Abstract
The bactericidal potency toward complement-resistant Escherichia coli of bactericidal/permeability-increasing protein (BPI) released from polymorphonuclear leukocytes (PMNs) in glycogen-induced inflammatory peritoneal exudates of rabbits is dependent on synergy with extracellular p15s. This synergy depends on the high molar ratio of p15s to BPI in the extracellular fluid (∼50:1), which greatly exceeds the intracellular ratio (∼5:1). To explore the possible basis of the greater accumulation of p15s in inflammatory fluid, we examined the subcellular localization of BPI and p15 in PMNs. Immunogold electron microscopy confirmed the storage of BPI in primary granules and showed that p15s are stored in secondary granules. Reverse-transcription polymerase chain reaction of density-fractionated rabbit bone marrow cells verified that p15s are expressed later than BPI during myeloid differentiation. As the inflammatory response evolves, p15 mRNA appears earlier in blood and exudate cells than mRNA for BPI, consistent with release of progressively less mature precursors from bone marrow. Finally, Ca2+-ionophore–mediated exocytosis of p15s occurs more readily than release of BPI. We therefore propose that localization of a synergistic partner of BPI (p15s) in more readily released secondary granules allows the neutrophil to mobilize potent BPI-dependent antibacterial activity extracellularly without significant depletion of intracellular BPI stores.
THE ESSENTIAL ROLE of polymorphonuclear leukocytes (PMNs) in host defense against bacterial infection depends on a complex arsenal that includes an array of cytotoxic polypeptides.1 Most of these agents are stored in the cytoplasmic granules of the PMNs and, in response to microbial and other inflammatory stimuli, may be released both into phagocytic vacuoles and extracellularly.2,3 Thus, in addition to the sequestration and destruction of pathogens by phagocytosis, PMNs can create an extracellular bactericidal environment by releasing antibacterial substances.4-10 Recently, we have shown that in a sterile inflammatory exudate generated in the peritoneal cavity of the rabbit, two antibacterial granule proteins, bactericidal/permeability-increasing protein (BPI) and p15s (15-kD antimicrobial proteins), reach concentrations in the cell-free inflammatory fluid (but not in plasma collected in parallel) that account for potent bactericidal activity against serum-resistant gram-negative bacteria.11 Although this activity was abolished by anti-BPI serum, the potency of the fluid could not be explained by the concentration of BPI alone, but required the additional presence of p15s. We had previously found that weakly antimicrobial concentrations of p15s act synergistically with BPI in both simple assay media and whole blood, thereby reducing the concentrations of BPI needed to kill the bacteria by up to 50-fold.12 In the inflammatory peritoneal fluid (ascitic fluid [AF ]), the amount of BPI was less than 3% of the BPI content of the PMNs, while up to 20% of the p15s in the PMNs accumulated in the AF.11 These findings raised questions about the location and secretion of these proteins during this experimental inflammatory process. In this study, we show that BPI and p15s reside in the primary and secondary granules, respectively, of rabbit PMNs. The different rates of exocytosis of these granules may account for the observed differences in BPI and p15 concentration seen in the AF and explain the potent antibacterial synergy observed in this inflammatory fluid.
MATERIALS AND METHODS
Cells.Sterile peritoneal exudates were elicited in New Zealand White rabbits by intraperitoneal injection of 300 mL of 2.5 mg/mL oyster-shell glycogen (USB, Cleveland, OH) dissolved in sterile saline.11 13 Peripheral blood leukocytes were prepared from fresh heparinized arterial blood by sedimentation in 2% dextran. Total and differential cell counts were performed by conventional procedures.
Electron microscopy.Cells were washed in Hanks' balanced salt solution without divalent cations (HBSS-; GIBCO-BRL, Grand Island, NY), centrifuged at 100g, then resuspended in freshly prepared fixative (4% paraformaldehyde, 0.4% glutaraldehyde, 2.5% sucrose in Dulbecco's phosphate-buffered saline, pH 7.0 [D-PBS]), and stored for 2 hours at 10°C. Samples were dehydrated through a series of graded alcohols then infiltrated and embedded in LR White (medium-grade resin). These fixation conditions were chosen to maximize antigenicity of intracellular BPI and p15 and retention of cellular architecture. Samples were polymerized at 50°C for 2 days. Ultrathin sections were cut with a diamond knife on an Ultracut E ultramicrotome (Reichert-Jung, Wien, Austria), and mounted on Formvar substrates carbon coated on slotted Athene nickel grids (Ladd Research, Burlington, VT). Grids were blocked for 10 minutes in 0.5% ovalbumin in PBS with 0.2% NaN3 . BPI was detected by incubating grids for 90 minutes at room temperature in 0.3 mg/mL monoclonal antibody (MoAb) 9(3/5) diluted in PBS with 0.1% Triton X-100, 0.1% Tween 20, 0.5% bovine serum albumin (BSA), and 0.2% NaN3 . This MoAb (generously provided by Xoma Corp, Berkeley, CA) was raised against recombinant human BPI but cross-reacts with rabbit BPI (C. Capodici, J. Weiss, K. Zarember; unpublished observations, July 1994). To label p15s, grids were incubated in a 1:100 dilution of guinea pig antirabbit p15 serum in the previous buffer. Grids were then rinsed two times for 2 minutes each in D-PBS. Primary antibodies were detected by incubation for 90 minutes in a 1:10 dilution of protein A conjugated to 15-nm gold particles (EY Lab, San Mateo, CA) in the previous antibody dilution buffer. Grids were rinsed twice for 2 minutes in D-PBS, rinsed in distilled water, stained in uranyl acetate and lead citrate, and examined with an EM 910 (Carl Zeiss, Thornwood, NY) or Elmiskop 1A (Siemens, Iselin, NJ).
Separation of PMN precursors from rabbit bone marrow.After euthanasia with Nembutal (Anpro Pharmaceuticals, Arcadia, CA), the femurs were rapidly dissected and the luminal contents mixed with 1/5 vol sodium citrate. This suspension was passed through cotton gauze and centrifuged for 10 minutes at 200g. Solutions of Percoll (Pharmacia, Piscataway, NJ) were mixed to 1.080 and 1.065 g/mL and rendered isotonic with concentrated PBS. Percoll at 1.080 g/mL was layered under an equal volume of 1.065 g/mL Percoll in a Corex tube (Corning, Corning, NY). After resuspension of the pellet in PBS, the cells were applied to the top of the gradient and centrifuged in a swinging-bucket rotor at 1,000g for 20 minutes at 4°C. Five bands resolved and were aspirated from the top using a Pasteur pipette. Fractions were diluted to 5 mL with PBS and spun at 200g for 10 minutes, washed again in 2 mL PBS, and resuspended in 1 mL PBS.
Purification of total RNA and mRNA.Total RNA was prepared using RNAzol B (Biotecx Laboratories, Houston, TX) following the manufacturer's instructions, except that cells were lysed by passing cells suspended in RNAzol B through successively smaller syringe needles (16 to 21 gauge). mRNA was isolated directly from cells using the Quick-Prep mRNA purification system (Pharmacia).
Semiquantitative reverse-transcription polymerase chain reaction (RT-PCR).RT of mRNA samples was initiated with a NotI/d[T]18 primer using the First-Strand cDNA Synthesis Kit (Pharmacia). PCRs were performed using Taq polymerase (Perkin Elmer, Norwalk, CT) and were overlayed with mineral oil. p15 cDNAs were amplified in a standard PCR mix that contained 3.5 mmol/L MgCl2 and primers (a) 5′-ACC ATG GCA GGA GTC TGG AAG GTA-3′, and (b) 5′-GCA GTG CAC CTG CTC CGT TGC CCA-3′. BPI cDNAs were amplified in 1 mmol/L MgCl2 with primers (c) 5′-CAG AAG GGC CTG GAC TAC GCC-3′ and (d) 5′-AGC GTG AGC TGC ACA TTC ATG-3′. Glyceraldehyde 3-phosphate dehydrogenase (G3PDH) was used as a housekeeping gene to control for equivalent loads. G3PDH was amplified in 3.5 mmol/L MgCl2 with primers (e) 5′-GAT CCA TTC ATT GAC-3′ and (f ) 5′-GAT CTC GCT CCT GGA-3′. After hot start (which was sometimes omitted), primers were annealed at 45 to 50°C for 1 minute and extended for 1 minute at 72°C followed by 30 cycles of 1 minute at 94°C, 30 seconds at 48 to 50°C, 1 minute at 72°C, and, finally, 15 minutes at 72°C. Products were extracted with CHCl3 to remove mineral oil, run on agarose gels, and visualized with ethidium bromide. A semiquantitative assay of mRNA content was performed by PCR of fivefold dilutions of cDNAs. For the analyses presented in the Results, a dilution for each cDNA was chosen that resulted in approximately half of the maximal band intensity in the cell fraction giving the most intense signal.
Double-immunofluorescent labeling of BPI and p15.After centrifugation in a Cytospin II (Shandon Southern Products, Astmoor, UK), cells were fixed for 1 minute at 4°C in buffered acetone formalin fixative (9.3% formalin [vol/vol]), 45% acetone, 1 mmol/L Na2KPO4 , 6 mmol/L KH2PO4 ). After rinsing with PBS and PBSA (PBS with 0.2% BSA), cells were soaked in 10% heat-treated rabbit serum for 10 minutes at room temperature. After a brief rinse with PBSA, cells were incubated in antisera diluted in PBSA (0.08% guinea pig anti-p15 serum and 0.4% goat anti-BPI serum or corresponding normal or preimmune serum controls) for 30 minutes at room temperature. After extensive washing with PBS and PBSA, cells were then incubated in 1 μg/mL fluorescein isothiocyanate (FITC)-conjugated rabbit antigoat IgG (Calbiochem, La Jolla, CA) for 30 minutes at room temperature followed by extensive washing and incubation in 10 μg/mL tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat antiguinea pig IgG (Kirkegaard and Perry Laboratories, Gaithersburg, MD) in PBSA for 30 mintes at room temperature. After extensive washing, cells were mounted in 9:1 glycerol:PBS supplemented with p-phenylenediamine and photographed on a Nikon (Melville, NY) Labophot microscope fitted with an epifluorescence attachment with a 100-W mercury lamp and a UFX-II photomicrography system. FITC was visualized using excitation wavelengths from 450 to 490 nm, a dichroic filter blocking wavelengths below 510 nm, and a barrier filter of 520 nm. TRITC was visualized with excitation between 510 and 560 nm, a dichroic mirror blocking below 580 nm, and a barrier filter of 590 nm.
Northern blotting.Total RNA was electrophoresed through a 1.2% agarose/formaldehyde gel and transferred to nitrocellulose (Schleicher and Schuell, Keene, NH) by capillary transfer.14 After baking, blots were blocked by prehybridization with denatured sonicated salmon sperm DNA (Sigma, St Louis, MO) and probed with 32P-dCTP–labeled (random primed method of Pharmacia) p15 or BPI PCR products. After washing, blots were exposed to either Kodak X-OMAT AR (Eastman-Kodak, Rochester, NY) or Fuji RX film (Fuji, Elmsford, NY).
Degranulation.The degranulation procedure was a modification of a published protocol.15 Peritoneal exudates were spun at 50g for 10 minutes at room temperature and the pellet was resuspended in an equal volume of HBSS- with 1 mmol/L EDTA. The suspension was spun again and resuspended to 107 cells/mL in HBSS- supplemented with 1 mmol/L MgCl2 (without EDTA or calcium). Aliquots were spun in siliconized polypropylene Eppendorf tubes and gently resuspended to 107/mL in HBSS with 1 mmol/L MgCl2 plus 0.5 mmol/L EDTA or CaCl2 at the appropriate concentration. After 2 to 3 minutes, ionomycin (from Streptomyces conglobatus; Sigma) dissolved in dimethylsulfoxide (DMSO) was added to achieve a final concentration of 1 μmol/L (final concentration of DMSO, ∼14 mmol/L). Suspensions were incubated at 37°C for 20 minutes with intermittent gentle agitation, then spun at 200g for 2 minutes. The supernatants were removed and assayed for BPI, p15s, and transcobalamin I as described later. Lactate dehydrogenase (LDH) and β-glucuronidase were detected as per the manufacturer's instructions (Sigma).
Assay for transcobalamin I (neutrophil vitamin B12-binding protein).Transcobalamin I was assayed by its ability to compete with protein-coated charcoal in adsorbing vitamin B12 .16 Protein-binding sites of charcoal (50- to 200-mesh; Fisher, Pittsburgh, PA) were blocked by suspension in 2 % BSA in PBS. Samples were diluted to 490-μL vol with PBS and approximately 1.5 ng of [58Co]-vitamin B12 (3.2 μCi/μg; Medi-Physics, Arlington Heights, IL) was added. After vortexing, the sample was incubated for at least 1 minute to allow vitamin B12 to interact with transcobalamin I. Samples were then vortexed for 10 seconds with 500 μL of 100-mg/mL protein-coated charcoal suspension. After 2 minutes at room temperature, samples were vortexed again for 10 seconds and spun in a microcentrifuge for 5 minutes at 14,000g. Recovery of counts in the supernatant (read in a 300- to 900-KeV window by a Pharmacia-LKB-Wallac 1282 Compugamma counter; Turku, Finland) was proportional to the amount of B12 -binding activity present.
Immunodetection of BPI and p15.Samples were applied to 0.45 μm nitrocellulose (Schleicher and Schuell) saturated with Tris-buffered saline (TBS). The membrane was air-dried, blocked for at least 30 minutes in 2% to 3% BSA dissolved in either PBS or TBS pH 7.4, then washed three to four times with TBS. Blots were then placed in antibody diluent (TBS supplemented with 0.014% BSA, 2.3 mmol/L EDTA, and 2.3% NP-40) and either goat anti-BPI (diluted 1:750) or guinea pig anti-p15 (diluted 1:150) and incubated at room temperature with shaking for at least 2 hours. Blots were washed for 15 minutes in TBS with at least four changes, placed in antibody diluent with a 1:1000 dilution of 125I-protein G (Amersham, Arlington Heights, IL), horseradish peroxidase (HRPO)-conjugated protein G, or an HRPO-conjugated mouse antigoat antibody (the latter two from Pierce, Rockford, IL) and incubated for at least 1 hour at room temperature with shaking. Blots were washed three times for 5 minutes each in TBS and two times for 5 minutes in PBS. Kodak X-OMAT AR film was used to detect 125I-protein G, and metal-enhanced diaminobenzidine (Pierce) was used to visualize HRPO conjugates. Normal goat serum (1:750) or normal guinea pig serum (1:150) did not significantly react with parallel blots. Dot-blots were quantitated using an Ambis radioanalytic system operating in optical mode (Ambis, San Diego, CA). Dose curves were linear from 4 to 400 ng, with a best fit line with an R2 value of greater than .95.
RESULTS
Immunoelectron microscopic evidence that p15s and BPI reside in different granule populations.PMNs, in response to many inflammatory agonists, more readily release the contents of the secondary granules than those of the primary granules into the extracellular space.2,3,15 17-20 We therefore explored the possibility that the greater accumulation of the p15s than of BPI in the inflammatory fluid reflects a different intracellular origin.
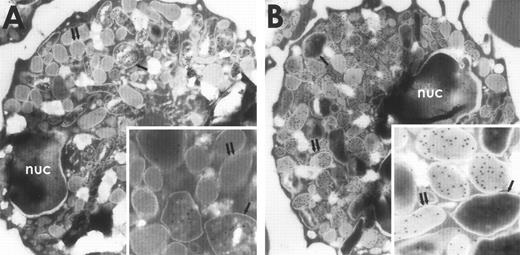
The subcellular localization of BPI and p15s in rabbit exudate PMNs was determined by immunoelectron microscopy using specific antibodies to these proteins visualized with protein A–coated gold particles (see Materials and Methods). After treatment of sections with the anti-BPI MoAb, most gold particles were seen in the larger more electron-dense primary granules (Fig 1A, inset), consistent with the localization of BPI in the primary granules of human PMNs.21-23 In contrast, antiserum raised against purified p15s localized to the secondary granules, which were distinguished from the primary granules by their low electron density, smaller size, and greater abundance24 (Fig 1B, inset). No appreciable labeling was observed either in other cells (erythrocytes, monocytes, and lymphocytes), in grids incubated with pooled normal guinea pig serum, or in grids incubated with the BPI MoAb that had been preincubated with BPI.
Immunogold electron microscopy of rabbit PMNs stained with antibodies against p15 or BPI and visualized with protein A–coated gold particles. Rabbit PMNs were incubated with an MoAb against BPI (A) or guinea pig antiserum raised against rabbit p15s (B) as described in Materials and Methods. Single arrows point to primary granules and double arrows point to secondary granules; nuc, nucleus.
Immunogold electron microscopy of rabbit PMNs stained with antibodies against p15 or BPI and visualized with protein A–coated gold particles. Rabbit PMNs were incubated with an MoAb against BPI (A) or guinea pig antiserum raised against rabbit p15s (B) as described in Materials and Methods. Single arrows point to primary granules and double arrows point to secondary granules; nuc, nucleus.
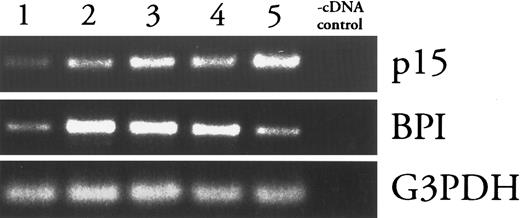
p15 (mRNA) appears later in PMN development than BPI (mRNA).Levels of mRNAs encoding primary granule proteins increase and decrease earlier during PMN differentiation than do those of secondary granule proteins, approximately coincident with the biogenesis of the respective granules.25 Thus, the presence of p15s and BPI in secondary and primary granules, respectively, predicts that p15 (mRNAs) should appear later than BPI (mRNAs) during PMN differentiation. This prediction was tested by the fractionation of rabbit bone marrow cells in isotonic Percoll density gradients followed by analysis of fractions for BPI and p15 mRNA content using RT-PCR. As described in studies with human bone marrow, density gradients can separate PMN precursors, which increase in density as they mature. Five bands of cells were detected and collected (fraction 1 is the least dense, fraction 5 is the most dense). Modified Wright-Giemsa staining showed that lighter fractions were enriched with less mature precursors and heavier fractions were enriched in more mature cells (data not shown). BPI message was most abundant in cells in fraction 2, whereas p15 mRNA was most abundant in cells from the most dense fraction (Fig 2). In contrast, levels of G3PDH mRNA were similar among all fractions.
Comparison of p15 and BPI mRNA levels in density-fractionated rabbit bone marrow cells. mRNAs from Percoll density gradient fractions of increasing density (1-5) were analyzed by RT-PCR as described in Materials and Methods. Absence of contamination (-cDNA control) was verified by subjecting a sample of the cDNA diluent (water) to PCR under each condition. Results shown are representative of two similar experiments.
Comparison of p15 and BPI mRNA levels in density-fractionated rabbit bone marrow cells. mRNAs from Percoll density gradient fractions of increasing density (1-5) were analyzed by RT-PCR as described in Materials and Methods. Absence of contamination (-cDNA control) was verified by subjecting a sample of the cDNA diluent (water) to PCR under each condition. Results shown are representative of two similar experiments.
Indirect double-immunofluorescent staining of BPI and p15 in these fractions showed BPI both in larger more irregularly-shaped promyelocytes and in more mature-looking myelocytes or band cells, whereas p15 staining was apparent only in more mature looking cells (Fig 3A and B). Thus, as judged both by RT-PCR of mRNA and indirect immunofluorescence of density-fractionated granulocyte precursors, p15s are produced later than BPI during PMN differentiation.
Indirect double-immunofluorescent labeling of BPI and p15 in rabbit bone marrow cells. Cells from Percoll density gradient-fractionated rabbit bone marrow cells were stained for BPI (A, FITC) and p15 (B, TRITC) as described in Materials and Methods.
Indirect double-immunofluorescent labeling of BPI and p15 in rabbit bone marrow cells. Cells from Percoll density gradient-fractionated rabbit bone marrow cells were stained for BPI (A, FITC) and p15 (B, TRITC) as described in Materials and Methods.
BPI and p15 mRNA content of circulating and exudate PMNs during evolution of the inflammatory exudate.Although the biosynthetic activities of the mature PMNs are thought to be limited, stimulated PMNs are capable of turning on the synthesis of certain gene products.26-32 It is therefore possible that de novo synthesis of BPI and p15 could contribute to the extracellular mobilization of these proteins in a local inflammatory setting. As a first approach to this question, we determined the mRNA content of peritoneal exudate cells and of peripheral blood cells collected in parallel.
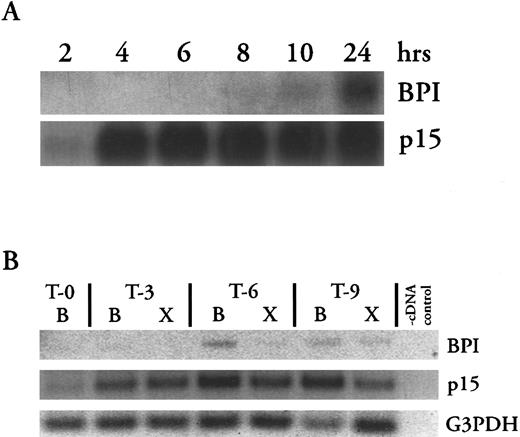
Northern blots (Fig 4A) show that cells collected from the peritoneum 2 hours after injection of inflammatory stimulus contained little p15 mRNA and no detectable BPI mRNA. However, by 4 hours, abundant p15 mRNA was detected at levels that persisted for at least 24 hours. In contrast, PMNs collected earlier than 8 hours after induction of inflammation contained no detectable BPI message, while cells collected thereafter contained increasing amounts of BPI mRNA.
Changes in p15 and BPI mRNA levels in blood and exudate cells after induction of inflammatory exudate. (A) Northern blots of p15 and BPI mRNA isolated from equivalent numbers of inflammatory exudate cells collected at indicated times after induction of the inflammatory response (see Materials and Methods). (B) RT-PCR analysis of mRNAs isolated from equivalent numbers of peripheral blood (B) or inflammatory exudate (X) cells as described in Materials and Methods and the legend to Fig 2. T-0 shows baseline mRNA levels in a blood sample collected immediately before injection of the inflammatory stimulus. Thereafter, blood and exudate samples were collected in parallel at the times indicated (in hours). Results shown are representative of two similar experiments.
Changes in p15 and BPI mRNA levels in blood and exudate cells after induction of inflammatory exudate. (A) Northern blots of p15 and BPI mRNA isolated from equivalent numbers of inflammatory exudate cells collected at indicated times after induction of the inflammatory response (see Materials and Methods). (B) RT-PCR analysis of mRNAs isolated from equivalent numbers of peripheral blood (B) or inflammatory exudate (X) cells as described in Materials and Methods and the legend to Fig 2. T-0 shows baseline mRNA levels in a blood sample collected immediately before injection of the inflammatory stimulus. Thereafter, blood and exudate samples were collected in parallel at the times indicated (in hours). Results shown are representative of two similar experiments.
RT-PCR was used to determine whether the appearance of message in exudate PMNs could be accounted for by migration from the bone marrow of less mature cells that retained mRNAs for these proteins. We compared the mRNA content of inflammatory exudate cells and peripheral blood cells collected at the same time intervals after the inflammatory stimulus (Fig 4B). Before induction of the inflammatory response, no p15 or BPI mRNAs were detected in circulating cells. Within 3 hours of the inflammatory stimulus, p15 message was detected both in circulating blood cells and in exudate cells. Levels of p15 mRNA increased from 3 to 9 hours after initiation of inflammation in blood cells (Fig 4B), but not in exudate cells (Fig 4A and B). In contrast, BPI mRNA was detected only at later times and in blood cells before exudate cells. These findings are consistent with the mobilization from the bone marrow of progressively less mature PMNs into the circulation in response to the inflammatory stimulus.
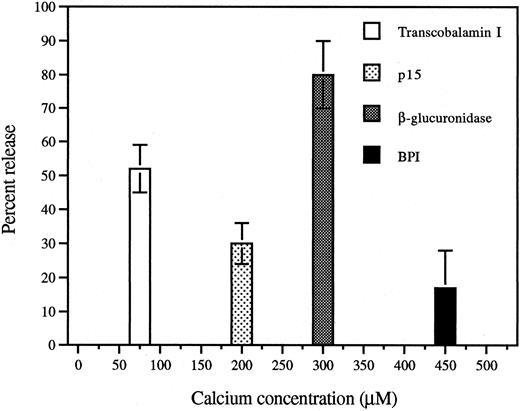
Differential release of p15s and BPI during Ca2+/ionomycin-induced degranulation of PMNs.To relate the subcellular localization shown in Fig 1 to the greater extracellular accumulation in vivo of the p15s than of BPI in AF,11 we examined the release of these proteins in vitro in response to a challenge known to induce graded and differential exocytosis of granules from human PMNs.15,20 Similarly, exposure of rabbit exudate PMNs to increasing extracellular concentrations of Ca2+ in the presence of the calcium ionophore ionomycin resulted in substantial release of the secondary granule marker transcobalamin I at low (half-maximal release at ∼75 μmol/L) calcium concentrations that induced little release of β-glucuronidase, a primary granule marker enzyme (Fig 5). BPI required a somewhat greater calcium dose than β-glucuronidase (Fig 5) and, at maximum, a much smaller fraction of cellular BPI was released similar to earlier observations in human PMNs.22 Significant release of p15 was observed at Ca2+ concentrations (50 to 200 μmol/L) that induced little or no release of β-glucuronidase or BPI (Fig 5). In comparison to BPI, p15s are more readily secreted from PMNs (Table 1), consistent with their greater accumulation in the AF11 and their different subcellular localization (Fig 1).
Calcium/ionomycin treatment of rabbit inflammatory exudate PMNs induces release of secondary granule contents at lower calcium doses than primary granule contents. Aliquots of EDTA-washed inflammatory exudate PMNs were incubated with increasing concentrations of calcium chloride. Suspensions were centrifuged and the supernatants analyzed as described in Materials and Methods. Release of each protein is shown at the calcium concentration at which half-maximal release occurred and is expressed as percent (mean ± SEM of 3 separate experiments) of the total content of each protein.
Calcium/ionomycin treatment of rabbit inflammatory exudate PMNs induces release of secondary granule contents at lower calcium doses than primary granule contents. Aliquots of EDTA-washed inflammatory exudate PMNs were incubated with increasing concentrations of calcium chloride. Suspensions were centrifuged and the supernatants analyzed as described in Materials and Methods. Release of each protein is shown at the calcium concentration at which half-maximal release occurred and is expressed as percent (mean ± SEM of 3 separate experiments) of the total content of each protein.
DISCUSSION
The existence of distinct populations of cytoplasmic granules in PMNs that differ in composition and response to defined secretagogues2,3,15,17,20 has led to the belief that the different granule species have distinct roles in PMN function. However, it has also been shown that important functions of the PMNs are regulated and coordinated by translocating and combining proteins that originate in different subcellular sites. Thus, the activation of the PMN oxidase system and production of cytotoxic oxidants requires the joining of components that in the resting cell are dispersed in the cytoplasm, primary and secondary granules, and other membrane-bounded compartments.33 In the case of a nonoxidative component of the microbicidal arsenal of PMNs, the inactive storage pro-form of bactenecin 5 is stored in the “large” granules of bovine PMNs and is cleaved into its active cytotoxic form by a proteinase contained in primary granules after exocytosis of these two granules into a common space.34
Our studies have provided another example of the mobilization of a potent PMN-derived microbicidal activity by the combination of proteins stored in different granule types. We have recently shown that the extracellular bactericidal activity against serum-resistant Escherichia coli that occurs in PMN-rich inflammatory exudates is due to the accumulation of two PMN granule proteins, BPI and the p15s.11 Although this activity requires BPI, the concentration of BPI in the inflammatory fluid, by itself, accounts for ≤ 20% of the observed antibacterial activity. Full activity requires p15s, which act synergistically with BPI against E coli.11,12 35 The extent of synergy depends on the molar ratio of the p15s and BPI.12 Thus, the potency of BPI-dependent bactericidal activity in the inflammatory fluid reflects extracellular accumulation of the p15s at high concentrations (≤1 μmol/L) relative to the concentration of BPI (10 to 20 nmol/L), a ratio that is at least 10 times greater than the molar ratio of these proteins within the PMNs.11 The greater extracellular accumulation of p15s during influx of PMNs to sites of inflammation may be accounted for by the storage of p15s in granules that are more readily released than those containing BPI (Figs 1 and 5, Table 1). This permits extracellular mobilization of potent BPI-dependent activity with only minimal depletion of the PMN stores of BPI needed for intracellular action.6
Ca2+/ionophore treatment of PMNs in vitro showed greater secretion of p15s than of BPI especially at Ca2+ concentrations that induce preferential release of secondary granules (Fig 5 and Table 1). However, the release of p15 and BPI was less than that of the corresponding granule markers, transcobalamin I (secondary) and β-glucuronidase (primary) (Fig 5). BPI associates with the primary granule membrane21 and plasma membrane,36 perhaps due to its hydrophobic properties. Similar structural features of the p15s might explain why their release is less prominent than that of transcobalamin I. The possibility that BPI and p15 undergo proteolysis in the extracellular fluid in this experiment, thereby accounting for the reduced recovery relative to the other granule markers, is unlikely, because Western analysis of inflammatory fluid reveals only intact BPI and p15.11
The enrichment of p15 (v BPI) mRNA in denser more mature cell subpopulations of bone marrow (Fig 2) and their absence from less mature myeloid precursors (Fig 3B) is consistent with the later biogenesis of secondary granules during myeloid maturation.24,37 The virtual absence of BPI or p15 mRNA in resting circulating blood cells (without inflammatory stimulus) is similar to reports for several other granule proteins.38,39 However, during inflammation, significantly elevated mRNA levels for p15 and BPI (to a lesser extent) were found. The earlier appearance of p15 mRNA than of BPI mRNA in both circulating and exudate cells may simply reflect the progressive release from bone marrow of relatively less mature myeloid elements as the inflammatory response evolves. However, the very early appearance of p15 mRNA within exudate cells (Fig 4) raises the possibility of local induction of p15 gene expression in mature PMNs within the inflammatory environment as has been described for several non–granule-associated gene products of PMNs.26,29 40 To our knowledge, these data are the first to indicate the presence of mRNAs encoding granule-associated antimicrobial proteins of PMNs in an inflammatory setting. Whether these mRNAs are translated during formation of the exudate remains to be explored.
p15s belong to the recently recognized cathelicidin family of antimicrobial proteins,41 which have been cloned from pig, cow, sheep, rabbit, and human bone marrow and have been isolated from PMNs and other tissues.3,39,42-46 In addition to the p15s, human CAP18 (pro-FALL-39) has been localized to the secondary granules.44 Other cathelicidins have also been identified in inflammatory fluids and/or in granules of PMNs that are subject to exocytosis during inflammation.34,47,48 There is growing evidence that these proteins have functions that extend beyond antimicrobial activity, possibly including a role in wound healing,47 chemotaxis, and protease regulation.48 These observations suggest that the cathelicidins may play important extracellular roles in inflammation including but not limited to antimicrobial action. If so, the substantial release of the p15s from PMNs serves not only to potentiate BPI action but may also serve other extracellular functions. Such functions are currently under investigation.
ACKNOWLEDGMENT
We wish to thank Drs O. Levy, Y. Weinrauch, C.E. Ooi, P. Glennon, M. Rosenfeld, and B. Goldschmidt, as well as S. Katz and Y.X. Yang, for helpful discussions and technical contributions to this study.
Supported in part by US Public Health Service Grant No. R37DK05472 and by the Xoma Corp.
Address reprint requests to Peter Elsbach, MD, Department of Medicine, New York University School of Medicine, 550 First Ave, New York, NY 10016.