone marrow (BM) and peripheral blood or from scraped BM smears or cytospin preparations.
Southern Blot Analysis of Gene Rearrangement and Zygosity Status
DNA was digested with the restriction enzymes stated in the text, electrophoresed through 0.7% agarose gels, transferred to Magnacharge Nylon (Sartorius, Goettingen, Germany) and hybridized to either a probe containing the constant region of the TCRβ gene,23 the TCRγ joining region,24 or the joining region of the IGH gene.25 Confirmation of monozygosity was achieved using the multilocus fingerprinting probe 33.15 (Cellmark Diagnostics, Abingdon, UK).
Polymerase Chain Reaction (PCR) Analysis of Gene Rearrangement
Antigen receptor gene rearrangements were also examined by PCR using primers complementary to the consensus sequences within the framework 3 region of the variable regions and the joining regions of the IGH gene; consensus sequences within the variable, diversity, and joining regions of the TCRβ chain genes and consensus sequences within the variable and joining regions of the TCRγ chain genes.26 Controls included omission of DNA, a B-lymphoma sample rearranged for IGH but not TCRβ and primers for BCL-2.27 Reaction conditions were as previously described.26 PCR products were separated by electrophoresis on 10% polyacrylamide gels, stained with ethidium bromide, and visualized under UV light.
Cloning and Sequencing of PCR Products
Fragments amplified using primers from the diversity and joining regions of the TCRβ gene were also separated through 2.5% Low Gel agarose (Severn Biotech, Kidderminster, UK). After electrophoresis, gels were photographed and the fragment migrating with identical mobility for each twin was cut out. The separate fragments from each twin were then cloned into the Sma I site of Puc18 using a Sureclone Ligation kit (Pharmacia, Uppsala, Sweden). After transformation, 10 colonies from each twin were picked and screened for the presence of inserts. Five positive colonies from each individual screen were then amplified, sequenced on an Applied Biosystems 373A automated DNA sequencer, and further analyzed by Geneworks 2.1 (Intelligenetics, California).
RESULTS
The Twin Pregnancy
The parents were unrelated Brazilians of Portuguese origin. There was no family history of leukemia or lymphoma and there are no other siblings. The twin pregnancy and delivery were normal. During the pregnancy, the mother did not smoke or take alcohol, any drugs or antibiotics. There were no known infections during pregnancy and no exposure to diagnostic x-rays. The twin placenta was single, ie, monochorionic. The twin boys were of identical appearance and their monozygosity has been confirmed using Southern blotting with variable numbers of random-repeat (units) probes (data not shown).
Clinical Presentation
Twin 1.A 9-year-old boy presented in August 1992 with unexpected dyspnea and was admitted to a hospital for clinical examination. He was in satisfactory general condition and physical examination was normal. Chest x-ray and computerized tomography (CT) scan revealed a large mediastinal mass. A biopsy of the mediastinal mass disclosed a lymphoblastic lymphoma. Blood counts and biochemistry were normal and there were no malignant cells in the BM and/or cerebrospinal fluid. Chemotherapy was given following the German protocol BFM-83 (for high-risk ALL) with prophylaxis of the central nervous system (CNS, 18 Gy divided in 10 fractions). In November 1993 (16 months from diagnosis), while still under maintenance, he presented with drowsiness and left facial palsies. A brain CT scan showed a mass in the skull base. Stereotactic biopsy was performed and histology was consistent with lymphoblastic lymphoma. He received further cranial radiotherapy (18 Gy) and chemotherapy using the BFM protocol for relapsed ALL. This treatment resulted in minor improvement but progression of neurological symptoms with partial left hemiplegia, swallowing difficulties and dysphasia. In April 1995, a white blood cell (WBC) count showed: WBC 9.3 × 109/L with 30% lymphocytes and 10% blasts; there was no organomegaly. The immunophenotype (%) of the circulating cells was consistent with T-ALL/lymphoblastic lymphoma: CD2: 100, CD7: 92, cytoplasmic CD3: 100, CD1a: 30, CD4: 78, CD8: 72, CD38: 86 TdT:10; cells were negative with CD34, CD25, HLADR, CD19, CD10, αβTCR (cell surface) and γδTCR (cell surface). Because of persistent CNS involvement, the patient received symptomatic and palliative treatment, and in April 1996 was alive in a stable clinical condition.
Twin 2.An 11-year-old boy was admitted to the Hospital do Cancer, Rio de Janeiro, in April 1995 for bone pain and atony. Physical examination showed hepatosplenomegaly (spleen and liver were palpable 8 cm and 4 cm below the costal margins, respectively). There was no lymphadenopathy and neurological examination was normal. Blood counts showed hemoglobin 14.0 g/dL, WBC 193.0 × 109/L with 84% of blasts and platelets 77.0 × 109/L. Biochemistry was normal except for a high level of lactic dehydrogenase (1.114 UI) and cerebrospinal fluid was normal. BM aspirate showed replacement of normal hematopoiesis by medium to large blasts with rather irregular chromatin, 1 to 2 nucleoli compatible with ALL-L2 (French-American-British criteria). Immunophenotyping of the blood and marrow cells performed by flow cytometry and immunocytochemistry showed the following (%) results: CD2: 100, CD7: 94, CD38: 99, cytoplasmic CD3: 100, cell surface CD3: 94, CD4: 92, CD8: 92, CD1a: 74 and CD10: 10; cells were negative with CD34, CD25, TdT, HLADR, CD19, αβTCR (cell surface), and γδTCR. Cytogenetic analysis of BM cells identified 20 (50%) normal cells and 50% that were 46XY del (4) (q12) i (14)(q10).28 The patient was treated according to the German protocol BFM-83 (for high-risk) achieving complete remission. He was given maintenance therapy with 6-mercaptopurine and methotrexate but relapsed in the BM and CNS. At present (August 1996) he is hospitalized with severe aplasia following reinduction chemotherapy.
These presentation features are compatible with the diagnoses of T-NHL and T-ALL with a similar immunophenotype corresponding to the subset of CD1a+, CD4+/CD8+ cortical thymocytes of intermediate differentiation status.29,30 Such cases can be TdT negative at initial diagnosis as described here although the majority are TdT positive. Mature T-cell lymphoproliferative disease or leukemia (eg, T-cell prolymphocytic leukemia31; or human T-cell lymphotropic virus 1 [HTLV-1] associated adult T-cell leukemia/lymphoma32 ) is excluded by age, clinical features, morphology, and immunophenotype. Both cases were seronegative for HTLV-1. In his recent relapse, twin 2 leukemic cells were 40% TdT positive.
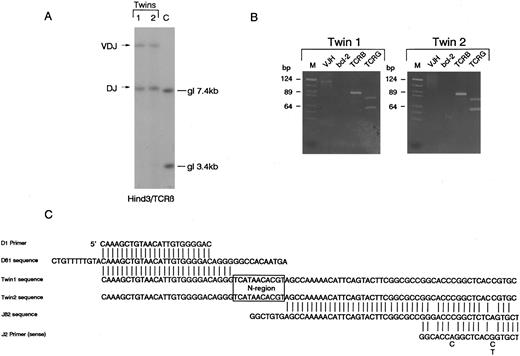
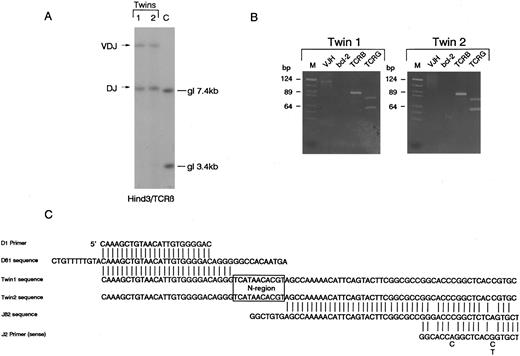
T-cell receptor β,γ gene and IGH gene rearrangements.Southern blot analysis of DNA from diagnostic samples (relapse blood in twin 1, pretreatment diagnostic blood in twin 2) revealed that the twins both had bi-allelic rearrangements of TCRβ with apparently identical sized restriction fragments suggesting similar or identical clonal rearrangements. The size of these fragments is indicative of one VDJβ and one DJβ rearrangement (Fig 1A). The identity of these rearrangements was analyzed by PCR. Using D and J region primers, a single identically sized fragment was amplified from the paired leukemic DNA. PCR was performed in laboratory conditions that did not provide any opportunity for cross-contamination and the same clonal rearrangements were amplified on a separate occasion from additional independent leukemic DNA samples retrieved from cytospin preparations (data not shown). The twin 1 DNA had a single V-JH rearrangement detectable by PCR which was absent in twin 2 DNA (Fig 1). The twin samples also appeared to have identical bi-allelic TCRγ rearrangements by both PCR (Fig 1B) and Southern blotting (data not shown). The PCR amplified TCRβ DJ rearrangement (Fig 1B) was then cloned independently from each twin sample as described in Materials and Methods. Sequencing of five independent clones from each twin revealed a single sequence (Fig 1C) which involved the joining of Dβ1 to Jβ2.4 with the deletion of the intervening sequences and addition of an identical N region of 11 nucleotides. Because one twin (2) was found to have isochromosome 14q, we attempted to verify this by FISH methodology and to similarly assess cells from twin 1. Unfortunately, the cell smears available were of inadequate quality for this purpose and no interpretable result was obtained.
Identical rearrangements of T-cell receptor genes in concordant twin T-cell malignancy. (A) Southern blot analysis of TCRβ rearrangements showing shared bi-allelic rearrangements (same sized restriction fragments). gl, germ line position. (B) PCR analysis of TCR and IGH gene rearrangements in the twin pair showing identical sized amplified bands for both TCRβ (TCRB) and TCRγ (TCRG) in the twin pair. Note twin 1 also has a weak clonal VDJ-IGH rearrangement that is absent in twin 2. It is not possible to say whether or not this clonal rearrangement was in the T-cell leukemic clone. bcl-2 primers were used as negative controls. M, markers. (C) Sequence comparison of cloned DJ-TCRβ amplicons showing identical N region (boxed).
Identical rearrangements of T-cell receptor genes in concordant twin T-cell malignancy. (A) Southern blot analysis of TCRβ rearrangements showing shared bi-allelic rearrangements (same sized restriction fragments). gl, germ line position. (B) PCR analysis of TCR and IGH gene rearrangements in the twin pair showing identical sized amplified bands for both TCRβ (TCRB) and TCRγ (TCRG) in the twin pair. Note twin 1 also has a weak clonal VDJ-IGH rearrangement that is absent in twin 2. It is not possible to say whether or not this clonal rearrangement was in the T-cell leukemic clone. bcl-2 primers were used as negative controls. M, markers. (C) Sequence comparison of cloned DJ-TCRβ amplicons showing identical N region (boxed).
DISCUSSION
These data provide compelling evidence that the T-lymphoblastic lymphoma and T-lymphoblastic leukemia clones from identical twins had originated from the same single cell. The probability of two independent clones of T cells sharing an identical 11 bp N region in a rearranged TCRβ gene by chance alone is 1× 4−11. That the same T-cell clone can be associated with a diagnosis of pediatric T-ALL or T-NHL accords with the notion that these divergent clinical diagnoses are somewhat artificial and may disguise a common biological origin in T-cell precursors.33
There are several theoretical possibilities to explain how this shared clonality might have arisen. One is that the twins became chimeric for normal T lymphocytes and other blood cells, before birth via their shared placental circulation and that cells belonging to the same T-cell clone were independently transformed postnatally — for example by mutation following extensive clonal expansion or proliferative stress via antigen recognition. This seems highly unlikely as it would require extraordinarily stringent immunological selection of cells from within a subclone of both twins, expressing not only the same VDJβ segments but the same N region. A second theoretical interpretation is that the leukemia developed postnatally in one twin and was somehow transferred to the other twin, who, being genetically identical, would not reject the accidental transplant. This would be operationally equivalent to the successful transplantation of animal leukemic cells between immunocompetent but inbred strains and similar to accidental transfer of cancer cells to immunosuppressed organ transplant recipients.34 The only documented “spontaneous” example of such a transfer of cancer cells, other than anecdotal reports on patients,35,36 is that of so-called infectious canine venereal sarcoma, the first cancer to be successfully transplanted 100 years ago, and which spreads via sexual contact.37 Successful, parasite-like, dissemination of malignant T cells between identical twins would require, we assume, body fluid and probably blood exchange. No blood transfusion between the twins has occurred and since we can think of no physical mechanism to facilitate such a transfer we consider it highly improbable, though not refuted.
The third and most plausible explanation is the same as that provided earlier for concordant leukemia in identical twin infants.17,18 This is that a single T cell in one twin in utero was subject to a transforming hit or mutation. This cell had already undergone stable allelic TCR β and γ rearrangements and these were preserved and present in the eventual leukemic and lymphoma subclones. Subsequently, but still prenatally, clonal descendants of this transformed cell metastasized to the other twin via the intraplacental anastomoses that commonly exist in monochorionic placentas.38 We cannot prove that this interpretation is correct in the absence of a unique, clonal mutation, equivalent to the MLL rearrangement in twin infant acute lymphoblastic leukemias; the T lymphoma/ALL cells were negative for all candidate genetic abnormalities assessed including TAL deletion, TCL-1 rearrangement and p53 mutations (exons 5-8) (data not shown).
What is extraordinary about this twin pair, compared with the infant twins, is that if the above interpretation is correct, then after birth the transformed or premalignant cells laid dormant or under some form of control many years before their eventual emergence as a dominant T lymphoma or T-cell acute lymphoblastic leukemia clone after 9 and 11 years, respectively, in the two twins. However, given the extended intervals between known genotoxic exposures and clinical leukemia,39,40 a latency of 9 or 11 years is not biologically implausible. Furthermore, a preclinical phase of clonal expansion of more than 10 years has been documented in serial studies on a patient with ataxia telangiectasia (AT) and T-cell leukemia.41 Because there is an increased concordance of T-cell leukemia in siblings with AT,41 we also considered the possibility that our twin pair might have had AT. Although both twins had hearing and balance problems from birth, in all other respects they lacked the clinical features of AT with no telangiectasias and no other neurological problems. Their development has been normal and they have had no signs of immunodeficiency (eg, recurrent infections).
More than 50 pairs of twins with concordant leukemia during childhood have been reported in the literature over the past 60 years but as far as we are aware, this is the first pair with T-cell leukemia. One adult pair of identical twins with concordant cutaneous T-cell lymphoma have been recorded.42 These had different TCRβ rearrangements. The concordance rate of T lymphoma/leukemia is unknown though probably significantly less than that of infants with ALL. We are aware of two examples of discordant T-ALL in identical twins (ie, where only one twin has T-ALL) and assume other such cases exist and are unreported. But lack of concordance in such pairs does not necessarily argue against a prenatal origin. First, only around 60% of monozygotic twins share a monochorionic placenta38 where the opportunities for interfetal migration of blood cells are optimized. Second, the twins could be discordant for some exposure or other postnatal event that is required for the preleukemia to leukemia conversion.
In the cases we report, a prenatal origin would never have been suspected were it not for the fact that two patients were identical twins. There is no reason to suppose that the etiological initiation of leukemia in a progenitor cell of a twin is any different in terms of mechanism and timing from that in non-twinned individuals. Therefore, we suggest that it is very likely that at least some other pediatric T malignancies are initiated in utero.
ACKNOWLEDGMENT
We thank Ramsa C. Harab and Marcelo H. Gomes for technical assistance, Dr G. Saglio for p53 mutation screening, Karin Gale for TAL-1 deletion screening, Dr Martin Yuille for TCl-1 rearrangement screening, Dr Elisabeth Vandenberghe for chromosome 14 FISH analysis, Dr Estela Matutes for review of the cases and Drs John Brown and Leanne Wiedemann for helpful suggestions, and Barbara Deverson with help in preparation of the manuscript.
Supported by the Leukaemia Research Fund (UK) and the Kay Kendall Leukaemia Fund.
Address reprint requests to Mel Greaves, PhD, Leukaemia Research Fund Centre, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Rd, London SW3 6JB UK.