Abstract
Methotrexate (MTX) transport was examined in 27 patients with untreated acute lymphocytic leukemia (ALL) and 31 patients with relapsed ALL using a previously described fluorescent MTX analog (PT430) displacement assay (Blood 80:1158, 1992). Only 13% of untreated patients were considered to have impaired MTX transport, whereas more than 70% of relapsed patients had evidence of impaired MTX transport. To further characterize the basis for this defect, Northern analyses for the reduced folate carrier (RFC) were performed on the RNA available from the leukemic blasts of 24 patients in whom MTX transport had been measured. Six of nine samples with impaired MTX transport had decreased RFC expression (one had no detectable RFC expression), while three had no decrease in RFC expression. None of 15 samples with normal MTX transport had decreased RFC expression. A reverse-transcriptase polymerase chain reaction (RT-PCR) assay was developed to quantitate RFC mRNA expression more accurately. Decreased RFC expression was demonstrated in six of the nine samples with impaired MTX transport, confirming the results obtained by Northern blot. These data indicate decreased RFC expression associated with impaired MTX transport is observed in relapsed ALL following treatment with MTX-containing therapy.
METHOTREXATE (MTX) continues to have an important role in the treatment of a variety of malignancies, including leukemia, lymphoma, choriocarcinoma, head and neck cancer, and osteogenic sarcoma.1 The usefulness of MTX in the treatment of these malignancies is limited by the development of drug resistance. In experimental tumor systems, the two common mechanisms of acquired resistance to MTX are decreased transport and amplification of the target gene for MTX, dihydrofolate reductase (DHFR).2-5 However, relatively little information is available on mechanisms of resistance to MTX in patient samples. We recently have shown that low-level gene amplification is present in approximately 30% of patients with acute lymphocytic leukemia (ALL) who relapse after treatment with this drug.6 In a previous study, blasts from two of four patients with ALL believed to be clinically resistant to MTX had evidence of impaired transport, allowing the suggestion that MTX transport impairment may also be a mechanism of acquired resistance in this disease.7
In this study, a competitive displacement flow cytometric assay using the fluorescent lysine analog of MTX, Nα-(4-amino - 4 -deoxy - N10 - methylpteroyl) - Nε - (4'flouresceinthio-carbamyl)-L-lysine(PT430) was used to measure MTX transport in blasts. This method is both sensitive and rapid, and is a useful method to detect MTX transport resistance.7 This assay has also been used by Matherly et al8 to demonstrate that decreased MTX transport may be a component of MTX resistance in childhood ALL.
In this report, we describe studies of MTX transport in the blasts of 74 leukemia patients, as measured by the PT430 competitive displacement assay. The availability of a human cDNA for the reduced folate carrier (RFC)9-12 has also allowed us to measure RFC mRNA expression by Northern blot analysis and to develop a reverse-transcriptase polymerase chain reaction (RT-PCR) assay to quantitate RFC gene expression.
MATERIALS AND METHODS
Preparation of patient samples.A total of 77 blast samples (27 with newly diagnosed ALL, 31 with relapsed ALL, and 19 with AML) from 74 patients were studied. The patients ranged in age from 2 to 73 years and included pre-B-, B-, and T-ALL, as well as M2, M4, and M5 AML. All relapsed patients with ALL had been treated with MTX as part of protocols (NYII or CCG-106 for children and ALL-1 or L-20 for adults13-15 ) used at Memorial Sloan-Kettering Cancer Center (New York, NY). Two milliliters of bone marrow aspirate or 10 mL of peripheral blood in preservative-free heparin was drawn after obtaining informed consent. Mononuclear cells were isolated from blood and bone marrow by Ficoll-Hypaque density centrifugation. All samples contained more than 70% leukemic blasts. Cells were washed twice with phosphate-buffered saline. Cells (2 to 5 × 107) were pelleted and used immediately for preparation of RNA. For use in the PT430 competitive displacement assay, 3 × 107 cells were added to RPMI-1640 media supplemented with 10% fetal calf serum.
RNA isolation.Total cellular RNA was isolated with RNAzol (BiotecX Laboratories, Houston, TX) according to the manufacturer's instructions. RNA was quantitated by determining the absorbance at 260 nm.
PT430 competitive displacement assay.PT430 was synthesized and kindly provided by Drs Joel Wright and Andre Rosowsky of the Dana-Farber Cancer Institute using previously described methods.16,17 The PT430 assay was performed as described previously.7,8,18 Briefly, 3 × 107 cells were incubated in the presence of 2 × 10−5 mol/L PT430 for 2 hours. After washing and allowing for efflux of unbound drug (30 minutes), displacement of PT430 was measured after incubation at 37°C in the presence of the competitors MTX and trimetrexate (TMTX) both at a concentration of 5 × 10−7 mol/L for 90 minutes. Changes in fluorescence were measured by flow cytometry. PT430 displacement was corrected for the nondisplaceable component. Less than 40% displacement of PT430 by MTX was considered indicative of defective transport as was defined in the previous study.7
Northern blotting.Thirty micrograms of total RNA was denatured in 3-(N-Morpholino)propanesulfonic acid (MOPS)-formaldehyde-formamide solution at 65°C for 15 minutes, then electrophoresed on a 1% agarose gel under denaturant conditions. After washing the gel with 10 × SSC (1 × SSC is 0.15 mol/L NaCl plus 0.015 mol/L sodium citrate) for 30 minutes at room temperature, RNA samples were transferred to an Optitran nitrocellulose membrane (Schleicher & Schuell, Keene, NH) according to a standard protocol.19 RNA was fixed to the nitrocellulose by UV irradiation using a Stratagene cross-linker (La Jolla, CA). The DNA for the probe was prepared by digesting pHuMtxT49 with BamH1 and purifying the 2.6-kb fragment using a Geneclean Kit (Bio101, Vista, CA). The probe was labeled with [α-32P]dCTP using a random-primed DNA labeling kit (Boehringer-Mannheim, Indianapolis, IN). The blots were prehybridized for 4 hours, hybridized with probe at 42°C overnight, and washed to a final stringency of 0.1 × SSC and 0.1% sodium dodecyl sulfate (SDS) at 65°C. Autoradiography was performed by overnight exposure to Kodak XAR-5 film (Eastman Kodak, Rochester, NY) with intensifying screens at −70°C. The Northern blots were stripped by boiling and reprobed with 36B420 to determine loading. Radioactivity was quantitated on a Betascope 603 blot analyzer (Betagen, Waltham, MA).
Quantitative RT-PCR.The method developed is based on a noncompetitive amplification of the target gene as compared with an internal standard (β-actin).21 Five to 10 μg of total RNA was incubated with 5 U of RNase-free DNase (Boehringer-Mannheim) for 15 minutes at 37°C, then extracted with phenol:chloroform, precipitated with 100% ethanol, washed with 75% ethanol, and resuspended in water. The RNA was denatured using methyl-mercury hydroxide and β-mercaptoethanol and converted to cDNA with random hexamers using a cDNA cycle kit (Invitrogen, San Diego, CA) as per the manufacturer's instructions, including a second-strand synthesis. RNA prepared from the CCRF-CEM cell line was used as an internal standard. The cDNA was stored at −80°C before use. The cDNA was serially diluted from 1:10 to 1:106 and each dilution was used for two PCR reactions: one using RFC primers and the other using β-actin primers. The oligonucleotides used as primers were synthesized by Operon Technologies, Alameda, CA. The β-actin primers were BA67 and BA68, which have been described previously.21 22 The RFC primers used were as follows: RFC617, 5′-CCAAGCGCAGCCTCTTCTTCAACC; and RFC949, 5′-CCAGCAGCGTGGAGGCAGCATCTGCC; which produce an approximately 300-bp PCR product. The PCR mixture contained 10 pmol of each oligonucleotide primer, 2 nmol of each of the four deoxyoligonucleotides, 5 μCi of [α-32P]dCTP, 0.125 U Taq polymerase, 2.5 μL of 10× CR buffer (Perkin-Elmer, Norwalk, CT), and 10 μL of cDNA diluted with water to a final reaction volume of 25 μL. Forty cycles of the PCR were performed at 94°C for 1 minute, 50°C for 1 minute, and 72°C for 1 minute in a Gene-Amp PCR system 9600 (Perkin-Elmer). The PCR reaction was optimized to produce the largest linear range in the samples that had low RFC expression. Twenty microliters of each PCR product was electrophoresed on a 4% acrylamide gel in 1 × TBE buffer (1 × TBE is 89 mmol/L Tris base, 89 mmol/L boric acid, 2 mmol/L EDTA) at 120 V for 1 hour. The gel was dried for 2 hours at 80°C under vacuum (Bio-Rad Model 583 gel dryer, Richmond, CA) and radioactivity was quantitated on a Betascope 603 blot analyzer (Betagen). RFC/β-actin ratios were calculated by determining the linear range for each sample, plotting the best fit line, and determining the RFC/β-actin ratio at the X intercept.
Biostatistical considerations.Equality of proportion of leukemic blasts obtained from patients with newly diagnosed ALL and from relapsed patients was tested using a binomial proportion test and two-sided P values are reported.23
RESULTS
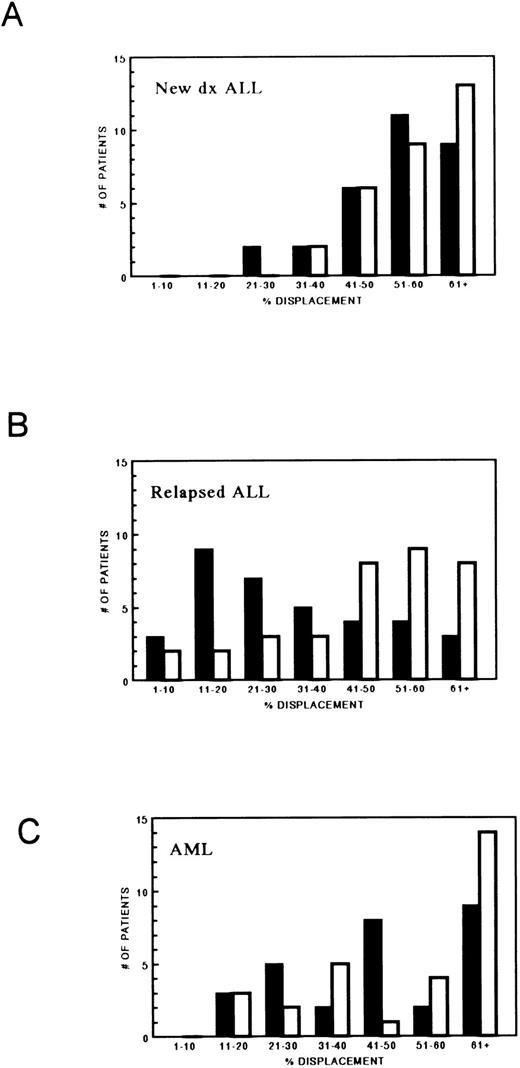
The displacement of PT430 by MTX, as well as TMTX, in the 77 blast samples in addition to the blasts from 17 patients previously reported7 is displayed as histograms in Fig 1. Only 4 of 30 (13%) leukemic blast samples obtained from patients with newly diagnosed ALL had less than 40% displacement of PT430 by MTX. In contrast, blasts from 25 of 35 patients (71%) with relapsed ALL had less than 40% displacement (P < .0001). In contrast, as expected, as TMTX does not use the RFC for uptake, only 10 of 35 patients (28%) with relapsed ALL had less than 40% displacement of PT430 by TMTX (compared with 2 of 27 untreated patients). In AML, 10 of 29 (34%) samples had less than 40% displacement of PT430 by MTX. Seven of 20 patients (35%) with newly diagnosed AML and 3 of 9 (33%) with relapsed AML had less than 40% displacement. In AML, failure to displace PT430 by TMTX (10 of 29, 34%) is more common than in ALL (2 of 27, 7%) (P = .032).
MTX transport in 94 leukemic blast samples as determined by PT430 (2 × 10−5 mol/L) displacement by MTX (5 × 10−7 mol/L) (▪) and TMTX (5 × 10−7 mol/L) (□). (A) Newly diagnosed ALL (n = 30); (B) relapsed ALL (n = 35); (C) acute myelogenous leukemia (AML) patients (n = 29).
MTX transport in 94 leukemic blast samples as determined by PT430 (2 × 10−5 mol/L) displacement by MTX (5 × 10−7 mol/L) (▪) and TMTX (5 × 10−7 mol/L) (□). (A) Newly diagnosed ALL (n = 30); (B) relapsed ALL (n = 35); (C) acute myelogenous leukemia (AML) patients (n = 29).
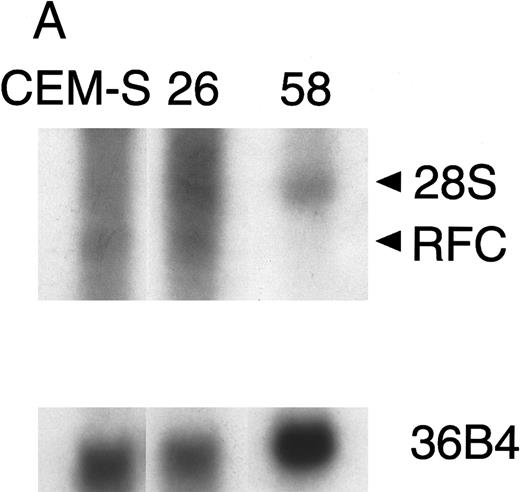
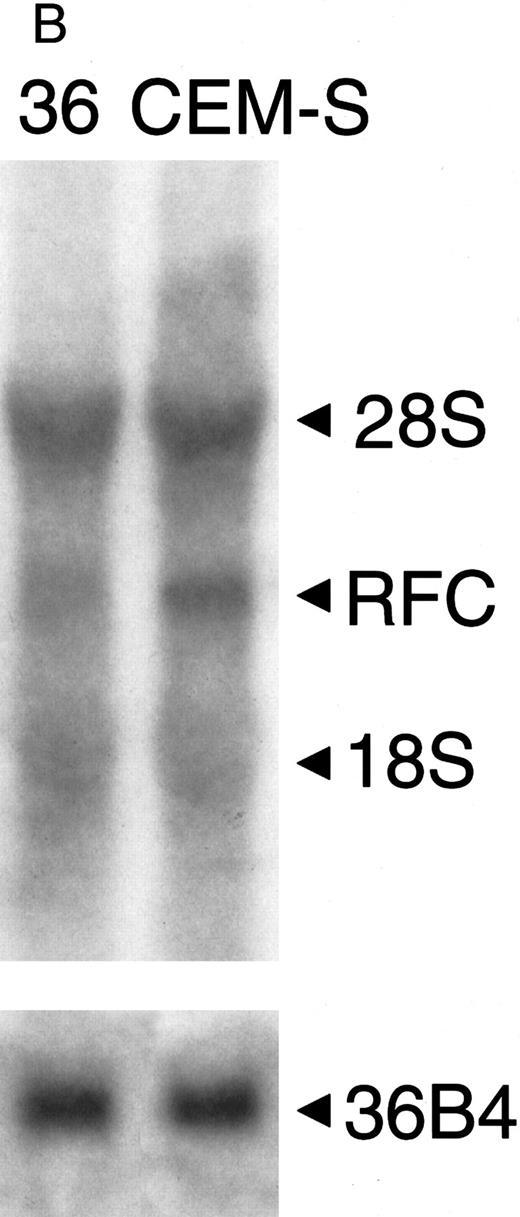
Northern blot of patient samples probed with RFC and 36B4 as a loading control. (A) Patient sample (no. 26) with normal RFC expression and patient sample (no. 58) with no detectable RFC expression relative to CCRF-CEM (CEM-S) cell line. (B) Patient sample (no. 36) with decreased RFC expression relative to the CCRF-CEM (CEM-S) cell line.
Northern blot of patient samples probed with RFC and 36B4 as a loading control. (A) Patient sample (no. 26) with normal RFC expression and patient sample (no. 58) with no detectable RFC expression relative to CCRF-CEM (CEM-S) cell line. (B) Patient sample (no. 36) with decreased RFC expression relative to the CCRF-CEM (CEM-S) cell line.
To determine the basis for the impairment of MTX transport in blasts from ALL patients in relapse, considered to be clinically resistant to this drug, we measured expression of the RFC. There was sufficient material available to perform Northern analysis on RNA obtained from blasts of 9 patients with impaired transport as measured by PT430 displacement and in 15 patients who did not show impaired transport of MTX. Representative Northern blots are shown in Fig 2, which illustrates patient samples with normal, decreased, and no detectable RFC expression relative to a human lymphoblast CCRF-CEM cell line, which is sensitive to MTX (EC50 = 15 nmol/L for 120-hour exposure).24 Of 9 transport-defective samples, 5 had decreased (30% to 90%) and 1 had no detectable RFC expression compared with the RNA from the CCRF-CEM cell line. The remaining 3 transport-defective samples tested had no decrease in RFC expression. None of the 15 samples with normal MTX transport had decreased RFC mRNA expression.
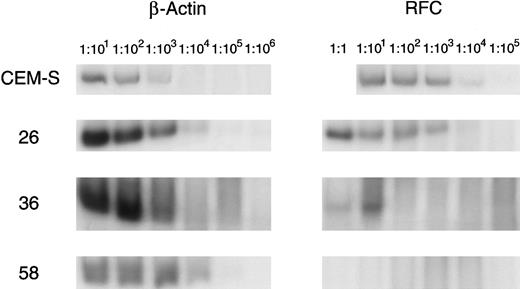
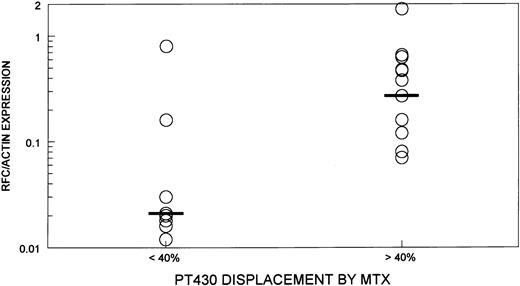
As the results of Northern analysis and quantitation of radioactivity using the Betascope were semiquantitative, a more sensitive method for quantitating RFC expression using RT-PCR was developed and standardized. This assay was performed on the 24 samples in which RFC expression had been measured by Northern analysis. A representative assay is shown in Fig 3 and the results are summarized in Fig 4. Six of nine transport-defective samples that had evidence of decreased RFC expression by Northern blot all had greater than a threefold reduction in RFC/actin ratios compared with a CCRF-CEM cell line. The remaining three transport-defective samples, as well as the 15 samples with normal MTX transport, had less than twofold reduction in the RFC/actin ratios as compared with the CCRF-CEM cell line. A wide range of RFC/actin ratios was observed in the transport normal samples (0.1 to 1.8).
Quantitative RT-PCR of patient samples for RFC expression. Serially diluted cDNA is amplified by PCR with primers for the RFC and β-actin, which is used as a standard. Patient samples with normal RFC expression (no. 26), no detectable RFC expression (no. 58), and decreased RFC expression (no. 36) are displayed with the CCRF-CEM (CEM-S) cell line.
Quantitative RT-PCR of patient samples for RFC expression. Serially diluted cDNA is amplified by PCR with primers for the RFC and β-actin, which is used as a standard. Patient samples with normal RFC expression (no. 26), no detectable RFC expression (no. 58), and decreased RFC expression (no. 36) are displayed with the CCRF-CEM (CEM-S) cell line.
The ratio of RFC/actin expression as determined by quantitative RT-PCR in transport-defective (PT430 displacement by MTX < 40%) (n = 9) and transport-normal samples (PT430 displacement by MTX < 40%) (n = 15). Black bar indicates the median of each group.
The ratio of RFC/actin expression as determined by quantitative RT-PCR in transport-defective (PT430 displacement by MTX < 40%) (n = 9) and transport-normal samples (PT430 displacement by MTX < 40%) (n = 15). Black bar indicates the median of each group.
DISCUSSION
Folates may be transported into cells by two systems: the folate receptors and the RFC. The folate receptors have a higher affinity for folic acid as compared with the reduced folates, while the RFC has a higher affinity for reduced folates and MTX as compared with folic acid.25 The RFC is the predominant transporter of MTX in most malignant cell types.26
The cDNA for the RFC was recently cloned from hamster cells by identifying a cDNA clone that complemented a MTX transport-defective Chinese hamster ovary cell line's ability to grow in media with low amounts of folinic acid (5-formyl tetrahydrofolate) as the sole folate source.27 A homologous cDNA was also identified from murine cDNA by identifying a clone that complemented a MTX transport-defective breast cancer cell line's ability to grow in the presence of folinic acid and TMTX.28 The murine and hamster cDNA sequences were subsequently used by investigators to identify the human homolog of the RFC.9-12
In this study, defective MTX transport (PT430 displacement by MTX < 40%) was found in 71% of blasts from patients with ALL who relapsed after treatments that contained MTX, while 13% of blast samples had evidence of low MTX transport at diagnosis. Evidence of decreased TMTX transport was found in 28% of relapsed ALL patients. As TMTX has been reported to share in the multidrug resistance (MDR) phenotype, it is possible that blasts from these patients were also expressing p-glycoprotein, the product of the MDR-1 gene, as all had received therapy that could result in increased MDR expression.29,30 Another theoretical possibility is a mutant DHFR that binds normally to MTX and PT430, but has decreased affinity for TMTX. This possibility is unlikely, as DHFR cDNA from blasts from relapsed ALL patients have been sequenced without evidence of mutations.31
A quantitative RT-PCR method based on the assay described by Horikoshi et al21 was developed to measure RFC expression. In six of nine transport-defective samples, decreased expression of the RFC gene was observed both by Northern analysis and quantitative RT-PCR. The association of RFC expression and MTX sensitivity, presumably secondary to alterations in transport, has been reported previously in cell lines.32 Three of nine transport-defective samples were found to have normal levels of RFC expression by Northern blot, as well as quantitative RT-PCR. Sequencing of the entire cDNA for the RFC in these three samples is currently in progress to determine if there are mutations in the coding sequence that might explain the low level of MTX transport in these samples. A mutation in the RFC of a murine L1210 leukemia cell line resulted in decreased MTX uptake and MTX resistance despite an approximately fivefold increase in RFC message.33 Defective MTX transport associated with decreased RFC expression is therefore common in relapsed ALL following treatment with MTX-containing therapy.
In AML, previous studies have attributed intrinsic resistance to MTX to defects in polyglutamylation of this drug.34,35 This current study provides evidence that defective MTX transport may also contribute to intrinsic resistance in AML, as impaired MTX transport was found in 35% of blasts from AML patients at the time of diagnosis. In addition, evidence of defective TMTX transport is also observed in 25% of AML patients at diagnosis. This also may be secondary to expression of p-glycoprotein, which has been reported to be capable of effluxing TMTX and has a high incidence of expression in untreated patients with this disease.36 37 We are currently testing the correlation of impaired displacement of PT430 by TMTX also with decreased daunorubicin uptake.
The high incidence of MTX transport resistance found in relapsed ALL patients allows the development of novel strategies to overcome this type of resistance. One such strategy that we are exploring is the use of TMTX with leucovorin protection, which may allow selective cell kill of MTX-resistant cells without significant toxicity.38
Supported by American Cancer Society Grant No. BC-561C, the Parker-Hughes Foundation, the Charles A. Dana Foundation, the Medical Research Council of Canada (W.F.F.), and Grant No. CA09512 from the National Cancer Institute. R.G. is the recipient of an American Society of Clinical Oncology Young Investigator Award. J.R.B. is an American Cancer Society Professor.
Address reprint requests to Joseph R. Bertino, MD, Program of Molecular Pharmacology and Therapeutics, Memorial Sloan-Kettering Cancer Center, New York, NY 10021.