Abstract
The p16 gene (MTS1, CDKN2, p16INK4A, CDKI) encoding an inhibitor of cyclin-dependent kinase 4 (cdk4) has been found to be deleted in various types of tumors, including leukemia, and is thought to code for a tumor suppressor gene. Our preliminary findings on eight pediatric patients with acute lymphoblastic leukemia (ALL) suggested that the survival of patients carrying a homozygous p16 gene deletion was significantly inferior to that of those without a deletion. The present study on 48 patients tested the hypothesis that the clinical outcome for pediatric ALL patients is correlated with the presence or absence of the p16 gene. Overall, nine of 48 children (18.3%) carried a homozygous p16 deletion. Such deletions were significantly more common (P = .003) among T-ALL patients (five of eight, 62.5%) than among precursor-B-ALL patients (four of 40, 10.0%). Of nine patients exhibiting p16 deletions, eight (88.9%) were classified as high-risk patients by the recognized prognostic factors of age, white blood cell count, and T-cell phenotype. The 4-year event-free survival in the study population as a whole was 72.7%. Without adjustment for other risk factors (univariate model), the presence of a homozygous p16 deletion was associated with a markedly increased probability of both relapse (P = .0003) and death (P = .002). These findings raise the question of whether the p16 deletion itself confers an increased risk of relapse after adjusting for the known risk factors. In this analysis, the estimated risk multiplier factor for relapse in patients carrying the p16 deletion was 14.0 (P = .0004) and for the risk of death 15.6 (P = .0008). We therefore conclude that the presence of a homozygous p16 deletion may well be an important risk factor for both relapse and death in childhood ALL, and that its prognostic effect is not a consequence of confounding by other factors already known to influence outcome in this disease.
SOMATICALLY ACQUIRED genetic damage leads to malignant transformation of cells. Among the specific genetic alterations thus far identified, the loss of tumor suppressor genes has been recognized as a critical factor. Cytogenetic deletions of chromosomal band 9p21 have been reported in 7% to 13% of acute lymphoblastic leukemia (ALL) cases,1,2 suggesting the presence of a tumor suppressor gene or genes in this region. The p16 gene (MTS1, CDKN2, p16INK4A, CDKI) located in 9p21 has been found to be homozygously deleted in many types of tumors, including melanomas, lung cancers, brain tumors, and leukemias.3,4 This gene encodes an inhibitor of cyclin-dependent kinase 4 (cdk4)5 and acts as a negative regulator of normal cellular proliferation in concert with cdk4, cyclin D, and the retinoblastoma protein, and hence scores highly as a putative tumor suppressor gene. More detailed studies have shown that homozygous or hemizygous deletions of the gene together with point mutations in the remaining allele are commonly observed in human cancers.6 Among ALLs, the T-cell phenotype (T-ALL) has been reported to show homozygous deletions in 25% to 83%7-12 of cases, whereas the frequency in precursor-B-ALL cases appears to be significantly lower.8,13-15 Evidence from many laboratories indicates that p16 point mutations are rare in ALL, including pediatric cases.9,11,13,14,16,17 Importantly, the recent generation of p16−/− mice by gene targeting provided direct evidence that p16 deficiency facilitates tumor development in a mammalian organism.18
There is conflicting evidence in the literature as to whether the presence or absence of the p16 gene is associated with particular risk groups among ALL patients.15,19 However, a large proportion of patients classified as high-risk leukemia have been found to show p16 gene deletion,10 and p16 gene loss has been reported to be associated with adverse prognostic features.15,20 We conducted a study comparing cultured cell lines and their corresponding primary leukemic cells and found that the presence or absence of the p16 gene was a feature of the primary leukemic cells; hence, deletion of the p16 gene was not an artifact of in vitro cell culture.21 Furthermore, the survival of ALL patients with the p16 gene deletion was significantly inferior to that of those without deletions. Because our observations were based on only eight patients, the relationship between p16 gene deletion and clinical outcome is now reexamined in a more comprehensive study with a larger number of patients. We focused on deletion analysis of the p16 gene, since it is evident that in ALL inactivation of this gene occurs mainly through homozygous deletions rather than mutations. The hypothesis to be tested was that the clinical outcome would be significantly inferior for ALL patients with the p16 gene deletion compared with those without the deletion.
SUBJECTS AND METHODS
Patient samples.We studied 48 children who were diagnosed with ALL between 1982 and 1994 at Princess Margaret Hospital, Perth, Australia. All children diagnosed with ALL in the state of Western Australia are treated in this hospital. The median age at diagnosis in the study population as a whole was 5.5 years (range, 1 week to 14.9 years). In patients homozygous for the p16 deletion the median age was 5.1 years, and in those without the deletion 5.8 years.
Clinical characteristics of the patients are listed in Table 1. The leukemia phenotype was determined by standard immunofluorescence analysis using a panel of monoclonal antibodies. Lymphoblasts were classified as precursor-B-ALL (CD19+CD22+/−CD10+/−) or T-ALL, provided at least two of the T-cell antigens CD2, CD5, and CD7 were expressed. Cases were studied based on the availability of cryopreserved Ficoll-Hypaque–enriched leukemic blasts from bone marrow aspirates. Informed consent was obtained from the patient or the patient's guardians. No significant differences were observed between our study group compared with large studies on pediatric ALL patients22 regarding demographic characteristics of the patients. The patients were risk-stratified according to recognized prognostic features, and therapy was administered according to Children's Cancer Group (CCG) protocols that were all based on full or modified Berlin-Frankfurt-Munster (BFM) protocols, and in one case on the New York regimen.23-25
Southern blot analysis.DNA of cryopreserved bone marrow specimens was extracted using standard methodology.26 In three cases, DNA was obtained from bone marrow slides, and destaining of the slides was performed according to a previously published method.27 DNA digestion using EcoRI, Southern blotting, labeling of probes, and hybridizations were performed according to previously published methods.26 The probes used in this study were generated by polymerase chain reaction (PCR) amplification using primers for exon 2 of the p16 gene. The blots were also hybridized to interleukin-2 receptor beta (IL-2RB) probe to accurately assess DNA loading in each lane.28
PCR.PCR to determine the presence of the p16 gene in three patient specimens were performed as described previously.21 A modified PCR protocol was established based on reactions terminated after various cycle numbers and graded mixes of control template consisting of DNA from cell lines with and without deletion of the p16 gene assayed in parallel with test samples. The oligonucleotide primers for exon 2 of the p16 gene were used as published by Kamb et al.4 A tube without template DNA was assayed as a control in each experiment, and primers for the β-globin gene29 were used to control for the PCR.
Scanning of autoradiographs.To determine p16 gene deletions, densitometric scanning of autoradiographs from Southern blot hybridizations was performed using a Vista-S6 Scanner (UMAX Technologies, Fremont, CA) in combination with Scan Analysis software (Biosoft, Ferguson, MO). Analysis was only performed on autoradiographs on which band intensity was in the linear range as a function of time of exposure. The intensity of the signal obtained for hybridization with the p16 probe was normalized to the DNA loading based on signal obtained for hybridization using the IL-2RB probe and DNA extracted from normal peripheral blood cells as control. Due to the presence of normal cells in all specimens, it was not possible to determine the incidence of hemizygous p16 deletions.
Univariate analysis.Binary data were analyzed using contingency tables. Formal tests of association were based on Fisher's exact test (two-tailed). Kaplan-Meier30 survival functions were used to display the univariate relationship between a single categorical variable of interest and time to first relapse (relapse-free survival [RFS]) and time to death (total survival [TS]). Event-free survival (EFS) was defined as the time to first relapse or death. Formal tests of statistical significance were based on the log-rank test (Mantel-Cox test).31 Standard errors for survivor functions were estimated using Greenwood's formula. All relapse and survival data were updated on March 11, 1996, and all follow-up data are censored at this point. One patient — an 8-year-old boy — died 119 days after diagnosis of precursor-B-ALL without attaining remission. He was included in the analysis of TS and EFS but not of RFS, because he was never at risk of relapse.
Multivariate analysis.Proportional hazards regression as described by Cox32 was used to analyze RFS and TS taking account of the risk factor profile of individual patients. The explanatory variable of primary interest was the binary covariate coding the presence or absence of the p16 deletion. The three risk factors considered in the consensus definition of high- versus standard-risk pediatric ALL patients33 are age and white blood cell (WBC) count at diagnosis and T-cell phenotype. The criteria for standard risk applied here are age between 1 and 9.99 years, WBC less than 50,000/μL, and B-cell lineage, and criteria for high risk include other age at diagnosis or WBC 50,000/μL or greater or T-cell lineage. To estimate the effect of the p16 deletion, having adjusted simultaneously for potential confounding variables, core Cox regression models (one for RFS and one for TS) were constructed containing covariates modeling age, WBC count, ALL phenotype, gender, and decade of diagnosis. Because the full core models were defined a priori, model construction procedures such as stepwise regression were not used. Finally, the binary covariate coding for the presence/absence of the p16 deletion was added to the core models.
Cox models were assessed comprehensively for goodness-of-fit.34 The need for interaction terms was investigated. The adequacy of the linear predictor was assessed by tabulating the mean value of Martingale residuals against quartiles of fitted values and against covariates. The proportional hazards assumption — that the relative risk associated with a given variable is constant throughout follow-up time — was tested for every covariate by adding appropriate time-dependent covariates. To ensure that the observed relationships between the p16 deletion and RFS and TS were not simply the result of a small number of unrepresentative patients, “local regression influence” was assessed by deleting patients one at a time from the data set, refitting the models, and reestimating the coefficient for the covariate coding for the p16 deletion.
RESULTS
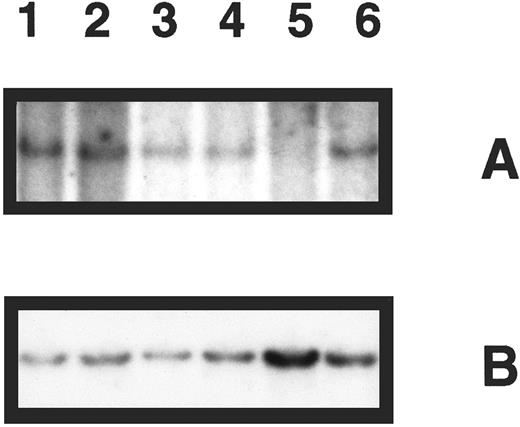
Deletion analysis of the p16 gene in primary bone marrow specimens was performed for 48 patients. Nine showed homozygous deletion (Table 1). A representative Southern blot showing five patient specimens hybridized with the probe for p16 and control hybridization for IL-2RB is shown in Fig 1. In the following analysis, we focused on deletion of the p16 gene and refer to homozygous p16 gene deletion as deletion of p16.
Southern blot analysis of p16 gene deletion in five patient specimens. (A) DNA samples digested with EcoRI and hybridized with p16 exon 2 probe revealing a band of 4.3 kb for normal DNA only (lane 1), which is not present in 1 patient specimen (lane 5). (B) The same blot was rehybridized with a probe for the IL-2RB gene, demonstrating the presence of DNA in all lanes.
Southern blot analysis of p16 gene deletion in five patient specimens. (A) DNA samples digested with EcoRI and hybridized with p16 exon 2 probe revealing a band of 4.3 kb for normal DNA only (lane 1), which is not present in 1 patient specimen (lane 5). (B) The same blot was rehybridized with a probe for the IL-2RB gene, demonstrating the presence of DNA in all lanes.
Table 1 details baseline clinical characteristics of the study population as a whole and distribution of the p16 deletion by relevant risk factors for childhood ALL. Deletion of the p16 gene was found in five of eight (62.5%) T-ALL patients, contrasting with four of 40 (10%) precursor-B-ALL patients (P = .003). The table also details outcome (relapse and death) by these same factors. High-risk patients have a markedly increased probability of carrying the p16 deletion (P = .01), emphasizing the importance of disentangling the true effect, if any, of the p16 deletion from the effect of risk factors already known to be critical in determining prognosis.
Table 2 details the relapse history and mortality by ALL phenotype and by risk status for patients who exhibited a p16 gene deletion and the equivalent data for patients who did not exhibit a p16 deletion. This comparison shows that high-risk patients who carry the p16 gene deletion have the expected high relapse rate (60% to 75%), and that T-ALL and precursor-B-ALL patients carrying the p16 gene deletion have similar frequencies of relapse. In contrast, patients not carrying the p16 deletion (Table 2) show lower relapse and death rates.
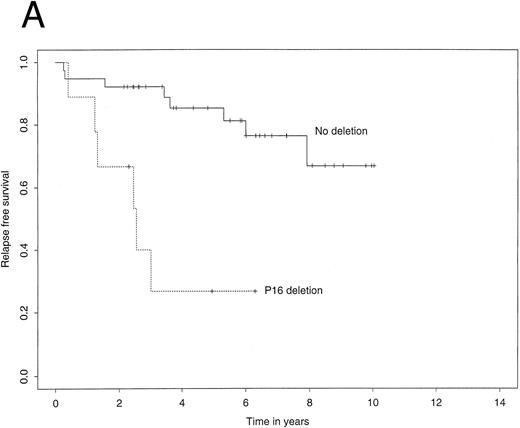
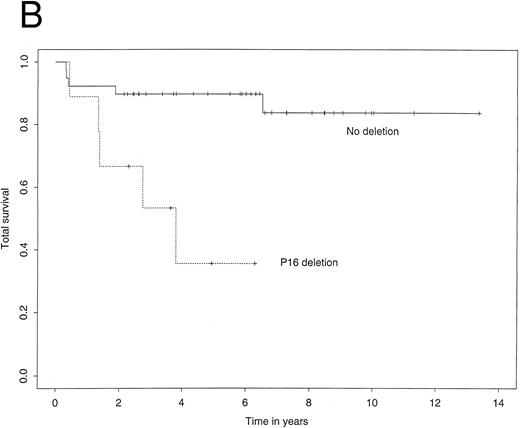
The 4-year EFS in the study population as a whole was 72.7% (95% confidence interval, 59.3% to 86.1%). The 4-year RFS was 74.3% (61.0% to 87.5%) and the 4-year TS 80.1% (66.8% to 93.3%). Figure 2A illustrates Kaplan-Meier curves for RFS stratified by p16 status. The corresponding curves for TS showed a similar difference (Fig 2B). Most importantly, among patients who did not carry a deletion of the p16 gene, the 4-year RFS was 85.3%, contrasting with 26.7% for those who did carry the deletion. Using the log-rank test, this difference was highly significant (P = .0003). Similarly, the 4-year TS was significantly worse for patients carrying the p16 gene deletion (35.6% v 89.7%, P = .002).
Kaplan-Meier survivor functions for (A) time to first relapse (RFS) and (B) time to death (TS) stratified by the presence or absence of a p16 deletion. Vertical bars denote censored observations.
Kaplan-Meier survivor functions for (A) time to first relapse (RFS) and (B) time to death (TS) stratified by the presence or absence of a p16 deletion. Vertical bars denote censored observations.
Cox regression models indicated that the known risk factors were associated with the increases in risk that would be expected (data not shown). Having adjusted simultaneously for the influence of age, WBC count, ALL phenotype, gender, and decade of diagnosis, the p16 deletion could still be shown to be a highly significant predictor of poor outcome. The adjusted risk multiplier for RFS was 14.01 (P = .0004) and for TS 15.6 (P = .0008).
DISCUSSION
This study determined the frequency of the p16 gene deletion in childhood ALL and correlated the results with clinical presentation features of the patients and clinical outcome. In a univariate analysis, the presence of a homozygous p16 deletion was associated with a markedly increased probability of both relapse (P = .0003) and death (P = .002). Furthermore, in a multivariate analysis that comprehensively adjusted for the influence of potential confounding variables, the estimated multiplier factor for the risk of relapse in patients carrying the p16 deletion was 14.0 (P = .0004) and for the risk of death 15.6 (P = .0008). We therefore conclude that the presence of a homozygous p16 deletion may well be an important risk factor for both relapse and death in childhood ALL, and that its prognostic effect is not a consequence of confounding by other factors already known to influence outcome in this disease. However, in any study of this type, one must consider whether any features of the study design or analysis could account for any positive or negative findings that are generated.
Formally, this study is a population-based historical cohort study. All cases of childhood ALL in Western Australia diagnosed after November 1982 were eligible for study. The only exclusion criterion was an inability to ascertain p16 status because of an inadequate sample of DNA. In consequence, serious selection bias is unlikely. Furthermore, comparison of patient characteristics in this study versus a study on 1,600 pediatric ALL patients showed that they do not differ.22
All patients studied here received treatment according to CCG protocols that changed over the period of study but were always based on the BFM regimen. Because the association between the year of diagnosis and the presence or absence of the p16 deletion was weak, any more subtle change in treatment protocol is unlikely to have confounded the estimated effect of the p16 deletion. However, the findings made in this study need to be confirmed in a prospective study on uniformly treated patients.
The study is relatively small and therefore has limited statistical power. However, our conclusions are based on positive findings, which are valid despite the small sample size. Furthermore, the positive findings pertain specifically to our primary hypothesis (stated before data collection) and therefore cannot be explained by a “lucky catch in a fishing expedition.” On the other hand, because of the low power, the 95% confidence intervals are very wide, and it is therefore possible that the true magnitude of the association of the p16 deletion with prognosis could be markedly stronger or weaker than we estimate.
A hypothesis generated post hoc from scrutiny of a set of preliminary data may well appear to be proven if some patients from the initial data set are included in the later analysis. In this case, five patients in the data set in which the association between the p16 deletion and prognosis was initially identified21 were also included in the current data set. To ensure that our results were not seriously biased by this issue, we repeated the analysis excluding these five cases, one of whom carried the p16 deletion. Substantive inferences were unchanged. For example, the adjusted estimate for the multiplicative effect of the p16 deletion for RFS changed from 14.01 to 13.95.
We report here a strong association between the presence of a homozygous p16 deletion and clinical prognosis, hence supporting the suggestions made by Heyman et al.19 Concern may be expressed that a larger independent study15 reported that the association between the presence or absence of a homozygous p16 deletion and the probability of nonresponse/relapse was nonsignificant. Analysis of the published results15 showed that of 27 patients who carried a homozygous p16 deletion, five (19%) had a relapse or nonresponse, as did 10 of 62 (16%) who did not carry the deletion. This equates to an unadjusted odds ratio of 1.18 (5/22 ÷ 10/52). Using the standard normal approximation to the distribution of the loge (odds ratio),35 the appropriate P value may be calculated as .79, which is indeed nonsignificant. However, this does not mean that the null hypothesis (no association) is true. The 95% confidence interval is 0.36 to 3.85, implying that these data are completely compatible with the p16 deletion having anything from a moderately strong beneficial effect on prognosis to a seriously detrimental effect. Our own data (Table 1) generated an unadjusted odds ratio of 7.5 (95% confidence interval, 1.53 to 36.8). A true unadjusted odds ratio anywhere in the range of 1.6 to 3.8 would therefore be consistent with both studies, and no conflict should be inferred to exist.
It is generally accepted that clinical features of pediatric ALL patients, including age and WBC count, are powerful predictors of outcome. However, despite risk stratification, up to 20% of newly diagnosed standard-risk patients are destined to relapse on current optimal therapy. More recently recognized biologic markers such as immunophenotype and genetic features36 and response to therapy24 have been shown to be independent of the traditional clinical features and provide additional markers of more aggressive disease. In this study, we have shown that deletion of the p16 gene is associated with enhanced risk of relapse and death independently of known prognostic factors. Because our study was relatively small and was conducted in a retrospective fashion on patients who did not receive identical therapy, it is essential that independent attempts are made to replicate our findings, ideally in a prospective study in a group of uniformly treated patients. Based on the findings presented here, we propose evaluation of the prognostic significance of p16 gene deletion in pediatric ALL in a prospective fashion in future CCG studies.
ACKNOWLEDGMENT
We thank Dr C. Cole for critically reading the manuscript.
Supported by the Children's Leukaemia and Cancer Research Foundation of Western Australia, the Cancer Foundation of Western Australia, and the Australia-China Council.
Address reprint requests to Ursula R. Kees, PhD, TVWT Institute for Child Health Research, PO Box 855, West Perth 6872 Australia.