Abstract
To elucidate the molecular basis of hereditary protein S (PS) deficiency and, in particular, type III or free PS deficiency, the allelic distribution and segregation patterns of the PS gene (PROS1) polymorphisms P626A/G and S460P (PS Heerlen) have been analyzed in a group of 45 proposita suffering from type I or type III PS deficiency. No differences between patients and controls were found in the frequency of the P626A/G alleles. By contrast, the frequency of the PS Heerlen allele in the group of patients with type III PS deficiency (9 of 46 chromosomes, P = .196) was significantly higher (P < .001) than in the control group (1 of 300 chromosomes, P = .003). The A allele of P626A/G was always associated with the P allele of S460P. However, this haplotype did not cosegregate with the type III PS-deficient phenotype in 3 of the families. Furthermore, multipoint linkage analysis excluded the whole PROS1 gene in 1 of these families, which is in agreement with the absence of mutations in the PROS1 gene, as determined by sequence analysis. Finally, linkage analysis with 4 microsatellite markers linked to the C4BPB and C4BPA loci also excluded these two genes. From these results we conclude that, at least in some families, the molecular basis of type III PS deficiency is not due to the Mendelian inheritance of a single defect in the PROS1 or in the C4BP genes.
THE IMPORTANCE OF protein S (PS) in the regulation of blood coagulation has been established by the identification of families with hereditary PS deficiency associated with an increased risk of developing venous thrombotic disease.1-3 PS is a vitamin K-dependent plasma glycoprotein that acts as a cofactor of activated protein C (APC) in the inactivation of the procoagulant factors Va and VIIIa.4-6 PS might also have APC-independent anticoagulant properties through direct inhibition of prothrombin and factor X activation.7-9 In human plasma, approximately 40% of PS circulates as free protein, whereas the remaining 60% forms a noncovalent 1:1 stoichiometric complex with the β-chain of the complement C4b-binding protein (C4BPβ+).10,11 This interaction is of high affinity and abolishes the anticoagulant properties of PS.12 Therefore, in vivo, all C4BPβ+ molecules circulate bound to PS and only the molar excess of PS over C4BPβ+ circulates in a free form and is active as a cofactor of APC.13
PS deficiency is inherited as an autosomal dominant disorder present in about 2% to 8% of families with hereditary thrombophilia.14,15 Three types of PS deficiency have been described.16 In type I or quantitative PS deficiency the plasma levels of total and free PS antigen are reduced together with reduced anticoagulant activity. Type II deficiency, which is quite uncommon, is characterized by normal concentrations of both total and free PS antigen, but low cofactor activity. Finally, in type III deficiency, with a prevalence similar to that of type I deficiency,17 18 free PS antigen and activity levels are reduced, whereas total PS antigen levels are normal.
Two highly homologous PS genes have been located near the centromere of chromosome 3: the active gene PROS1 and a transcriptionally inactive pseudogene, PROS2.19 20 The PROS1 gene contains 15 exons, spanning about 80 kb of genomic DNA, with a transcript of about 3.5 kb.21-24 A few diallelic polymorphisms have been identified in the PROS1 gene, the best known of which are the presence of adenine or guanine at the proline codon 626 (P626A/G) in exon 1525 and the uncommon serine to proline substitution at codon 460 (S460P) in exon 13, which results in the PS Heerlen variant.26 In addition, several mutations, most of them point mutations or short deletions or insertions, have been found associated with the PS-deficient phenotype. However, most of these mutations have been identified in type I or quantitative PS-deficient pedigrees27-38 and only 3 of them (ivs E + 5,G → A; Asn217 → Ser; and Met570 → Thr) have been associated to type III or free PS deficiency, although there is no clear explanation as to how these particular mutations affect only the free and not the total PS concentration.32,35 In another study, Duchemin et al39 found the P or PS Heerlen allele of the rare, and apparently neutral, S460P polymorphism as the only PROS1 sequence abnormality detected in several type III-deficient probands, and from protein binding studies they suggested a causative role of this variant PS molecule in the type III-deficient phenotype. Furthermore, from the study of several Swedish families in which there is coexistence of type I and type III deficiencies, Zöller et al40 concluded that these two types of PS deficiency are phenotypic variants of the same genetic disease. This has been confirmed in a couple of families by the identification of the PROS1 mutation cosegregating with both phenotypes.41 The molecular basis of type III PS deficiency is therefore quite unclear and, given its characteristics, the possibility that it is also caused by mutations in genes other than PROS1 cannot be excluded.
In a preliminary study of the allele frequencies of the S460P and the P626A/G PROS1 polymorphisms in a group of 38 Spanish patients with different types of PS deficiency and in a control group, we reported the existence of linkage disequilibrium between the PS Heerlen allele and type III PS deficiency.42 To further confirm and analyze this association, in the present study we describe the results obtained when these two polymorphisms were analyzed in 45 probands with different types of PS deficiency and in a larger control group. Because linkage disequilibrium was confirmed, we used these two polymorphisms as gene markers for segregation analysis in the type III-deficient families that carry the Heerlen allele. The results obtained not only indicated absence of cosegregation of the PS Heerlen allele with type III PS deficiency in some families, but also excluded linkage with the PROS1 gene in one of them. Furthermore, sequence analysis of all the PROS1 exons and intron flanking regions in these type III PS-deficient families showed no other abnormality than the described polymorphisms.
MATERIALS AND METHODS
Patients and Controls
The genotypes for the P626A/G and S460P polymorphisms were determined in 45 unrelated patients with PS deficiency and thrombosis, in their relatives, and in a control group of 122 healthy adults (244 unrelated chromosomes). According to the plasma values of total PS antigen, free PS antigen, and functional PS in the proposita and in their family members, the families were classified as follows: 13 as having PS deficiency type I, 23 with PS deficiency type III, and 9 families were unclassified and enclosed in a group that we named type I/III, because some members had PS values compatible with a type I deficiency, whereas others had type III deficiency. With the exception of one type III propositus who died as a consequence of a cerebrovascular accident, the remaining had developed at least one of the following venous thrombotic events: deep vein thrombosis, pulmonary embolism, or superficial thrombophlebitis. Arterial thrombosis was also found in three proposita carrying the PS Heerlen allele. Familial history of thrombotic disease was positive in 20 pedigrees (7 with type I PS deficiency, 9 with type III deficiency, and 4 with type I/III deficiency), although in 5 of them the presence of thrombosis was not always associated with the PS-deficient phenotype. These five families belonged to the type III or I/III deficiency groups and four of them carried the PS Heerlen allele. The mean age of the proposita was 49 years (range, 25 to 79 years) and their mean age at the first thrombotic event was 40 years (range, 16 to 74 years).
PS Assays
In most individuals (proposita and family members), plasma levels of total and free PS antigen (Asserachrom Protein S; Diagnostica Stago, Asnières, France) and PS activity (Staclot Protein S; Diagnostica Stago) were determined as previously described.43 Free PS antigen was measured in the plasma supernatant after complex precipitation with PEG-6000 (Asserachrom Protein S). The reference values obtained from a control group of 100 healthy blood donors and expressed as a percentage of the value obtained for each assay in a normal plasma pool were 75% to 140% for total and free PS antigen and for PS activity in all men and in women older than 46 years of age. For women less than 46 years of age, the reference values were 65% to 120% for total PS antigen and for PS activity and 60% to 100% for free PS antigen. The diagnosis of PS deficiency in each propositus was made after the finding of PS values lower than the normal range in at least two different plasma samples. In some cases, total and free PS antigen levels were also determined with the new Diagnostica Stago enzyme-linked immunosorbent assay methods that use monoclonal antibodies specific for both the free and complexed forms of PS (Asserachrom Total Protein S) and only for the free form of PS (Asserachrom Free Protein S). These new kits were used to confirm previous diagnosis in a few families (ie, families PS6 and PS10) and to diagnose PS deficiency in a few proposita or proposita relatives. Agreement between both methods was good in all but one case, which was then excluded from the study.
Genetic Analysis
Numbering system for amino acids and nucelotides.Amino acids and nucleotides are numbered according to Schmidel et al.23 Exons and introns are numbered 1 through 15 and 1 through 14, respectively. Numbering of the position of the primers is relative to either the 5′ or 3′ boundary of each exon.29
PROS1 amplifications by polymerase chain reaction (PCR). Genomic DNA was isolated from peripheral blood lymphocytes according to standard procedures.44 All 15 exons of the PROS1 gene as well as their 5′ and 3′ flanking intronic regions were amplified by the PCR in a final volume of 100 μL containing 0.5 μg of genomic DNA, 0.5 μmol/L of each primer, 0.2 mmol/L of each dNTP (Pharmacia, Uppsala, Sweden) and 2 U of Taq-polymerase (Perkin-Elmer Cetus, Norwalk, CT), in 10 mmol/L Tris HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2 , and 0.01 mg/mL gelatin. After an initial denaturation step of 5 minutes at 95°C or 6 minutes at 98°C (exons 1, 4, 7, and 11), 35 PCR cycles were performed. Cycling parameters were 30 seconds to 1 minute of denaturation at 94°C; 30 seconds to 1 minute of annealing at 58°C (exons 1 and 5 to 6), at 55°C (exons 2, 10, and 12), at 50°C (exons 3 and 9), 56°C (exon 4), 54°C (exons 7 and 15), 57°C (exon 8), 53°C (exon 11), 52°C (exon 13), and 62°C (exon 14); and 1 to 2 minutes of extension at 72°C. The amplification primers used were those described by Reitsma et al29 with the exception of the forward primers for exons 1 (PS1.1a, 5′TGTTATCACTTCCCCTCTCG3′, −96 to −77), 8 (PS8.1a, 5′GATGTCATAGTATTCTTCCC3′, −134 to −115), and 13 (PS13.1a: 5′ATCATTGAGAAAGGGAATGG3′, −134 to −115); the reverse primer for exon 14 (PS14.2a, 5′AAATGTCGGTACTAGCCCTAG3′, 147 to 127); and the forward and reverse primers for exon 4 (PS4.1a, 5′TTGGGACAGTTCCTACC ATG3′, −78 to −59; and PS4.2a, 5′CTTTACCTACAGAGTTTTTG3′, 54 to 35). Except for the PCR fragment that includes exon 3, where the differences between the active gene and the pseudogene are unknown, the amplification of only PROS1 gene sequences was confirmed by restriction analysis with endonucleases that specifically cleaved either PROS1 or PROS2 sequences.
Polymorphism Analysis
The genotypes for the P626A/G and the S460P polymorphisms were respectively determined by restriction analysis of exon 15 with BstXI and of exon 13 with Rsa I, using the digestion conditions recommended by the suppliers (Boehringer Mannheim, GmbH, Mannheim, Germany). Digestion products were electrophoretically resolved on 2.5% agarose gels. Genotypes for the P626A/G polymorphism were further confirmed by single-strand conformation analysis with silver staining, using the PhastSystem Unit and PhastGel Homogeneous 12.5% polyacrylamide gels (Pharmacia).
DNA Sequencing
Amplified fragments from all exons and intron/exon boundaries were purified with the use of QIAquick-spin PCR purification columns (Qiagen, Hilden, Germany) and directly sequenced using an Applied Biosystems protocol for Taq cycle-sequencing with dye terminators and an Applied Biosystems 373A DNA sequencer (Applied Biosystems, Foster City, CA). Both the sense and the antisense DNA strands were sequenced using the same primers as those used for the amplification with the exception of exon 14, for which a new reverse primer was synthezised (R8, 5′TTCTATTTTAAGTGGTGCG3′, position 1925 to 1906 in the cDNA).
Regulator of Complement Activation (RCA) Gene Cluster Analysis
Possible genetic linkage of type III PS deficiency with the C4BP (C4b-binding protein) genes in the RCA gene cluster was determined by typing for 4 microsatellites linked to C4BPB and C4BPA (the genes for the C4BPβ+ and C4BPα chains of C4BP, respectively). These microsatellites are RCA5, C4BPB, RCA8, and REN. C4BPB and REN map within or near the genes of the same name. The C4BPB microsatellite is located 29 bp downstream from the polyadenylation site of the C4BPB gene.45 REN is located 2 cM centromeric of C4BPB.46 RCA8 is located 20 kb upstream from C4BPB. RCA5 is located 30 kb downstream C4BPB, within the C4BPA gene (Heine et al, Immunogenetics, in press). Families PS10 and PS6 were typed for these 4 microsatellites by PCR. Amplification was performed in a total volume of 10 μL containing 50 ng of genomic DNA; 0.5 μmol/L of each primer; 1 U of Taq-polymerase (Perkin-Elmer Cetus); 250 μmol/L of dATP, dGTP, and dTTP; 10 μmol/L of dCTP; 0.1 μL of dCTP α32P at 3,000 Ci/mmol/L; 1.5 mmol/L MgCl2 ; 50 mmol/L KCl; and 10 mmol/L, Tris-HCl (pH 8.3). Amplification conditions were 1 cycle at 94°C for 4 minutes, 55°C for 1 minute, and 72°C for 1 minute, followed by 30 cycles of 94°C for 45 seconds and 55°C for 40 seconds. Samples were resolved on 6% polyacrylamide sequencing gels.
Statistical and Linkage Analysis
Statistical differences between the allele frequencies in the patients' group and in the control group were analyzed with the χ2 method, with Yates corrections when necessary. Two-point and multipoint linkage analysis was performed by calculation of the Lod Scores at various recombination fractions, using the 2.2 version of FASTLINK software (A. Schäffer, National Institutes of Health).47
RESULTS
P626A/G and S460P Allele Frequencies
As shown in Table 1, the frequencies of the A (0.722) and G (0.278) alleles of the P626A/G polymorphism in the group of 45 PS-deficient proposita were not statistically different from those found in the control group of 122 healthy individuals (A = 0.623 and G = 0.377). Furthermore, no differences were found when the PS-deficient type I, type III, or type I/IIII groups were separately compared with the control group.
Analysis of the S460P polymorphism showed a significantly (P < .001) higher frequency of the P (PS Heerlen) allele in the group of patients with PS deficiency than in the control group (Table 2), thereby indicating linkage disequilibrium between the P allele of S460P and PS deficiency. The P allele was only found in 1 of the 300 control chromosomes, its estimated frequency being 0.003. By contrast, the frequency of the P allele in the group of patients with type III PS deficiency, with 9 of 23 proposita being heterozygotes for this polymorphism, was estimated to be 0.196 (P < .001). The frequency of the P allele in the two other groups of PS-deficient patients (type I and type I/III) was not significantly different from that in the control group (Table 2). Therefore, the linkage disequilibrium observed with PS deficiency was in fact due to linkage disequilibrium between the PS Heerlen allele and type III PS deficiency.
Segregation and Linkage Analysis
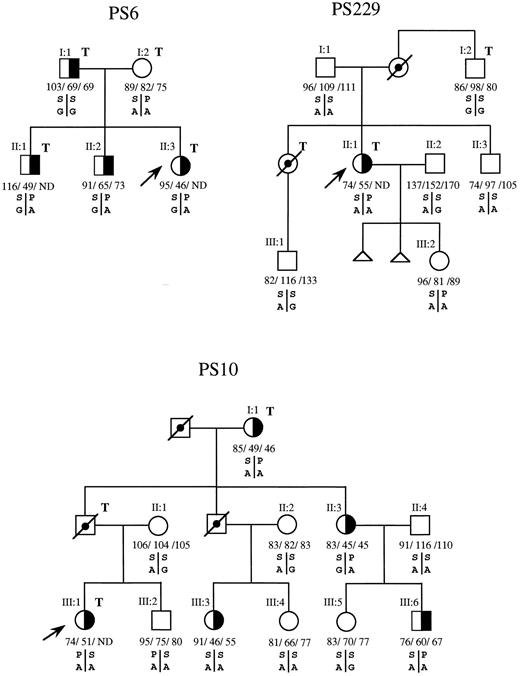
Segregation analysis of the PS-deficient phenotype and the S460P and P626A/G polymorphisms in each family showed that the P allele of S460P was associated with the A allele of P626A/G (PA haplotype) in all except 3 families in which the phase could not be established. When we analyzed whether the PS-deficient phenotype cosegregated with the PA haplotype of these two polymorphisms, it was seen that in family PS6 (Fig 1) this haplotype in the three PS-deficient offspring had been inherited from their mother, who has repeatedly been proven to have normal plasma values of PS. Absence of cosegregation of the PS Heerlen allele and type III PS deficiency was also found in family PS229, in which the PA haplotype but not the PS-deficient phenotype of the propositus (PS229, II:1, Fig 1) has been inherited by her daughther (PS229, III:2). In family PS10, the propositus and her brother (III:1 and III:2, Fig 1) have identical PROS1 genotypes (the PA haplotype inherited from the father and the SA from the mother), but have different protein S phenotypes. Furthermore, their two cousins (PS10, III:3 and III:4) also have different PS phenotypes, despite sharing the same PROS1 genotypes and not carrying the PS Heerlen allele. With respect to the other 8 proposita with PS deficiency that carried the PS Heerlen allele and the remaining proposita with type I, type III, or type I/III deficiencies that did not carry the PS Heerlen allele, family studies were either not possible (6 cases) or uninformative for linkage analysis despite the fact that, in 4 of these families, the PS Heerlen allele cosegregated with PS deficiency.
Segregation patterns of the P626A/G and S460P polymorphisms in the PROS1 gene in families PS6, PS229, and PS10, indicating absence of cosegregation between the P or PS Heerlen allele of S460P and type III PS deficiency in families PS6 and PS229 and absence of linkage in family PS10. (╞, ◑) Heterozygosis for type III PS deficiency; (, ) not tested; (□, ○) deceased; (↗) propositus; T, thrombotic disease. Individual values for total PS antigen, free PS antigen, and PS activity are expressed as the percentage of normal and from left to right, respectively, below the corresponding symbol. ND, not determined. S or P are the serine or proline alleles at codon 460; A or G are the alleles for adenine or guanine at codon for proline 626.
Segregation patterns of the P626A/G and S460P polymorphisms in the PROS1 gene in families PS6, PS229, and PS10, indicating absence of cosegregation between the P or PS Heerlen allele of S460P and type III PS deficiency in families PS6 and PS229 and absence of linkage in family PS10. (╞, ◑) Heterozygosis for type III PS deficiency; (, ) not tested; (□, ○) deceased; (↗) propositus; T, thrombotic disease. Individual values for total PS antigen, free PS antigen, and PS activity are expressed as the percentage of normal and from left to right, respectively, below the corresponding symbol. ND, not determined. S or P are the serine or proline alleles at codon 460; A or G are the alleles for adenine or guanine at codon for proline 626.
Because the segregation patterns of the PS-deficient phenotype and the S460P and P626A/G polymorphisms in family PS10 indicated absence of linkage between type III PS deficiency and the PROS1 gene, we proceeded to confirm this point by performing a multipoint linkage analysis after a further confirmation of the PS phenotype in the family members. With this purpose in mind, PS was measured in new plasma samples from all the available PS10 members by using a new enzyme-linked immunosorbent assay method that includes monoclonal antibodies for the determination of both free and total PS directly from plasma. The results obtained from these new determinations were in good agreement with previous data, thus confirming the diagnosed phenotype of the different family members. Multipoint linkage analysis confirmed the absence of linkage between the deficient phenotype and the PROS1 gene. As shown in Table 3, a Lod Score of −2.02 was estimated at recombination fractions equal to 0.0016, both upstream from S460P and downstream from P626A/G. Because, by convention, a Lod Score equal to or less than −2 is enough to exclude the analyzed genomic region as the locus for the disease phenotype, our results exclude about 160 kb upstream from S460P and another 160 kb downstream from P626A/G, which is far more than the 80 kb of the PROS1 gene.
PROS1 Sequence Analysis
Apart from the sequence differences due to the S460P and P626A/G polymorphisms, no other sequence abnormalities were identified in any of the 15 exons and intron boundaries of the PROS1 gene from the proposita of families PS10 and PS6 or from the proposita of 3 other type III-deficient families, one of which did not carry the PS Heerlen allele. The absence of mutations in the PROS1 sequences was confirmed by sequencing both strands of the DNA and careful comparison of subjects and controls. Furthermore, the presence of any doubtful base change was excluded either by restriction analysis or DNA sequencing of a newly synthesized PCR product. By contrast, sequence analysis in 7 families with a clear type I PS deficiency and in one family with both type I and type III-deficient patients, none of which carried the PS Heerlen allele, showed the presence of a cosegregating PROS1 mutation in each family (manuscript in preparation).
Analysis of the RCA Loci
Segregation analysis in families PS10 and PS6 of the RCA5, C4BPB, RCA8, and REN microsatellites, which are closely linked to the C4BPB and C4BPA genes, also suggested absence of linkage between the type III PS-deficient phenotype and the RCA loci containing these two genes. In family PS6, the propositus (II:3, Fig 2) carries different RCA alleles from those inherited by her two affected sibs (II:1 and II:2, Fig 2). Similarly, in family PS10, the affected members do not share a common RCA haplotype and two sibs (III:3 and III:4, Fig 2) that inherited the same RCA alleles from their parents have different PS phenotypes. Linkage analysis with the informative markers in each of these two families confirmed the exclusion of the C4BPB and C4BPA genes as being the cause for the deficient phenotype. In family PS6, two-point analysis between the PS deficiency locus and each of the RCA5, RCA8, and REN markers gave an estimated Lod Score of −2.08 at a recombination fraction of 0.002 for each marker. This excludes a region of 0.2 cM (≈200 kb) around each marker as that containing the deficiency locus. A larger region was excluded in family PS10 using the C4BPB, RCA8, and REN markers. In this family, the best results were obtained with the multipoint analysis, which gave an estimated Lod Score of −2.19 at a recombination fraction of 0.075 (≈7.5 cM), both upstream from REN and downstream from C4BPB.
Segregation patterns in families PS6 and PS10 of four microsatellites closely linked to the C4BPB and C4BPA genes, indicating no linkage between these genes and type III PS deficiency. (╞, ◑) Heterozygosis for type III PS deficiency; (, ) not tested; (□, ○) deceased; (↗) propositus; T, thrombotic disease. The genotype of each individual for these microsatellites is indicated below each symbol as basepair fragments for the RCA5, C4BPB, RCA8, and REN alleles, from top to bottom, respectively. The corresponding haplotype, in lowercase letters, is indicated below. Haplotypes between parentheses are the inferred ones.
Segregation patterns in families PS6 and PS10 of four microsatellites closely linked to the C4BPB and C4BPA genes, indicating no linkage between these genes and type III PS deficiency. (╞, ◑) Heterozygosis for type III PS deficiency; (, ) not tested; (□, ○) deceased; (↗) propositus; T, thrombotic disease. The genotype of each individual for these microsatellites is indicated below each symbol as basepair fragments for the RCA5, C4BPB, RCA8, and REN alleles, from top to bottom, respectively. The corresponding haplotype, in lowercase letters, is indicated below. Haplotypes between parentheses are the inferred ones.
DISCUSSION
Because the characterization of the PS gene (PROS1) and the recognition of hereditary PS deficiency as an important predisposition factor to develop venous thrombosis, several different mutations, most of them point mutations, short deletions, or insertions, have been identified in the PROS1 gene and are considered causative of a PS-deficient phenotype. However, most of these mutations have been identified in type I or quantitative PS-deficient pedigrees and only a few of them, one of which is the previously described PS Heerlen allele, have been associated to type III or free PS deficiency, coexisting with type I PS deficiency in at least 2 families.32,35,39 41 This is in part due to the fact that most investigators have essentially focused their research on type I PS deficiency, but, as our sequencing results show, it may well happen that some patients suffering from type III deficiency have no PROS1 mutations. Because of the fact that free PS results from the molar excess of PS over the C4BPβ+ isoform of C4BP protein and because the mechanisms regulating this interaction are still unclear, the possibility that type III PS deficiency may also result from abnormalities in genes other than PROS1 should therefore be considered.
The analysis of two intragenic polymorphisms described in PROS1 (S460P and the P626A/G) could be helpful as a first approach to understanding the molecular basis of PS deficiency and, as a previous step to the classical but laborious DNA sequence analysis, it may be particularly useful to exclude or confirm linkage of this gene with the PS-deficient phenotype in type III families. With this purpose, we first calculated and compared the allele frequencies of the P626A/G and S460P polymorphisms in two different populations: one of the patients with thrombosis and PS deficiency, which was further classified into three different groups according to the deficiency type (I, III, and I/III), and a healthy control population. The results obtained indicated that the allele frequencies of the P626A/G polymorphism in the patient groups were not significantly different from those in the control group that were also not significantly different from those found in other European Caucasian populations.25,32,48 On the contrary, and in agreement with the results reported by Duchemin et al39 in a French population, comparison of the frequencies of the S460P alleles between the control and the PS-deficient groups indicated a clear linkage disequilibrium between the PS Heerlen allele and type III PS deficiency, with 20% of the type III PS-deficient proposita and only 0.3% of the controls being heterozygotes for this polymorphism.
Although the PS Heerlen had been previously described to be a rare but neutral polymorphism,26 most likely identical to the PS variant identified in an American family,49 the presence of such a strong linkage disequilibrium between the Heerlen allele and type III PS deficiency could indicate that this amino acid variant or another mutation linked to this allele may result in type III PS deficiency. In fact, Duchemin et al,39 by analyzing the binding of wild-type and Heerlen PS molecules to C4BP, suggest that the binding of two PS Heerlen molecules to one C4BP molecule may explain the reduced levels of free PS in these patients. If this was the case, then the PS Heerlen allele would cosegregate with the deficient phenotype in these families and this was not found in 3 of the 11 families analyzed in this study. In these families, we found not only PS Heerlen carriers with normal PS values (which could be explained by an incomplete penetrance of the Heerlen allele), but also affected members that did not carry the Heerlen allele, therefore indicating that neither the Heerlen mutation nor another mutation in this allele of the PROS1 gene cause, by themselves, type III PS deficiency in these families. Furthermore, multipoint linkage analysis using both the S460P and the P626A/G polymorphisms as genetic markers allowed for the exclusion of the whole PROS1 gene in one of these families (PS10). This absence of linkage, which indicates that a mutation in the PROS1 gene is not responsible for the PS-deficient phenotype in this family, is also in agreement with the absence of any specific mutation in the DNA sequences of all PROS1 exons and intron/exon boundaries of the propositus of family PS10. No specific PROS1 mutation could be identified in the proposita of another 4 type III PS-deficient families, including the other family (PS6) in which the PS Heerlen allele does not cosegregate with the deficient phenotype. Unfortunately these families were uninformative for linkage analysis.
The finding of linkage disequilibrium between the PS Heerlen allele and type III PS deficiency, together with the absence of linkage between this deficiency state and the PROS1 gene, could be explained by different mechanisms. One explanation could be that a mutation responsible for type III PS deficiency originated in a PS Heerlen carrier, in a locus not linked to the PS gene, in such a way that the allelic equilibrium has not yet been established. Candidate genes could be C4BPB, those regulating its expression, and those for other proteins that may regulate PS concentration in plasma.50 Variations in the concentration of the C4BPβ+ isoform of C4BP would alter the equilibrium between bound and free PS, and it has recently been shown that the C4BP isoform pattern is genetically determined and inherited with the C4BPB and C4BPA genes.51 For this reason, we thought that C4BPB and C4BPA would be good candidates to be associated with type III PS deficiency. Linkage analysis in families PS10 and PS6 using four different microsatellite markers closely linked to the C4BPB and C4BPA genes also confirmed the absence of linkage between these genes and the type III PS-deficient phenotype. Therefore, if the type III-deficient phenotype in these families is due to an alteration in the concentration of C4BPβ+, the defect must lie in other genes that influence the expression of C4BPB.52 Secondly, the possibility of type III PS deficiency being a complex disease that results from the interaction of several factors, some of which may be genetic, cannot be excluded and furthermore could explain a contribution of the PS Heerlen allele to the disease phenotype.
To study which of these hypotheses is the correct one would require the analysis of more and larger families, which is beyond the scope of this work. Nevertheless, from our results we conclude that, at least in some families, the molecular basis of type III PS deficiency is not the Mendelian inheritance of a single defect in the PROS1 or in the C4BP genes. This fact, together with the identification of PROS1 mutations associated with type III or type I/III PS deficiency in other families, indicates that type III PS deficiency is likely to be a genetically heterogeneous disease that results from either a single mutation in a major gene such as PROS1 or from a more complex mechanism that involves the interaction of different factors.
ACKNOWLEDGMENT
We thank all Estudio Multicéntrico Español de Trombofilia (EMET) participants for providing us with the patients' samples, Dr S. Rodrı́guez de Córdoba for helpful suggestions and critical review of the manuscript, and Helena Kruyer for her helpful assistance with the manuscript.
Supported by Dirección General de Investigación Cientı́fica y Tècnica (DGICYT, PB94-1233), Fondo de Investigación Sanitaria (FIS-94/0039), and Servei Català de la Salut, Generalitat de Catalunya. D.H.-S. was supported by the North Atlantic Treaty Organization under a fellowship awarded in 1996.
Address reprint requests to Núria Sala, PhD, Departament de Genètica and Molecular, IRO, Hospital Duran i Reynals, Autovia de Castelldefels km 2.7, E-08907-Hospitalet de LLobregat, Barcelona, Spain.