Abstract
Recent findings have shown that the expression of the seven trans-membrane G-protein–coupled CXCR4 (the receptor for the stromal cell-derived factor [SDF]-1 chemokine) is necessary for the entry of T-lymphotropic human immunodeficiency virus (HIV) strains, acting as a coreceptor of the CD4 molecule. In the human system, the role of CXCR4 in HIV infection has been determined through env-mediated cell fusion assays and confirmed by blocking viral entry in CD4+/CXCR4+ cells by SDF-1 pretreatment. We observed that the human megakaryoblastic CD4+ UT-7 cell line fails to express CXCR4 RNA and is fully resistant to HIV entry. Transfection of an expression vector containing the CXCR4 c-DNA rendered UT-7 cells readily infectable by different T-lymphotropic syncytium-inducing HIV-1 and HIV-2 isolates. Interestingly, HIV-1 infection of CXCR4 expressing UT-7 cells (named UT-7/fus) induces the formation of polynucleated cells through a process highly reminiscent of megakaryocytic differentiation and maturation. On the contrary, no morphologic changes were observed in HIV-2–infected UT-7/fus cells. These findings further strengthen the role of CXCR4 as a molecule necessary for the replication of T-lymphotropic HIV-1 and HIV-2 isolates and provide a useful model to study the functional role of CD4 coreceptors in HIV infection.
THE BINDING OF CD4 receptor with viral gp120 envelope protein is the first step in human immunodeficiency virus (HIV) infection. However, as widely shown in human and nonhuman cells, CD4 exposition is not per se sufficient to allow viral entry.1,2 Recent studies have demonstrated the role that CXCR4 (also called LESTR, HUMSTR, or fusin)3 and CC-CKR54 play as coreceptors of syncytium-inducing (SI) T-lymphotropic5 and non–syncytium-inducing (NSI) macrophage (M)-tropic6-10 HIV strains, respectively. Both molecules are receptors for chemokines; CXCR4 is the stromal cell-derived factor (SDF )-111,12 receptor, whereas MIP-1α, MIP-1β, and RANTES bind CC-CKR5 receptor.4 These pioneristic data were obtained by transiently transfecting target cells with different molecular constructs and measuring the activity of reporter genes (ie, luciferase, cloramphenicol-acetyl-transferase, or β-galactosidase) in env-mediated cell fusion experiments or inserted in HIV env-defective molecular clones in env-complementation assays.
UT-7 is a cell line established from a patient with acute megakaryoblastic leukemia and is strictly dependent for growth on either granulocyte-macrophage colony-stimulating factor (GM-CSF ), interlukine-3 (IL-3), or erythropoietin (Epo).13 These cells express specific megakaryocytic markers such as glycoprotein (GP) IIb and GPIIIa as well as the erythroid marker glycophorin A. These features, together with the possibility of inducing UT-7 cells to differentiate toward erythroid (by Epo treatment)14 or megakaryocytic (by phorbol myristate acetate induction)13 differentiation, indicate that UT-7 cells are phenotypically related to a common hematopoietic progenitor pertaining both to the megakaryocytic and erythroid cell lineages.15
In this report we show that, despite the exposure of the CD4 receptor, UT-7 cells are totally resistant to HIV infection. The inability of HIV to entry and replicate directly correlates with the lack of CXCR4 gene expression. In fact, transfection of a vector expressing the CXCR4 gene renders UT-7 cells highly susceptible to infection by different SI T-lymphotropic HIV-1 and HIV-2 strains. In addition, HIV-1 infection of UT-7 cells stably expressing CXCR4 c-DNA (named UT-7/fus cells) induces morphologic changes highly reminiscent of the megakaryocytic differentiation process.
MATERIALS AND METHODS
Cell cultures.UT-7 cells were grown in Iscove's modified Dulbecco's medium (GIBCO, Grand Island, NY) supplemented with 10% decomplemented fetal calf serum (FCS) and 1 ng/mL of recombinant human GM-CSF (Boehringer Mannheim, Mannheim, Germany) or IL-3 (Genetics Istitute, Cambridge, MA). UT-7/fus cells were cultivated in the same medium supplemented with 1 mg/mL of G418 antibiotic (GIBCO-BRL, Gaithersburg, MD; 70% of activity). CEMss and C8166 cells were maintained in RPMI 1640 medium supplemented with 10% decomplemented FCS.
Construction of CXCR4-expressing vector and transfection.Total RNA was extracted from quiescent human peripheral blood lymphocytes according to the manufacturer's instructions (RNA-FastI kit; Molecular System, San Diego, CA). Five micrograms of RNA was reverse transcribed in a final volume of 50 mL using oligodT as primer. Ten milliliters of reverse transcribed RNA was then subjected to polymerase chain reaction (PCR) using oligoprimers (whose sequence was reported by Dragic et al9 ) overlapping the start and stop codons of the CXCR4 open reading frame and carrying HindIII (oligo forward) and EcoRI (oligo reverse) restriction sites at the respective 5′ ends. The reaction was performed as previously described16 in a total volume of 50 mL. The product of amplification was digested with HindIII and EcoRI enzymes and inserted in pcDNA3 (Invitrogen, San Diego, CA) expression vector previously digested with the same restriction enzymes.
UT-7 cells were transfected by the electroporation method by adding 10 μg of linearized plasmidic DNA at 5 × 106 cells pulsed at 200 V, 950 μF through a Gene Pulser apparatus (BioRad, Richmond, CA).
HIV infection and detection.Culture supernatants from acutely infected CEMss cells were used as a source of HIV (NL4-3, LAV-1, NDK, and HIV-2ROD ) preparations. Titers of SI strains (ranging from 3 × 106 to 107 50% tissue culture infective dose [TCID50] per milliliter) were measured as previously described17 by scoring the syncytia number on C8166 cells 5 days after infection with serially diluted virus preparations. HIV Bal-1 virus preparations were obtained as described18 and titrated as picograms per milliliter of p24 protein through quantitative enzyme-linked immunosorbent assay (Abbott, North Chicago, IL). Infections were performed by adsorbing the virus for 1 hour at 37°C. After infection, cells were washed and fed. HIV release was monitored by reverse transcriptase (RT) assay on culture supernatants.19
In experiments of PCR on infected UT-7 cells, only HIV strains (ie, NL4-3 and HIV-2ROD ) free of intravirionic DNA detectable by PCR were used.
DNA and RNA analyses.Early intracellular HIV retrotranscription in infected UT-7 cells was assessed by DNA-PCR using primers specific for the long terminal repeat (LTR) region of both HIV-1 and HIV-2 already described.16 Viral preparations were treated with DNAase (Boehringer, GmbH, Mannheim, Germany) for 2 hours at room temperature (rt) in the presence of 1 mmol/L of MgCl2 . Samples were prepared by lysing 105 cells in 50 μL of tridistillated DNA-free water at 95°C for 5 minutes and subjected to PCR as described.16 Amplified products were run on 1.8% agarose gel electrophoresis, transferred onto nitrocellulose membrane, and hybridized using a 32P end-labeled specific oligonucleotides16 as probe. Cell lysates were contemporary amplified by using primers specific for the human β-globin.17
RT-PCR for the detection of CXCR4 expression was performed by using the same oligoprimers employed for molecular cloning (see above) and probed with an oligoprimer internal of the CXCR4 c-DNA (from nt 961 to nt 989).3 RNA were normalized by using primers specific for the human β-2 microglobulin gene.20
Northern blot was performed as previously described.21 Filters were hybridized with 32P random primed labeled CXCR4 c-DNA or a 316-bp fragment of the human glyceraldehyde-3-phosphodehydrogenase (GAPDH) c-DNA (Ambion, Austin, TX) derived from exons 5 through 8.
Amplified CXCR4 c-DNA used for the UT-7 transfection was sequenced by the dideoxy chain termination method using the SequenaseII kit (US Biochemical Corp, Cleveland, OH).
Gp120 competition assay and antigenic and morphologic analyses.Gp120 competition assay was performed by incubating 5 × 105 cells with 2 μg of LAV-1 gp120 recombinant protein (Intracel, Cambridge, MA) for 1 hour at 4°C. Cells were washed three times with phosphate-buffered saline (PBS) and then incubated with 1:20 diluted phycoerythrine (PE) Leu3a monoclonal antibody (MoAb; Becton Dickinson, Mountain View, CA) for 1 additional hour at 4°C and finally analyzed with a cytofluorimeter (FAC-scan; Becton Dickinson).
PE-conjugated MoAbs to CD4 (Leu3a), CD31, CD41a, and glycophorin A (Becton Dickinson) were used at appropriate dilutions. After incubation at 4°C for 1 hour, cells were washed and analyzed by a cytofluorimeter. HIV gag-related proteins were detected by intracellular direct immunofluorescence. Cells (3 × 105) were washed three times with PBS, resuspended in 200 μL of Permafix (Ortho Diagnostics, Raritan, NJ), and incubated for 30 minutes at rt. After two additional washes, cells were incubated 1 hour at rt with 1:20 diluted KC57-RD1 (Coulter Corp, Hialeah, FL) anti-p24 HIV MoAb conjugated with PE, washed three times with PBS, and finally analyzed by a cytofluorimeter.
Uninfected and HIV infected cells cytocentrifuged onto glass slides were stained with May-Grünwald Giemsa and morphologically analyzed.
RESULTS
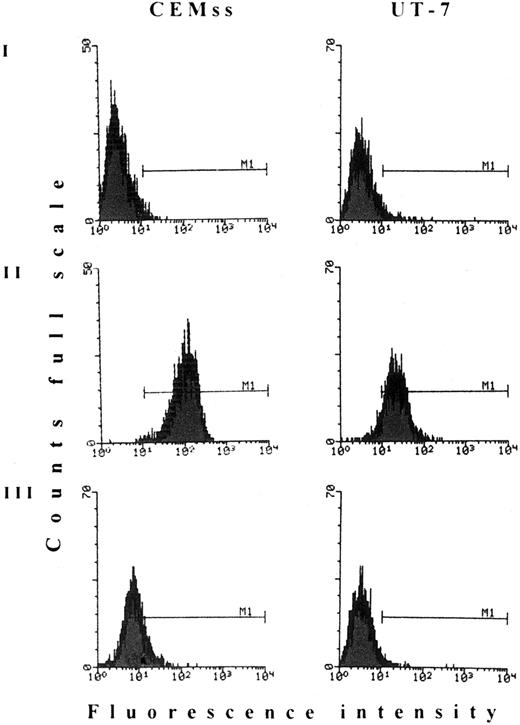
HIV does not penetrate into UT-7 cells, despite the exposure of CD4 receptors able to bind HIV gp120.In a first set of experiments we observed that, although UT-7 cells homogenously expose the CD4 receptors, infection with different HIV-1 (either SI or NSI) or HIV-2 strains repeatedly failed. Thus, to assess whether CD4 receptors were able to efficiently bind the HIV env glycoprotein, gp120/CD4 competition assay was performed. As shown in Fig 1, gp120 HIV protein efficiently competes for the binding with the Leu3A MoAb, suggesting that the CD4 molecules exposed on UT-7 cell membrane are able to bind HIV virions. In this assay, CEMss, a T-lymphoblastoid human cell line exposing high levels of CD4 receptors, was used as a positive control. To determine at which step the HIV infection is blocked, we analyzed the earliest events of retrotransciption. DNA-PCR analysis on infected cell lysates was performed by using oligonucleotides specific for HIV-LTR sequences immediately 5′ to primer binding-site16 that is the genomic region firstly retrotranscribed. As shown in Fig 2, no signals were detected in UT-7 cells at any time after infection with HIV-1 or HIV-2. These data showed that the block of HIV infection occurs in a step between viral binding and RNA retrotranscription.
HIV-1 gp120 competition assay. Both CEMss and UT-7 cells were labeled with (I) PE-conjugated unspecific mouse IgG; (II) Leu3A anti-CD4 MoAb; and (III) Leu3A MoAb after pretreatment with saturating amounts of HIV-1 recombinant gp120 protein. M1, marker 1.
HIV-1 gp120 competition assay. Both CEMss and UT-7 cells were labeled with (I) PE-conjugated unspecific mouse IgG; (II) Leu3A anti-CD4 MoAb; and (III) Leu3A MoAb after pretreatment with saturating amounts of HIV-1 recombinant gp120 protein. M1, marker 1.
PCR analysis of HIV-1– or HIV-2–infected UT-7 cells. Both UT-7 and CEMss cells were infected with (A) NL4-3 HIV-1 or (B) HIV-2ROD strains (moi 1); cells were collected 8, 24, and 48 hours postinfection; and cell lysates were analyzed by PCR. Lysates from CEMss cells 5 days after infection with NL4-3 HIV-1 or HIV-2ROD were used as positive controls (c+). Cell lysates were also amplified with β-globin–specific primers as control.
PCR analysis of HIV-1– or HIV-2–infected UT-7 cells. Both UT-7 and CEMss cells were infected with (A) NL4-3 HIV-1 or (B) HIV-2ROD strains (moi 1); cells were collected 8, 24, and 48 hours postinfection; and cell lysates were analyzed by PCR. Lysates from CEMss cells 5 days after infection with NL4-3 HIV-1 or HIV-2ROD were used as positive controls (c+). Cell lysates were also amplified with β-globin–specific primers as control.
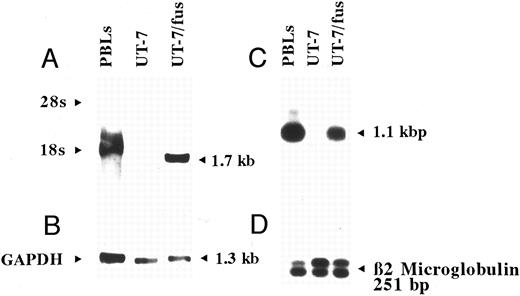
Transfection of the UT-7 cells with CXCR4 expressing vector: characterization of UT-7/fus cells.We first examined whether the inability of HIV to penetrate into UT-7 cells may be correlated to the lack of expression of CXCR4, the coreceptor for T-lymphotropic SI HIV strains. Both Northern blot and RT-PCR analyses showed that UT-7 cells do not express CXCR4 mRNA (Fig 3). We then transfected UT-7 cells with an expression vector containing the human CXCR4 c-DNA under the control of the immediate-early cytomegalovirus promoter.
Analysis of CXCR4 expression in UT-7 and UT-7/fus cells. Northern blot (A and B) and RT-PCR (C and D) were performed on total RNA obtained from human peripheral blood lymphocytes and parental UT-7 and UT-7/fus cells. Northern blot was hybridized with CXCR4 c-DNA (A) or GAPDH (B) probes. RT-PCR products were hybridized with an end-labeled oligoprimer internal to CXCR4 c-DNA (C). As control, RT-PCR for β2-microglobulin19 was performed (D).
Analysis of CXCR4 expression in UT-7 and UT-7/fus cells. Northern blot (A and B) and RT-PCR (C and D) were performed on total RNA obtained from human peripheral blood lymphocytes and parental UT-7 and UT-7/fus cells. Northern blot was hybridized with CXCR4 c-DNA (A) or GAPDH (B) probes. RT-PCR products were hybridized with an end-labeled oligoprimer internal to CXCR4 c-DNA (C). As control, RT-PCR for β2-microglobulin19 was performed (D).
The G418-resistant cell population (UT-7/fus cells) was characterized in terms of CXCR4 RNA expression, growth, morphology, and antigenic phenotype. As expected, UT-7/fus cells expressed high levels of CXCR4 mRNA (Fig 3). No significant differences in the rate of cell growth between UT-7 and UT-7/fus cells were detected in the presence of IL-3 or GM-CSF (not shown). Similarly, the analysis of membrane antigens (CD4, CD31, CD41a, and glycophorin A) failed to evidentiate any modification in UT-7/fus with respect to UT-7 cells (not shown). However, a slight increase in polyploidy was detectable in UT-7/fus with respect to parental cells (see below). As a control, we used parental or UT-7 cells transfected with empty pcDNA3 expressing vector, whose presence did not modify the morphology, the antigen expression, and cell growth.
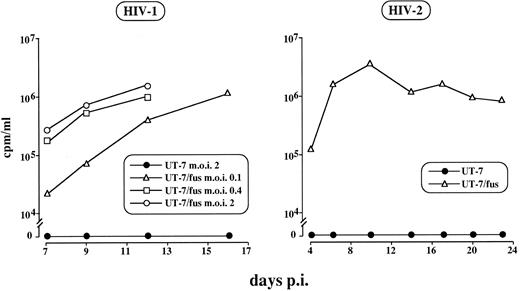
UT-7/fus cells are highly susceptible to HIV infection.To assess whether the expression of the transfected CXCR4 gene was sufficient to render UT-7 cells susceptible to HIV infection, UT-7/fus cells were challenged with the NL4-3 strain at different multiplicity of infection (moi; ie, 0.1, 0.4, and 2) and the presence of virions in culture supernatants was tested by RT activity. The results shown in Fig 4 indicate that UT-7/fus cells were readily infectable at any moi used and that the kinetics of viral release strictly depends on viral load. In addition, the values of RT activity were as high as those detected in highly HIV-sensitive CEMss cells (not shown). Conversely, no virus release was observed after infection of either UT-7– (Fig 4) or pcDNA3-transfected UT-7 cells (not shown). Similar results were obtained on UT-7 and UT-7/fus cells infected with LAV-1, NDK (not shown), or HIV-2ROD strains (Fig 4).
HIV-1 and HIV-2 infection of UT-7 and UT-7/fus cells. Cells were infected with NL4-3 HIV-1 or with HIV-2ROD strains and RT activity in supernatant cultures was assayed at different time points. RT activity below the background was detected in any sample derived from UT-7 infected supernatant, regardless of moi or HIV strain used.
HIV-1 and HIV-2 infection of UT-7 and UT-7/fus cells. Cells were infected with NL4-3 HIV-1 or with HIV-2ROD strains and RT activity in supernatant cultures was assayed at different time points. RT activity below the background was detected in any sample derived from UT-7 infected supernatant, regardless of moi or HIV strain used.
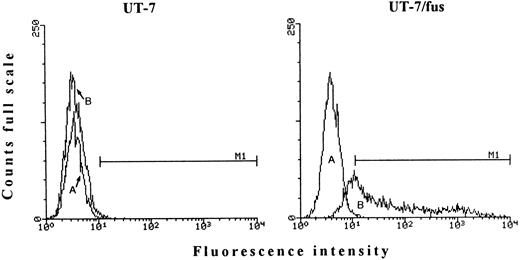
To evaluate the proportion of HIV susceptible UT-7/fus cells, immunofluorescence analysis for the production of intracellular p24 protein was performed 7 days after HIV-1 infection (moi, 0.4). Figure 5 shows that virtually all UT-7/fus cells were HIV+. We also observed that, at different days after infection, depending on the moi used (ie, 10 to 14 days after infection with the higher moi), UT-7/fus cells died.
FACS analysis for the presence of intracytoplasmic HIV-1 gag protein. Both UT-7 and UT-7/fus cells were labeled with PE-conjugated unspecific mouse IgG (A) or PE-conjugated anti–HIV-1 p24 MoAb (B) 7 days after infection (moi 0.4). M1, marker 1.
FACS analysis for the presence of intracytoplasmic HIV-1 gag protein. Both UT-7 and UT-7/fus cells were labeled with PE-conjugated unspecific mouse IgG (A) or PE-conjugated anti–HIV-1 p24 MoAb (B) 7 days after infection (moi 0.4). M1, marker 1.
In contrast to HIV-1, HIV-2 infection of UT-7/fus cells did not lead to any evident morphologic alterations, even though the virus was able to enter and replicate, as shown in Fig 4. Furthermore, upon HIV-2 infection, chronically infected UT-7/fus cells could be readily obtained. As expected, UT-7/fus cells remain resistant to infection with Bal-1, a M-tropic NSI HIV strain (data not shown).
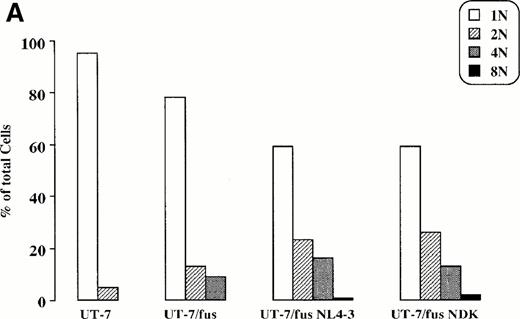
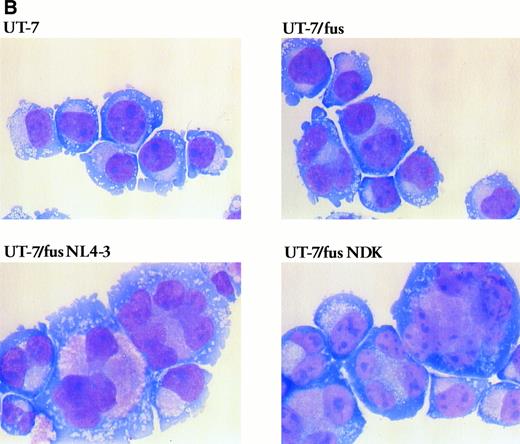
HIV infection induces modifications in UT-7/fus cell morphology and phenotype.As reported above, in UT-7/fus cells a slight increase of polyployidy with respect to the parental cells was observed (Fig 6A). However, upon HIV-1 infection, UT-7/fus cells greatly modified their morphology, progressively involving the entire cell population. Morphological analysis (Fig 6B) shows (1) a generalized and homogeneous enlargement of cell size, characteristic of differentiation process in normal megakaryocytes. This is also detectable in dot plots (Fig 7), where a clear difference between UT-7/fus (R1) and HIV-infected UT-7/fus (R2) cell populations could be observed. Also shown by morphological analysis are (2) an increase of polyploidy as shown by the presence of cells with up to 8 nuclei (as also shown in Fig 6A); and (3) a cytoplasm less azzurophilic than parental UT-7 or UT-7/fus cells and some large eosinophilic areas containing several purple punctuations.
Morphologic analysis of UT-7, UT-7/fus, and HIV-infected UT-7/fus cells. (A) Analysis of UT-7, UT-7/fus, and NL4-3 or NDK HIV-1–infected UT-7/fus cells on the basis of nuclei number (N) as evaluated by optical microscopy. Results from a representative experiment are shown. (B) Cytospin of uninfected UT-7 and UT-7/fus cells compared with UT-7/fus cells infected with NL4-3 or NDK HIV-1 strains. (Original magnification × 400.)
Morphologic analysis of UT-7, UT-7/fus, and HIV-infected UT-7/fus cells. (A) Analysis of UT-7, UT-7/fus, and NL4-3 or NDK HIV-1–infected UT-7/fus cells on the basis of nuclei number (N) as evaluated by optical microscopy. Results from a representative experiment are shown. (B) Cytospin of uninfected UT-7 and UT-7/fus cells compared with UT-7/fus cells infected with NL4-3 or NDK HIV-1 strains. (Original magnification × 400.)
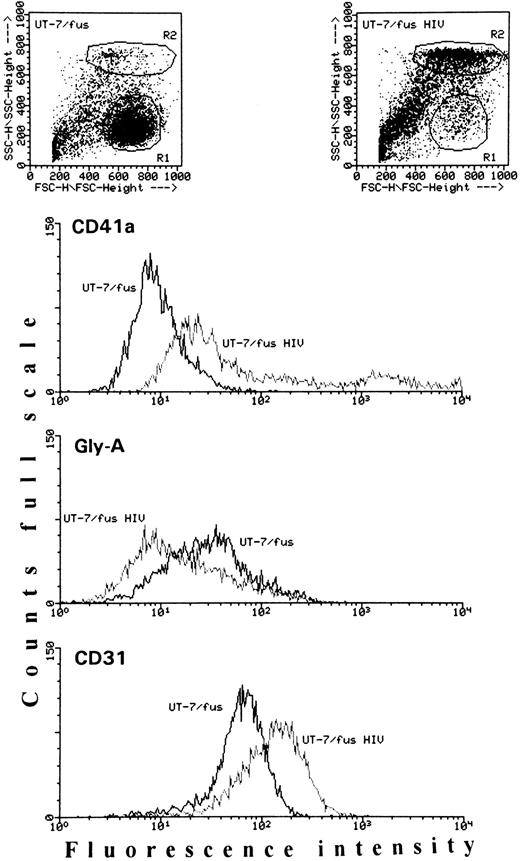
Antigenic analysis of uninfected or HIV-1–infected UT-7/fus cells. Cells were analyzed for the presence of CD41a, glycophorin A, and CD31 antigens. In each panel, slopes from uninfected and HIV-1–infected UT-7/fus cells are compared. The mean fluorescence intensity of both uninfected and HIV-1–infected UT-7/fus cells labeled with unspecific mouse IgG was always less than 5. In addition, on the top, the dot plots of each cell population are shown. The uninfected and HIV-1–infected UT-7/fus cell populations analyzed are, respectively, included in R1 and R2 areas.
Antigenic analysis of uninfected or HIV-1–infected UT-7/fus cells. Cells were analyzed for the presence of CD41a, glycophorin A, and CD31 antigens. In each panel, slopes from uninfected and HIV-1–infected UT-7/fus cells are compared. The mean fluorescence intensity of both uninfected and HIV-1–infected UT-7/fus cells labeled with unspecific mouse IgG was always less than 5. In addition, on the top, the dot plots of each cell population are shown. The uninfected and HIV-1–infected UT-7/fus cell populations analyzed are, respectively, included in R1 and R2 areas.
We next characterized HIV-infected UT-7/fus cells in terms of CD4 (the HIV receptor), CD31 (the platelet endothelial cell adhesion molecule [PECAM-1] expressed on platelets, but also on monocytes, granulocytes, and endothelial and T cells),22 CD41a (the GPIIb and GPIIIa complex, specifically expressed on megakaryocytes and platelets),23 and glycophorin A (an erythroid marker) membrane antigen exposure. When the majority of UT-7/fus cells was infected by HIV-1 (ie, 7 days after infection with moi 0.4), a clear increase of both CD31 and CD41a was detected with respect to uninfected UT-7/fus cells (Fig 7). At the same time, a general decrease in both glycophorin A (Fig 7) and CD4 exposure (not shown) was observed. Conversely, no variations in CD4, CD31, CD41a, and glycophorin A were detected in UT-7 cells challenged with HIV-1 or HIV-2 (not shown). Similarly, in HIV-2 chronically infected UT-7/fus cells, the exposure of these antigens was not modified, except for CD4, which, as expected, was fully downmodulated as in HIV-1–infected UT-7/fus cells (data not shown).
DISCUSSION
The demonstration that the pretreatment of CD4+ cells with certain β-chemokines induces inhibition of HIV replication24 has been rapidly followed by the identification of chemokine receptors as molecules involved in HIV entry1,6-10 and by the demonstration of their indispensable role as coreceptors for HIV entry. In particular, it has recently been shown that NSI M-tropic HIV strains (whose replication could be inhibited by pretreatment with chemokine MIP-1α, MIP-1β, and RANTES)24 use CC-CKR56-10 and, in a lesser extent, CC-CKR36,7 and CC-CKR2b6 as coreceptors. Conversely, SI T-lymphotropic HIV strains need the presence of CXCR4 (the receptor for the SDF-1 chemokine) to enter the cell.5 The CXCR4 molecule is a seven-transmembrane G-protein–coupled receptor, exhibiting approximately 30% homology with members of both α- and β-chemokine receptor families.4 The CXCR4 ligand (SDF-1) has chemoattractive activity for hematopoietic stem cells, monocytes, B lymphocytes, and, in a stronger fashion, T lymphocytes.11 Furthermore, it has proved to be a pre-B lymphocyte growth-stimulating factor.25 CXCR4 mRNA is widely expressed in hematopoietic and nonhematopoietic tissues.26 27 Thus, the lack of CXCR4 expression in the UT-7 cell line renders a unique model of human CD4+ hematopoietic cells that has no counterparts thus far.
We show here that, in UT-7 cells, the presence of CD4 receptors fully active in binding the viral gp120 is not sufficient to allow the entrance of HIV-1 or HIV-2 SI isolates, as suggested by the DNA-PCR analysis performed shortly after HIV challenge. We showed that resistance to HIV is due to the lack of CXCR4 expression. In fact, transfection of a vector expressing the human CXCR4 c-DNA renders UT-7 cells highly susceptible to infection by SI HIV-1 isolates. We also show that the CXCR4 molecule is functional as a coreceptor for HIV-2. The observation that UT-7/fus cells are fully resistant to infection with M-tropic NSI HIV strains was in line with previous studies on the inability of CXCR4 to allow the fusion and entry of M-tropic isolates.5
It has been reported that normal human megakaryocytes express CD4 on their surface and represent one of the targets of in vivo viral infection.28-30 Previous in vitro studies on megakaryocytic cell lines such as CMK and DAMI cells showed that HIV infection does not induce modifications on cell morphology. However, productive infection of CMK cells was obtained only with the HIV-2ROD strain,31 whereas DAMI cells, although fully CD4−, could be productively infected by the HTLV-IIIB HIV-1 strain.32 Nevertheless, the viral outcome appeared very delayed (considering that, in the most efficient of 26 different infection experiments and at the highest moi, the earliest peak of viral production was detected only 30 days postinfection) and levels of viral production appeared somewhat inconsistent. In addition, a relevant fraction of cells (from 40% to 80%) remained uninfected even after months of culture. Thus, the interaction between DAMI cells and the T-tropic, syncytium-inducing HTLV-IIIB strain led to a chronic, slow-low infective cycle.
It is conceivable that, in infected DAMI cultures, an HIV quasispecies able to replicate in CD4− cells had been selected (HIV strains able to infect CD4− cells have already been reported)33 whose expression level, however, was not strong enough to induce the overcoming of the differentiative block in DAMI cells. In addition, the more advanced stage of differentiation at which DAMI cells appeared blocked34 with respect to UT-7 cells may render more difficult the evaluation of a possibly small differentiation progress that HIV expression could induce on infected cells. Thus, considering that both the CD4 receptor28-30 and the CXCR-4 HIV coreceptor (C. Chelucci, unpublished data) are expressed by a consistent fraction of normal human cells of megakaryocytic lineage, UT-7/fus cells represent the model that could better reproduce the interactions occurring in vivo between HIV and partially or fully differentiated megakaryocytic cells.
Two additional human megakaryocytic cell lines (MEG-01 and CHFR-288) were shown to be both CD4+ and HIV-1 susceptible,35,36 but no detailed data on the cellular response after infection were reported. Conversely, we show that infection of UT-7/fus cells with SI HIV-1 isolates induces evident morphologic alterations and, finally, cell death. These morphologic changes (ie, increase in cell size, induction of polyploidy, and appearance in cytoplasm of eosinophilic areas containing multiple purple granules) are compatible with those described for normal in vitro megakaryocytic differentiation.37 This evidence was also supported by the increase in HIV-1–infected UT-7/fus cells of a specific megakaryocytic marker as GpIIb/IIIa complex and the contemporary increase of PECAM-1, coupled with a clear decrease in the exposure of the glycophorin A, a marker specific for erythroid differentiation. Taken together, these findings strongly suggest that a progression of UT-7/fus cells toward megakaryocytic differentiation could be achieved through the expression of HIV-1. Conversely, no morphologic modifications were observed in HIV-1–infected UT-7 cells (thus excluding any unspecific effect due to contaminants possibly present in HIV-1 preparations) or after HIV-2 infection of UT-7/fus cells. The evidence that HIV-2–infected UT-7/fus cells were fully CD4 downregulated strongly suggests that a large majority of them was infected. Thus, the lack of any evident multinuclearization among the HIV-2–infected cultures could not be considered as the consequence of the infection occurring in only few UT-7/fus cells.
The evidence that differentiation was induced by HIV-1 and not by HIV-2 infection suggests that the intracellular expression of some HIV-1 proteins, rather than a possible CXCR4 stimulation induced after viral binding, may be directly or indirectly involved in the induction of megakaryocytic differentiation. It has been shown that TE761 human rhabdomyosarcoma cells can be induced to differentiate after transfection with an HIV-1 molecular clone or with the vpr gene alone.38 Similarly, it is conceivable that the expression of the vpr gene (or eventually other HIV-1 genes) could induce UT-7/fus cells to escape the block of differentiation typical of UT-7 cells. Recently, the possibility to partially overcome the differentiation block in UT-7 cells has been demonstrated both in a thrombopoietin (TPO)-dependent subclone of UT-7 cells39 and in UT-7 cells transfected with mouse TPO receptor (mpl) and treated with TPO.40
There is no reason to believe that the polynucleated HIV-infected UT-7/fus cells are derived from the formation of HIV-induced syncytia, because several morphologic and antigenic features suggesting megakaryocytic maturation were displayed by multinucleated cells. Furthermore, syncytia formation has never been described in both productively HIV-infected CD4 positive megakaryocytic cell lines31,32 or in CD34-derived human megakaryocytes infected with SI HIV strains.41 The inability of UT-7/fus cells to form syncytia was also deduced from the lack of any polynucleated cells in cultures infected with HIV-2ROD , a viral strain able to induce large syncytia in any CD4- and CXCR4-coexpressing cells, such as T lymphocytes or HeLaCD4 cells. Finally, the evidence that all the HIV-1–infected UT-7/fus cells appear homogenously enlarged is a strong argument against the virus-induced formation of syncytia as the cause of UT-7/fus multinuclearization.
To our knowledge, UT-7 cells represent the first reported example of a naturally CD4+ human cell line that is not HIV infectable. The uniqueness of UT-7 cells under this aspect may be coherent with the wide expression of CXCR4 among different cell types. In this context, it is tempting to suggest that the UT-7 cells may be blocked at an early differentiative stage in which CXCR4 is not yet expressed and that this phenomenon may be related to an event physiologically occurring during megakaryocyte differentiation. Experiments are in progress to evaluate this hypothesis in unilineage cultures of human megakaryocytic cells derived from highly purified hematopoietic progenitors.
Furthermore, we believe that UT-7 and UT-7/fus cells represent a unique tool in the study of early events of viral infection, such as binding, envelope fusion with the target membrane, and viral penetration. Comparative studies between UT-7 and UT-7/fus cells may also provide new insights into the role of G protein activation in HIV entry and in HIV-induced cell pathogenesis.
ACKNOWLEDGMENT
We thank Dr A.R. Migliaccio for critical reading of the manuscript and A. Lippa for editorial assistance.
Supported by grants from AIDS Projects of the Ministry of Health, Rome, Italy.
Address reprint requests to Maurizio Federico, PhD, Laboratory of Virology, Istituto Superiore di Sanità, Viale Regina Elena 299, 00161 Rome, Italy.