Abstract
Ohnuma et al reported a series of methotrexate-resistant MOLT-3 human T-cell acute lymphoblastic leukemia cell lines that showed decreasing methotrexate (MTX) uptake as the sublines acquired increasing MTX resistance (Cancer Res 45:1815, 1985). The alteration of MTX uptake kinetics in these cells, the intermediately resistant MOLT-3/MTX200 and the highly resistant MOLT-3/MTX10,000 cell lines, was attributed to a change in Vmax for methotrexate transport, without an apparent change in affinity of the transporter for MTX. We studied these cell lines to determine whether alteration of transcription or translation of the recently isolated reduced folate carrier gene (RFC1) was the cause of MTX transport deficiency in these cell lines. Reconstitution of RFC activity in MOLT-3/MTX10,000 cells by transduction with a murine RFC retroviral vector reversed MTX resistance and trimetrexate sensitivity. Although RFC1 RNA levels were unchanged in the resistant cell lines, FACS analysis using a polyclonal anti-RFCl antibody showed no detectable RFCl protein in the MOLT-3/MTX10,000 cells. Determination of the nucleotide sequence of RFC1 genes from MOLT-3/MTX10,000 cells revealed that this cell line contained 3 RFC1 alleles: a wild-type allele, an allele containing the premature stop codon at codon 40 and a third allele containing another mutation, which resulted in a premature stop codon at codon 25. We examined the relative expression of these alleles by determining the nucleotide sequence of 24 RFC1 cDNA subclones from MOLT-3/MTX10,000 cells and found that only one-third of these clones contained the wild-type sequence. Determination of the genomic sequence of RFC1 in MOLT-3/MTX200 cells demonstrated that these cells were heterozygous for a mutation at codon 40, but were homozygous for the wild-type sequence at codon 25. Thus, the acquisition of MTX transport-deficiency in MOLT-3/MTX10,000 cells results from inactivating mutations of RFC1 gene alleles.
FOLATE ANTAGONISTS are effective anticancer drugs in the treatment of malignancies such as acute lymphoblastic leukemia, lymphoma, and breast cancer. Some antifolate drugs, most notably methotrexate, use specific uptake mechanisms to gain intracellular access. Two major types of folate-transport proteins have been described, the folate receptor1-4 and the reduced folate carrier (RFC).5-10 The two types of folate uptake mechanisms have activities that are classically distinguished by different affinities for various folate compounds and by characteristic patterns of uptake inhibition by folic acid and folinic acid.
RFC activity was first shown and characterized in methotrexate (MTX)-resistant cell lines.5-10 These cell lines had in common decreased MTX uptake and loss of activity of a transporter that possessed transport characteristics which distinguished it from the family of folate receptors. Several laboratories, including our own, have recently isolated a gene that encodes a reduced folate carrier, RFC1.11-14 Studies of RFC1 transgenes have shown that the gene product transports MTX and reverses MTX-resistance and trimetrexate (TMTX)-sensitivity in transport-deficient cell lines.14 15
Molecular probes for RFC1 now enable examination of transport-deficient cell lines to determine whether the decrease in RFC functional activity is, in fact, due to changes in RFCl. Characterization of these cell lines is an essential initial step in determining the actual role of RFCl. Therefore, we examined RFC1 expression and genotype in the well-characterized, transport-deficient MTX-resistant MOLT human T-cell lymphoblastic cell line, MOLT-3/MTX10,000,6 to determine whether the previously described reduction in reduced folate carrier functional activity was the result of changes in the regulation of expression or in the nucleotide sequence of RFCl.
MATERIALS AND METHODS
Cell lines.Establishment and characterization of WT MOLT-3, MOLT-3/MTX200, and MOLT-3/MTX10,000 cell lines has been previously described.6 MOLT-3/MTX200 and MOLT-3/MTX10,000 cell lines are 200- and 10,000-fold resistant to MTX, respectively, relative to WT MOLT-3 at the IC50 level of toxicity. WT MOLT-3 cells were also obtained from ATCC (Rockville, MD). The cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), and continuously selected in the presence of MTX. The cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
Determination of the transport kinetics of MTX and 5-methyl THF.The measurements of transport kinetics of MTX and 5-methyl tetrahydrofolate (5-methyl THF) were described previously.6 Briefly, the parent and MTX-resistant cells were washed twice and resuspended in Earl's balanced salt solution (GIBCO-BRL, Gaithersburg, MD) as a cell concentration of 2 × 107/mL. After preincubation of the cells for 15 minutes at 37°C, [3H]MTX or [5-14C]-methyl-THF (59 μCi/mmol; Amersham, Arlington Heights, IL) at graded concentrations (final concentrations of MTX, 5, 10, 20, and 33 μmol/L; of methyl-THF, 2, 2.5, 5, and 10 μmol/L) was added to the medium to initiate the reaction. At various time intervals (0 to 15 minutes) the reaction was terminated by pipeting out a 0.2 mL aliquot of cell suspension into a 15-fold dilution of ice-cold 0.9% NaCl solution. The cells were washed 3 times by centrifugation and then the cell pellet was dissolved in 0.5 mL NCS (Amersham) overnight at 37°C. The samples were counted in a Beckman model 250 liquid scintillation counter (Beckman Instruments, Fullerton, CA) as 30% counting efficiency. The net uptake value represented an average from 3 separate experiments performed in duplicate. In this assay, uptake of MTX and methyl-THF were found to be linear up to 10 minutes. The initial uptake values at 2.5 minutes for MTX or methyl-THF were determined graphically and were plotted in the Lineweaver-Burk plot.
The equations with the least square fit were generated with a computer program (CA-Cricket-Graph III, Computer Associates International, Islandia, NY). The straight line intercepting X-axis represented −1/Kt and the one intercepting Y-axis 1/Vmax.
mRFC retroviral transduction.The murine RFC cDNA16 was subcloned into the pLXSN retroviral backbone and the orientation of the insert was confirmed by nucleic acid sequencing. The pLXSN/mRFC plasmid was transfected into the retroviral packaging cell lines PA317 and GP+E-86 by a standard calcium phosphate precipitation method, according to manufacturer's instructions (GIBCO-BRL). Transfected clones were selected by limiting dilution in 96-well plates by incubation in medium containing G418 (1 mg/mL). Viral titers of surviving clones were determined by the ability of supernatants to confer G418 resistance to NIH3T3 cells. Several clones were identified with titers > 104 neoR colonies/mL and supernatants from these clones were then screened for their ability to increase MTX uptake in transport-deficient MTXRZR-75-1 cells8 as described below. MOLT-3/MTX10,000 cells were incubated in supernatant from one of the mRFC producer clones for 3 days at 32°C and then selected for G418 resistance. Other MOLT-3/MTX10,000 cells were transduced with an empty virus control vector supernatant (pLXSN) and also selected for G418 resistance. The transduced cells were maintained in medium containing 1.0 mg/mL G418.
MTX uptake studies of transduced cells.Transduced MOLT-3/MTX10,000 cells were washed three times with phosphate-buffered saline and resuspended in folate-free improved modified essential medium (IMEM; Life Technologies, Inc, Gaithersburg, MD) at a concentration of 2 × 107 cells/mL. An equal volume of medium containing 6 μmol/L [3H]-labeled methotrexate (Moravek Biochemicals, Brea, CA) was added at time 0. At 5 and 15 minutes, cells were pipeted quickly into ice-cold normal saline and spun 3 seconds in a microcentrifuge. The cells were then twice resuspended and washed in ice-cold normal saline. The supernatant from mRFC retroviral producer cell line clones were screened by incubating MTXRZR-75-1 human breast cancer cells in supernatant for 24 hours on day 1 and incubating the cells for 24 hours on day 3 in 1 μmol/L [3H]-labeled methotrexate. The cells were solubilized by overnight incubation in 0.2 N NaOH, neutralized with 0.2 N HCl, and radioactivity was determined by liquid scintillation counting. Protein concentrations were determined by Bradford assay according to manufacturer's instructions (BioRad, Hercules, CA).
Cytotoxicity studies.MOLT-3/MTX10,000 cell lines transduced with either the mRFC gene (pLXSN/mRFC) or with an empty vector control (pLXSN) were plated in triplicate in 96-well microtiter plates in folate-free IMEM containing 5% FBS at a density of 1,000 cells/well. After 24 hours, serial dilutions of drugs were added to the cells in folate-free IMEM with 5% FBS and either 2 μmol/L folic acid (for MTX cytotoxicity) or 2 μmol/L folinic acid (for TMTX cytotoxicity). After another 6 days, the cells were fixed in 10% tricarboxylic acid, rinsed with water and dried. The cells were then stained with sulforhodamine in 1% acetic acid, washed with 1% acetic acid, and dried again as described. The stained cells were solubilized in 1 mol/L Tris base and the absorbance at 540 nm was determined on a microplate reader. Results are reported as the average of at least three independent determinations performed in triplicate.
Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR).Total RNA (1 μg) from each cell line was reverse transcribed in 20 μL of RT buffer (10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 5 mmol/L MgCl2, 1 mmol/L each of dATP, dGTP, dCTP, dTTP) containing 2 U/mL RNase inhibitor, 0.003 A260 units random hexanucleotides (both from Boehringer Mannheim, Indianapolis, IN) and 0.4 U/mL AMV reverse transcriptase (Promega, Madison, WI). The mixture was incubated at 25°C for 10 minutes, then at 42°C for 30 minutes, and finally at 99°C for 5 minutes. A dilution series for RFC1 and the control gene glyceraldehyde-3-phosphate dehydrogenase (G3PDH) were prepared to determine optimal dilution of cDNA after the RT reaction that would result in PCR amplification in a linear range given the range of constitutive levels of expression of the genes (data not shown). For the RFC1 PCR, the cDNA was not diluted, as the RFC1 cDNA level was within the linear range of the assay. For G3PDH, the RT reaction was diluted 1:800 in RT buffer. Target sequences for RFC1 and G3PDH were separately amplified for 28 cycles (95°C for 10 seconds, 60°C for 30 seconds and 72°C for 15 seconds in a Perkin-Elmer 9600 Thermocycler [Foster, CA]) by the PCR with specific primers for each gene, using the hot start modification. DIG-dUTP (0.1 mL, Boehringer Mannheim) was added to each tube of PCR mixture containing 12.5 mmol/L Tris, pH 8.3, 62.5 mmol/L KCl, 2.5 mmol/L MgCl2, 0.2 mmol/L each of dATP, dGTP dCTP, dTTP, 2.5 U/mL Taq polymerase (Boehringer Mannheim) and 0.3 mmol/L of each specific primer pair in a total volume of 100 μL. An aliquot (15 μL) of the PCR products was separated on a 2% agarose gel in Tris-Borate-EDTA buffer. The gels were denatured, neutralized and the DNA transferred in 10X standard saline citrate (SSC) onto a nylon membrane (Boehringer Mannheim). The membranes were then processed for chemiluminescence detection of the PCR products by the Genius system according to the manufacturer's instructions (Boehringer Mannheim). The relative amounts of each PCR product for RFC1 were quantitated by densitometry and were normalized relative to the amount of PCR product for G3PDH that had been amplified in parallel reactions. The primers used for RFC1 were RFCl-1: at nt +119 from the translation start site (5′-AGATACGGCCAGGGGAGAGCTTCAT-3′) and RFCl-2: at nt +394 (5′-GTAGGAGGAATAGGCGATGCGCGC-3′). These primers span an intron-exon border at nt 190 (data not shown). The primers for G3PDH were G3PDH-1: at nt 75 (5′-GGTCGGAGTCAACGGATTTGGTCGTA-3′) and G3PDH-2: at nt 670 (5′-CCAGTAGAGGCAGGGATGATGTTCTG-3′).
Fluorescence-activated cell sorter (FACS) analysis.A rabbit polyclonal antibody was generated using a peptide sequence leu-phe-phe-asn-arg-asp-asp-arg-gly-arg-cys-glu-thr-ser-ala-ser-glu-leu as previously described.14 WT and MOLT-3/MTX10,000 cells grown in log phase were obtained and washed twice in RPMI 1640 medium and resuspended at a concentration of 1 × 107 cells/mL in medium. Preimmune and immune serum were diluted 1:5 in FACS wash and 25 μL was used for staining. Cells were incubated with 30 μL of human IgG for 15 minutes at room temperature and then in a primary antibody for 60 minutes at 4°C. The cells were then washed, resuspended in 200 μL FACS wash, and aliquots of 2.5 × 105 cells were incubated with the second antibody by the addition of 10 μL goat antirabbit Ig phycoerythrin 0.5 mg/mL (Southern Biotechnology, Birmingham, AL), which had been diluted 1:20 in FACS wash. The cells were washed twice in FACS wash and run on the Becton Dickinson FACStar Plus flow cytometer (San Jose, CA), with a forward scatter and side scatter gate around the population. The data captured for use occurred after the channel number was converted to a linear value. The linear value referred to the dynamic range of signal intensities (1 to 10,000).
Nucleic acid sequencing.The entire coding region of RFC1 was cloned by RT-PCR using primers, which flanked the coding region (5′ primer: at nt −52 from start of translation 5′-CAGGCACAGTGTCACCTTCGTCCCCT-3′ and 3′ primer: at nt +2365: GCCACATGCAGTTCTTCATTCTAC-3′). The PCR products were subcloned into the Sma I-digested and alkaline phophatase-treated pGEM-3Z (Promega). The nucleotide sequences of the cloned cDNAs were determined by dideoxynucleotide sequencing using Sequenase (United States Biochemical, Cleveland, OH) according to the manufacturer's instructions. The reactions were size-fractionated on a 6% denaturing acrylamide gel (National Diagnostics, Atlanta, GA). Custom oligonucleotide primers for sequencing were purchased from the Midland Certified Reagent Co (Midland, TX). To confirm the cloned cDNA sequence, genomic DNA (1 μg) extracted from the cell lines was amplified by PCR using standard methods as previously described17 with primers from the flanking introns (5′ primer 5′-TGTGGCTGGAGACTTCCCTCAGCCT-3′ and 3′ primer 5′-ATGCCTCGTCCCGCGTGAAGTTCT-3′). After treatment with a DNA purification system (Promega) the nucleotide sequence of the PCR fragments was determined using primers end-labeled with [γ-33P]-ATP and T4 DNA polynucleotide kinase (Promega). The sequencing reactions were performed in a Perkin Elmer 9600 thermal cycler using a thermal cycle sequencing kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions and size-fractionated on a 6% polyacrylamide gel. To determine nucleotide sequences of individual RFC1 clones in the region of codons 25 and 40, fragments of RFC1 cDNA amplified by RT-PCR (5′ primer: at nt −52 from the translation start site 5′-CAGGCACAGTGTCACCTTCGTCCCCT-3′ and 3′ primer: +395 from start of translation 5′-ATGTAGGAGGAATAGGCGATGCGC-3′) were gel-purified and subcloned by blunt-end cloning into pCR-Script SK(+) vector (Stratagene). Mini-preps of individual clones were then analyzed by nucleic acid sequencing using the 5′ primer and the Sequenase sequencing system as described above.
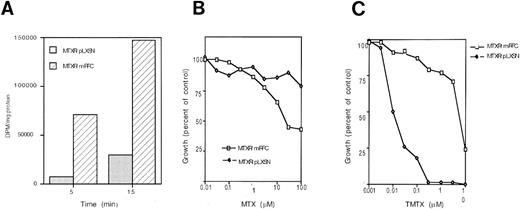
Characterization of mRFC-transduced MOLT-3/MTX10,000 cells. (A) MTX uptake in mRFC-transduced MOLT-3/MTX10,000 cells (MTXR/mRFC) and control cells transduced with empty vector (MTXR/pLXSN). (B) Cytotoxicity curves of mRFC-transduced MOLT-3/MTX10,000 cells (MTXR/mRFC) against MTX in medium containing folic acid compared with MOLT-3/MTX10,000 cells transduced with an empty vector (MTXR/pLXSN). (C) Cytotoxicity curves of mRFC-transduced MOLT-3/MTX10,000 cells against TMTX in medium containing folinic acid compared with MOLT-3/MTX10,000 cells transduced with an empty vector. Each point represents the average of two to three separate experiments performed in triplicate.
Characterization of mRFC-transduced MOLT-3/MTX10,000 cells. (A) MTX uptake in mRFC-transduced MOLT-3/MTX10,000 cells (MTXR/mRFC) and control cells transduced with empty vector (MTXR/pLXSN). (B) Cytotoxicity curves of mRFC-transduced MOLT-3/MTX10,000 cells (MTXR/mRFC) against MTX in medium containing folic acid compared with MOLT-3/MTX10,000 cells transduced with an empty vector (MTXR/pLXSN). (C) Cytotoxicity curves of mRFC-transduced MOLT-3/MTX10,000 cells against TMTX in medium containing folinic acid compared with MOLT-3/MTX10,000 cells transduced with an empty vector. Each point represents the average of two to three separate experiments performed in triplicate.
RESULTS
The transport characteristics of the WT MOLT-3, the intermediately resistant MOLT-3/MTX200 cell line and the highly resistant MOLT-3/MTX10,000 cell line for MTX and the RFC substrate 5-methyl THF are shown in Table 1. As previously reported by Ohnuma et al,6 there appears to be a decrease in the Vmax for MTX as the level of MTX resistance increases, whereas the Kt remains essentially unchanged. Similarly, the Vmax for 5-methyl THF has decreased in the resistant cell lines, whereas the Kt is also essentially unaffected for this substrate. For MTX, the Vmax of the intermediately resistant cell line MOLT-3/MTX200 is 25% of the wild-type value (6.9 v 1.7 nmol/2.5 min/107 cells), and it further decreases to 16% of the WT MOLT3 value in the MOLT-3/MTX10,000 cell line (6.9 v 1.1 nmol/2.5 min/107 cells).
We first determined whether transduction of the resistant cells with an RFC gene would reverse the resistant phenotype. MOLT-3/MTX10,000 cells transduced with the murine RFC expression vector pLXSN/mRFC were selected for G418 resistance, and resistant clones were pooled and analyzed for MTX and TMTX sensitivity. The G418-resistant clones showed an increase in MTX uptake (Fig 1A). Reconstitution of RFC activity restored MTX sensitivity (Fig 1B) and resistance to TMTX (Fig 1C).
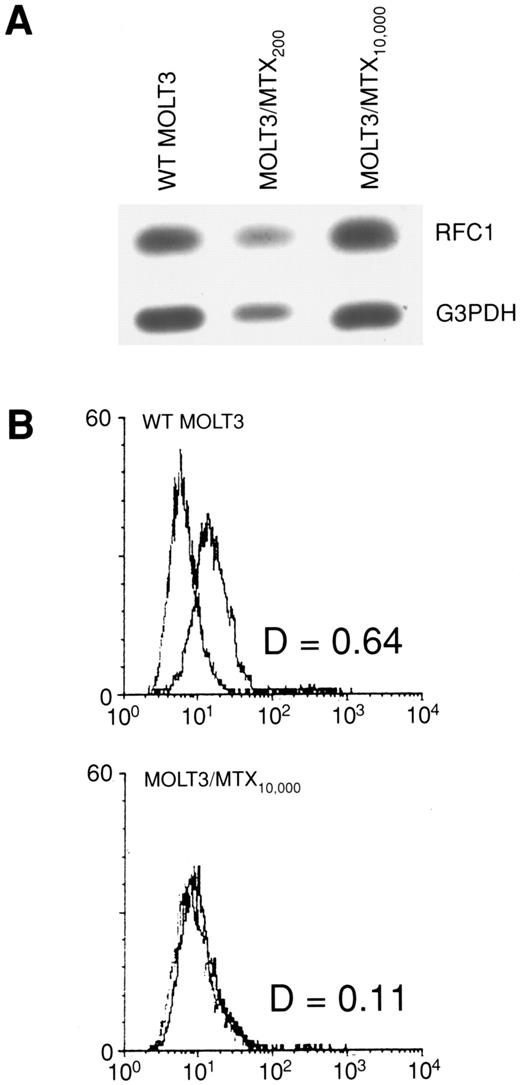
To ascertain whether changes in RFC1 expression accounted for the loss of RFC activity in MOLT-3/MTX10,000 cells, we examined RFC1 RNA and protein levels. As shown in Fig 2A, a quantitative RT-PCR assay showed that there was no significant difference in RFC1 RNA expression in the MTX-resistant cell lines. To determine protein expression, we analyzed the WT MOLT-3 and MOLT-3/MTX10,000 cells by FACS using a polyclonal antibody generated against an RFC1 peptide sequence. As shown in Fig 2B, the RFC1 polyclonal antibody recognized a protein on WT MOLT-3 cells that is not seen in the MOLT-3/MTX10,000 cell line. Thus, these studies indicate that RFC1 protein was decreased below the level of detection in MOLT-3/MTX10,000 cells even though RFC1 RNA levels were apparently unaltered.
(A) Quantitative RT-PCR of RFC1 expression in WT MOLT-3 cells, MOLT-3/MTX200 cells and MOLT-3/MTX10,000 cells. G3PDH expression is used as a control. (B) Flow cytometric analysis of WT MOLT-3 (top) and MOLT-3/MTX10,000 cells (bottom) with anti-RFCl polyclonal antiserum and control preimmune serum. The X-axis is fluorescence intensity and the Y-axis is cell number.
(A) Quantitative RT-PCR of RFC1 expression in WT MOLT-3 cells, MOLT-3/MTX200 cells and MOLT-3/MTX10,000 cells. G3PDH expression is used as a control. (B) Flow cytometric analysis of WT MOLT-3 (top) and MOLT-3/MTX10,000 cells (bottom) with anti-RFCl polyclonal antiserum and control preimmune serum. The X-axis is fluorescence intensity and the Y-axis is cell number.
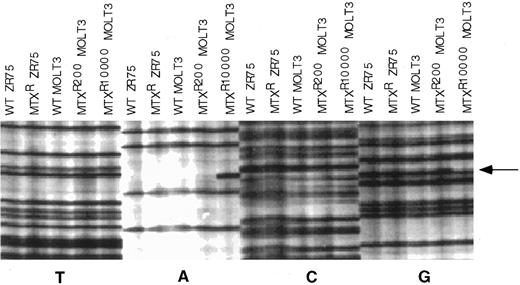
We then examined the nucleotide sequence of the entire protein coding region of RFC1 alleles in the MTX-resistant MOLT-3 sublines to attempt to identify the molecular mechanism that would account for these observations. First, we determined the sequence of a single cDNA clone from WT MOLT-3, MOLT-3/MTX200, and MOLT-3/MTX10,000 cell lines. As shown in Fig 3, a sequencing gel showed the presence of a mutation at codon 25 in the RFC1 cDNA clone from the MOLT-3/MTX10,000 cell line. This mutation changed a TGG codon encoding tryptophan into TAG, a premature stop codon. There were no mutations in this region in the single RFC1 cDNA clone isolated from MOLT-3/MTX200 cells, and no other mutations were identified in the remainder of the coding region in any of the MOLT-3 cDNA clones analyzed.
Nucleotide sequencing gel of individual cDNA clones of the nucleotide sequence in the region of RFC1 codon 25. The sequencing gel was arranged so that all dideoxy termination reactions for the different templates were grouped together, which facilitated recognition of aberrant sequences. cDNAs included in the gel were from WT ZR-75 human breast cancer cell line, a transport-deficient MTX-resistant ZR-75 cell line,8 WT MOLT-3, MOLT-3/MTX200, and MOLT-3/MTX10,000 cells. Arrow designates site of mutation in the MOLT-3/MTX10,000 cDNA clone.
Nucleotide sequencing gel of individual cDNA clones of the nucleotide sequence in the region of RFC1 codon 25. The sequencing gel was arranged so that all dideoxy termination reactions for the different templates were grouped together, which facilitated recognition of aberrant sequences. cDNAs included in the gel were from WT ZR-75 human breast cancer cell line, a transport-deficient MTX-resistant ZR-75 cell line,8 WT MOLT-3, MOLT-3/MTX200, and MOLT-3/MTX10,000 cells. Arrow designates site of mutation in the MOLT-3/MTX10,000 cDNA clone.
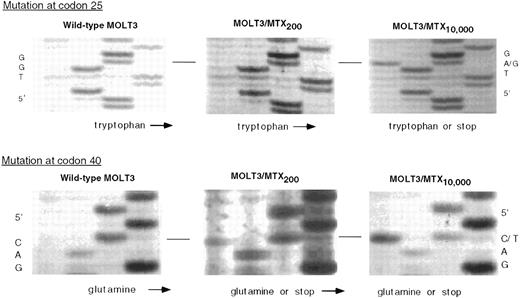
RFC1 sequences from the genomic gene were then amplified and the nucleotide sequence determined to confirm this mutation. As shown in Fig 4, the genomic sequence around codon 25 shows MOLT-3/MTX10,000 cells are heterozygous at this position, with both TGG and TAG sequences present, whereas the nucleotide sequence of the WT MOLT-3 and MOLT-3/MTX200 genomic sequence is homozygous for TGG. Furthermore, another mutation was evident at codon 40, with both the MOLT-3/MTX10,000 and MOLT-3/MTX200 cells showing heterozygosity for a mutation which results in another premature stop codon. The sequences at codon 40, CAG and TAG, encode glutamine and stop, respectively, whereas only the sequence encoding glutamine is present in the WT MOLT-3 cells.
Genomic DNA sequence of WT MOLT-3, MOLT-3/MTX200, and MOLT-3/MTX10,000 cells in the region of codon 25 (top) and codon 40 (bottom). Thermal cycle sequencing reactions on PCR-amplified genomic DNA were performed as described in Materials and Methods and size-fractionated on a 6% polyacrylamide gel.
Genomic DNA sequence of WT MOLT-3, MOLT-3/MTX200, and MOLT-3/MTX10,000 cells in the region of codon 25 (top) and codon 40 (bottom). Thermal cycle sequencing reactions on PCR-amplified genomic DNA were performed as described in Materials and Methods and size-fractionated on a 6% polyacrylamide gel.
Thus, a single inactivating mutation is present in the MOLT-3/MTX200 cells and an additional inactivating mutation is present in the RFC1 genomic sequence of MOLT-3/MTX10,000. The mutation at codon 40 resulting in a premature stop codon was not present on the single cDNA clone isolated from MOLT-3/MTX10,000 cells, indicating that the two mutations may reside on different alleles. To determine whether these mutations resided on different alleles, and to determine their relative expression, we determined the nucleotide sequence of 24 partial RFC1 cDNA clones from MOLT-3/MTX10,000 cells. To our surprise, the sequencing data showed that there are three RFC1 alleles present in the MOLT-3/MTX10,000 cell lines, one allele each with the inactivating mutations at either codon 25 or codon 40, and a third allele with the wild-type sequence. Of 24 clones, two-thirds of the alleles expressed contained inactivating mutations of RFC1 (Table 2). Sequencing of individual clones of genomic DNA from this exon also show the existence of the wild-type allele in MOLT-3/MTX10,000 cells (data not shown).
DISCUSSION
MTX resistance in MOLT-3/MTX10,000 cells is due, in part, to decreased uptake of MTX (Table 1 and ref. 6). The data shown in Table 1 presents kinetic evidence that the decrease in MTX uptake in the resistant cells was caused by a stepwise decrease in the Vmax for the transporter as opposed to a change of affinity of the transporter for MTX. In the original report characterizing these cell lines, Ohnuma et al6 suggested that the decrease in MTX uptake in these cells was caused by the downregulation of a reduced folate carrier activity. Our data support these transport kinetic studies and present a molecular explanation for them.
Transduction of MOLT-3/MTX10,000 cells with the murine RFC gene increased MTX uptake and reversed MTX resistance and TMTX sensitivity in these cells, showing that reconstitution of RFC activity could at least partially reverse the resistant phenotype. Full reversal would not be expected in these cells, as they also have increased dihydrofolate reductase activity.6
The quantitative RT-PCR and FACS analysis shown in Fig 1 strongly suggest that RFC1 is involved in the decrease in RFC activity in MOLT-3/MTX10,000 cells. Taken together, these data indicate that although RFC1 is transcribed, its translation is decreased in the resistant cells. To determine the genetic mechanisms that would account for these results, we analyzed the nucleotide sequence RFC1 alleles in MOLT-3/MTX10,000 cells. By screening a single cDNA clone from MOLT-3/MTX10,000 cells, we initially identified a premature stop codon that was confirmed on genomic DNA. Further analysis of the genomic DNA sequence revealed another premature stop codon (at codon 40), providing an explanation for the loss of activity of two alleles.
Nucleic acid sequencing of RFC1 cDNA clones showed conclusively that alleles with two different inactivating mutations of RFC1 are expressed in MOLT-3/MTX10,000 cells. However, the results from the sequencing also indicate that a third allele with a wild-type sequence is also present and expressed. These results are not inconsistent with the genomic sequencing data, and point out the importance of sequencing several individual clones before making conclusions of heterozygosity based on genomic sequencing.
Fortunately, we were able to examine a subline with an intermediate level of resistance, the MOLT-3/MTX200 cells, to gain insight into the evolution of the mutations. The genomic sequence of MOLT-3/MTX200 cells contains the codon 40 mutation, but not the codon 25 mutation; thus, it appears that the codon 40 mutation occurred during the first stages of selection for MTX resistance, and that the codon 25 mutation on the other allele occurred later in selection.
These studies show that decreased expression of the RFC1 gene product contributes to decreased MTX uptake and MTX resistance in the MOLT-3/MTX10,000 cell line. The fact that two RFCl alleles acquire mutations suggests the importance of this gene product in selection for MTX resistance. The residual MTX uptake in MOLT-3/MTX10,000 cells may be due to expression of the wild-type allele. It is of some interest whether expression of the two inactivated alleles affects the translation of the wild-type allele. These data also suggest that there is no evidence for a feedback mechanism that might have resulted in increased RFC1 transcription in the face of lessened activity, as the RNA levels between the resistant and parental cell lines are equivalent.
The in vivo role of RFC1 in antifolate drug resistance and in normal folate metabolism remains to be described. These studies have practical implications for further evaluation of the role of RFC1 in clinical drug resistance. If we had screened MOLT-3/MTX10,000 cells by RNA levels alone, we would have falsely assumed that RFC1 was not involved as a mechanism of resistance. Another cell line with decreased RFC activity, an L1210 murine leukemia cell line, exhibited only a 30% decrease in RFC RNA transcript levels; however, molecular analysis demonstrated a mutation in the RFC coding region resulting in an amino acid substitution.18 Clearly, screening tissues and tumors for the presence of protein, or with functional assays, is likely to be more predictive of RFC activity than screening for RNA levels alone. In addition, identification of RFC1 as a mechanism of MTX resistance now opens the door for consideration of novel methods, besides the administration of high-dose MTX, to overcome MTX transport resistance.
Supported in part by the T.J. Martell Foundation for Leukemia, Cancer and AIDS Research (New York, NY) (T.O.), and by the United Leukemia Fund (New York, NY) (T.O.).
Address reprint requests to Jeffrey A. Moscow, MD, Medicine Branch, National Cancer Institute, Bldg 10, Room 12N226, National Institutes of Health, Bethesda, MD 20892.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore by hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.