Abstract
Adenosine deaminase (ADA) deficiency typically causes severe combined immunodeficiency (SCID) in infants. We report metabolic, immunologic, and genetic findings in two ADA-deficient adults with distinct phenotypes. Patient no. 1 (39 years of age) had combined immunodeficiency. She had frequent infections, lymphopenia, and recurrent hepatitis as a child but did relatively well in her second and third decades. Then she developed chronic sinopulmonary infections, including tuberculosis, and hepatobiliary disease; she died of viral leukoencephalopathy at 40 years of age. Patient no. 2, a healthy 28-year-old man with normal immune function, was identified after his niece died of SCID. Both patients lacked erythrocyte ADA activity but had only modestly elevated deoxyadenosine nucleotides. Both were heteroallelic for missense mutations: patient no. 1, G216R and P126Q (novel); patient no. 2, R101Q and A215T. Three of these mutations eliminated ADA activity, but A215T reduced activity by only 85%. Owing to a single nucleotide change in the middle of exon 7, A215T also appeared to induce exon 7 skipping. ADA deficiency is treatable and should be considered in older patients with unexplained lymphopenia and immune deficiency, who may also manifest autoimmunity or unexplained hepatobiliary disease. Metabolic status and genotype may help in assessing prognosis of more mildly affected patients.
DEFICIENCY OF adenosine deaminase (ADA) typically causes Severe Combined Immune Deficiency (SCID) in infants, who present with growth failure, opportunistic infections, lymphopenia, and defective cellular and humoral immune function.1,2 Immunodeficiency results from toxic effects of ADA substrates, including apoptosis induced by deoxyadenosine triphosphate (dATP) pool expansion3-5 and inhibition of transmethylation reactions caused by impaired catabolism of S-adenosylhomocysteine6-8 (reviewed in Hershfield et al2 ). ADA− SCID is treatable by marrow transplantation and enzyme replacement; gene therapy trials are underway.
After the discovery of ADA− SCID in 1972, screening programs identified rare healthy children whose red blood cells (RBCs) lacked ADA but whose nucleated cells had 5% to 70% of normal activity (partial deficiency).9-13 More recently, ADA deficiency has been discovered in several older children and in two sisters in their mid-30s with late or adult onset of immunodeficiency.14-17 More than 40 mutant ADA alleles have been identified.2,18 In SCID patients, neither allele encodes an active enzyme, whereas in more mildly affected patients, at least one ADA gene product appears to have some function. The extent of deoxyadenosine (dAdo) induced dATP accumulation and S-adenosylhomocysteine hydrolase inactivation in RBCs reflect the degree of ADA deficiency.19-21
Whether some children with partial ADA deficiency progress to an immune deficient state is unclear. To date, ADA deficiency has been identified as a cause of adult-onset immunodeficiency in only a single family.17 Because of atypical features, older patients may be misdiagnosed with a disease of unknown cause, such as common variable immunodeficiency or idiopathic CD4+ T lymphocytopenia.22 23 To better define and increase awareness of the spectrum of manifestations in adults, we report the metabolic, immunologic, and genetic findings in two patients diagnosed with ADA deficiency in adult life: one overtly immunodeficient and the other still in good health.
MATERIALS AND METHODS
Study Subjects
Patient no. 1.This cachectic 39-year-old woman was referred to the University of Zurich pediatric clinic for immunologic evaluation. She had been hospitalized eight times for pneumonitis in the previous year. Her family history was unremarkable and included a healthy 13-year-old daughter. In early childhood she had eczematoid dermatitis and frequent febrile convulsions, otitis media, pneumonitis, hepatitis, furunculosis, and diarrhea. At 5 years of age, perforating appendicitis was complicated by abdominal wall necrosis; her granulocyte count was 1,100/μL and her lymphocyte count was 900/μL. An extensive search, including spleen biopsy, failed to account for neutropenia, lymphopenia, thrombocytopenia, hepatitis, and splenomegaly.
Pulmonary infections were frequent until age 11; leukopenia persisted. The patient then did fairly well until age 28, when she became septic after cesarian section. A year later she developed hepatomegaly, cholecystitis, and cholelithiasis. Liver biopsy specimens showed granulomas of undetermined cause. She developed pancytopenia and chronic sinopulmonary infections. Asthma and serum IgE of 1,780 International Units (IU)/mL (normal, 47 to 200 IU/mL) raised suspicion of hyper-IgE syndrome. At age 30 she was treated for pulmonary tuberculosis; at age 34 she underwent lobectomy for hemoptysis. At age 38, computerized tomography (CT) showed diffuse pulmonary miliary infiltrates with fibrosis and bronchiectasis, hepatosplenomegaly, and cholelithiasis.
Immunologic evaluation at age 39 showed a total lymphocyte count of 275/μL (CD4+ T lymphocytes, 190/μL; CD8+ T lymphocytes, 60/μL; and CD19+ B lymphocytes, 20/μL). Lymphoproliferative responses to mitogens were diminished (phytohemagglutinin, 34,051 disintigrations per minute (dpm); pokeweed mitogen, 10,917 dpm; and anti-CD3 34,947 dpm) and were absent to antigens (candidin, diphtheria, cytomegalovirus, and tetanus). IgG measured 1 week after an intravenous Ig infusion was normal at 14.7 g/L (IgG1, 12.0 g/L; IgG2, 0.61 g/L). IgA (1.05 g/L) and IgM (0.3 g/L) were both less than the 10th percentile. IgE was 1,232 IU/mL. No antibody response could be detected to pneumococcal polysaccharide, diphtheria, or tetanus toxoids. Rheumatoid factor (RF), antinuclear antibody (ANA), antinative DNA, and Coombs' tests were negative. Human immunodeficiency virus was excluded by polymerase chain reaction (PCR). Because these findings pointed to combined T- and B-cell immunodeficiency, metabolic studies were performed, which led to the diagnosis of ADA deficiency.
Five months after diagnosis and while receiving corticosteroid therapy for chronic obstructive pulmonary disease, patient no. 1 developed gastrointestinal candidiasis and progressive hemiplegia. Brain CT and magnetic resonance imaging (MRI) studies showed the evolution over several weeks of multiple hypodense ischemic foci and central pontine myelinosis. Brain biopsy specimens established the diagnosis of progressive multifocal leukoencephalopathy caused by JC virus. She died after developing tetraplegia and bulbar paralysis.
Patient no. 2.This 28-year-old man was found to lack erythrocyte ADA activity while screening the family of his infant niece, who had died after a haploidentical bone marrow transplant for ADA− SCID. His past history was unremarkable except for recurrent tonsillitis beginning at age 10 years, which ceased after a tonsillectomy at age 23. He had no history of recurrent otitis, bronchitis, pneumonia, sepsis, meningitis, or hepatitis.
Hematologic and immunologic evaluation showed a white blood cell (WBC) count of 5,700/μL with 25% lymphocytes (1,420/μL), 66% neutrophils, 5% monocytes, 3% eosinophils, and 1% basophils. He had 82.9% CD3+ T cells, 64.7% CD4+ T cells (906/μL), and 17.4% CD8+ T cells (CD4/CD8 ratio, 3.72). Serum IgG was 708 mg/dL, IgA was 246 mg/dL, and IgM was 65 mg/dL. He had protective levels of serum antibody to rubella, rubeola, mumps, tetanus, and Haemophilus influenzae type b. Lymphoproliferative responses to mitogens, tetanus toxoids, and allogeneic cells were normal.
Mutational Analysis
ADA cDNA and genomic sequences are as reported.24,25 General PCR methods for amplifying ADA genomic and cDNA segments (the latter numbered from the start of transcription, with translation starting at position 96) and for cloning and sequencing PCR products are as described.16,26 27 Specific target sequences amplified for this study included (primers listed in Table 1): cDNA nucleotide (nt) 16 to 1,497 (full length), primers 1 and 2; cDNA nt 96 to 1,188 (coding region), primers 3 and 4; genomic exon 7 (nt 28,376 to 29,115 spanning introns 6 to 9), primers 5 and 6; and genomic exon 5 (nt 25,794 to 26,169), primers 7 and 8.
Single-Strand Conformational Polymorphism (SSCP) Analysis
The amplified genomic exon 5 product was subjected to nested PCR of nt 25,861 to 26,031 with primer 9 (5′ end-labeled with 32P-γdATP) and primer 10. The product was denatured and electrophoresed on a 0.5× MDETM gel containing 10% glycerol, followed by autoradiography, as recommended by the gel manufacturer (AT Biochem, Malvern, PA). For dideoxy sequencing, a separate nested PCR product (nt 25,794 to 26,169) was generated using unlabeled primers 9 and 10.
Screening cDNA Subclones for the R101Q Mutation and Retention of Exon 7
cDNA nt 96 to 872 (ATG start codon through exon 8) was amplified with primers 3 and 11. Bsg I digestion then distinguished exon 4 wild type (resistant) from R101Q mutant clones (sensitive). cDNA nt 471 to 1,126 (exons 5 to 11) was amplified using primers 12 and 13. Digestion with Nci I then distinguished clones containing exon 7 (sensitive) from those lacking exon 7 (resistant). The normal exon 7 Nci I site is unaffected by the A215T mutation.
Construction of ADA cDNA Containing the P126Q or A215T Mutations
P1260 cDNA.Using recombinant PCR,28 two primary PCR reactions were performed with wild-type ADA cDNA as template and the primer pairs (mutation underlined): (1) P126Q (+), 5′GAAGGGGACCTCACCCAAGACGA and primer 4; and (2) primer 3 and P126Q (−) 5′CCACCTCGTCTTGGGTGAGGT. The two PCR products (having a central 15-bp overlap) were gel purified, combined, annealed, and amplified with primers 3 and 4. The final product was gel purified, cut with EcoRI/HindIII, and cloned into pBluescript (Stratagene, La Jolla, CA). Sequencing identified the P126Q mutation and no other change.
A215T cDNA.Primers for the primary reactions were (1) primer 2 and A215T (+) 5′ACTGTCCACACCGGGGAGGT; and (2) primer 1 and A215T (−) 5′CCTCCCCGGTGTGGACAG. The products were purified, annealed, extended, amplified with primers 1 and 2, cut with EcoRI/HindIII, and cloned into pBluescript. Sequencing showed the A215T mutation and no other change.
Effect of Mutations on ADA Catalytic Activity
As reported,16,27 29 mRNA transcribed in vitro from wild-type and mutant ADA cDNA subclones was translated in a rabbit reticulocyte lysate in the presence of [35S]methionine. Aliquots containing equal amounts of wild-type and mutant translation products (shown by 10% sodium dodecyl sulfate-mercaptoethanol polyacrylamide gel electrophoresis and fluorography) were electrophoresed on cellulose acetate and stained for ADA activity in situ.
RESULTS
Patient No. 1 (CKu) and Family
In RBC of patient no. 1, ADA activity was 0.2 nmol/h/mg protein (normal ± 1 standard deviation [SD], 80.4 ± 40.2); S-adenosylhomocysteine hydrolase activity was 0.59 nmol/h/mg (normal, 4.2 ± 1.9); and total deoxyadenosine nucleotides (dAXP) were 28 nmol/mL (normal, <2). Blood mononuclear cells were not available for analysis.
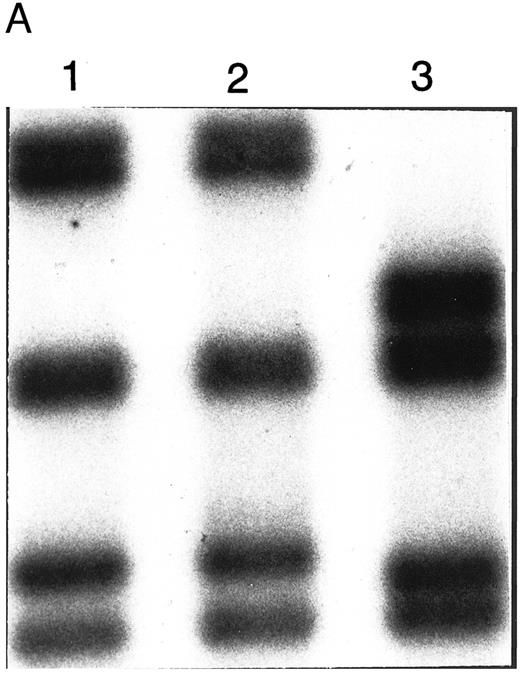
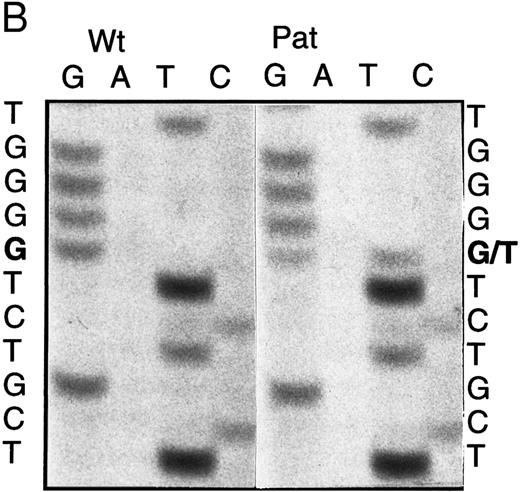
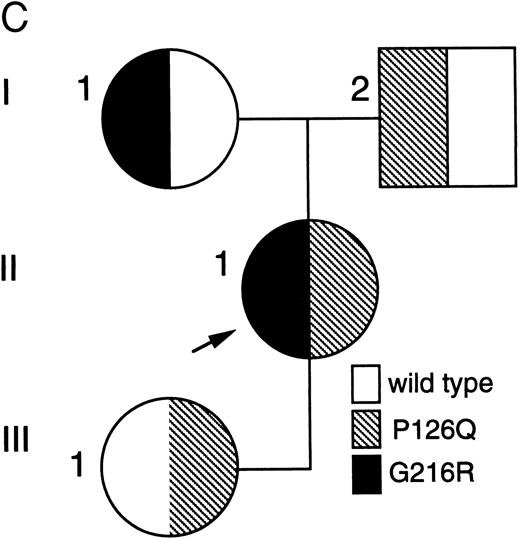
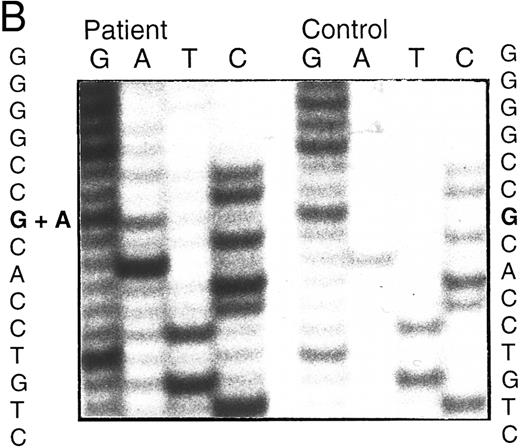
BstXI digestion of exon 7 amplified from genomic DNA of patient no. 1 indicated heterozygosity for the mutation, codon 216 GGGGlycine to AGGArginine (G216R) (data not shown), which occurs in 10% to 15% of ADA-deficient patients16 30 (and unpublished data). SSCP analysis suggested that her second ADA allele had a mutation in exon 5 (Fig 1A). Sequencing of amplified genomic exon 5 showed heterozygosity for a C > A transversion of genomic nt 25902 (mRNA nt 472; Fig 1B). This mutation, codon 126 CCAProline to CAAGlutamine (P126Q), has not been previously reported. Patient no. 1 had inherited G216R from her mother and P126Q from her father; she transmitted P126Q to her daughter (data not shown, Fig 1C).
Analysis of ADA genotype of patient no. 1 and family. (A) SSCP analysis of genomic exon 5. Lanes 1 and 2, controls; lane 3, patient no. 1. (B) Sequencing gel showing heterozygosity of patient no. 1 for the P126Q mutation in genomic exon 5 DNA. (C) Pedigree of family 1. Arrow indicates patient no. 1 (II-1).
Analysis of ADA genotype of patient no. 1 and family. (A) SSCP analysis of genomic exon 5. Lanes 1 and 2, controls; lane 3, patient no. 1. (B) Sequencing gel showing heterozygosity of patient no. 1 for the P126Q mutation in genomic exon 5 DNA. (C) Pedigree of family 1. Arrow indicates patient no. 1 (II-1).
Patient No. 2 and Family
In the SCID proband (III-1, Fig 2A), RBC ADA was 0.1 nmol/h/mg protein, and dAXP were 508 nmol/mL. These values for patient no. 2, her healthy maternal uncle (II-3, Fig 2A), were <0.1 nmol/h/mg and 15 nmol/ml, respectively. All other family members studied had normal or heterozygous range ADA activity and undetectable dAXP. ADA activity in cultured T cells from patient no. 2 was 170 nmol/h/mg protein versus 1,435 and 1,918 for T cells of his sister and her husband, the parents of the SCID proband (normal, 2,047 ± 1,36016).
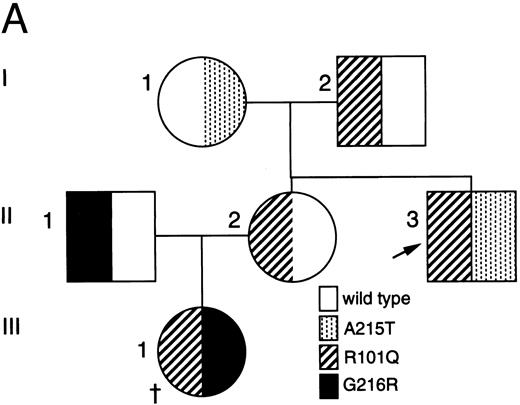
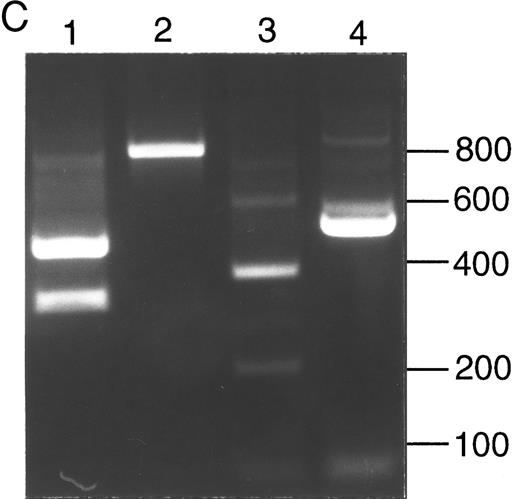
Analysis of ADA cDNA and genomic DNA of patient no. 2 and family. (A) Pedigree of family 2. Arrow indicates patient no. 2 (II-3); † indicates the deceased proband (III-1), an infant with SCID whose genotype is presumed. (B) Sequence of genomic exon 7 DNA (patient no. 2, left; control, right). Patient no. 2 is heterozygous for the A215T mutation. (C) Ethidium-stained agarose gel illustrating the method of screening cDNA clones for the R101Q mutation and for retention of exon 7 (see the Materials and Methods). Numbers at right indicate length in nucleotides of DNA markers. Lanes 1 and 2, PCR fragments (exons 1 to 8) digested with Bsg I. Lane 1, clone bearing R101Q mutation (cuts with Bsg I); lane 2, clone with wild-type (R101) exon 4 (lacks Bsg I site). Lanes 3 and 4, PCR fragments (exons 5 to 11) digested with Nci I. Lane 3, clone containing exon 7 (cuts with Nci I within exon 7); lane 4, clone with a deletion of exon 7 (lacks exon 7 Nci I site).
Analysis of ADA cDNA and genomic DNA of patient no. 2 and family. (A) Pedigree of family 2. Arrow indicates patient no. 2 (II-3); † indicates the deceased proband (III-1), an infant with SCID whose genotype is presumed. (B) Sequence of genomic exon 7 DNA (patient no. 2, left; control, right). Patient no. 2 is heterozygous for the A215T mutation. (C) Ethidium-stained agarose gel illustrating the method of screening cDNA clones for the R101Q mutation and for retention of exon 7 (see the Materials and Methods). Numbers at right indicate length in nucleotides of DNA markers. Lanes 1 and 2, PCR fragments (exons 1 to 8) digested with Bsg I. Lane 1, clone bearing R101Q mutation (cuts with Bsg I); lane 2, clone with wild-type (R101) exon 4 (lacks Bsg I site). Lanes 3 and 4, PCR fragments (exons 5 to 11) digested with Nci I. Lane 3, clone containing exon 7 (cuts with Nci I within exon 7); lane 4, clone with a deletion of exon 7 (lacks exon 7 Nci I site).
Five ADA cDNA subclones from T cells of patient no. 2 had a previously reported exon 4 mutation, codon 101 CGGArginine to CAGGlutamine (R101Q).31 Amplified genomic DNA was heterozygous for a Bsg I site created by R101Q. Three cDNA subclones lacking this Bsg I site had a deletion of exon 7, but no other mutation, suggesting that his second allele might have a deletion or splice site mutation of exon 7. Sequencing of his amplified genomic DNA showed normal exon 7 splice junctions, but heterozygosity for a previously reported exon 7 missense mutation, codon 215 GCCAlanine to ACCThreonine (A215T; Fig 2B).13 Analysis of family members indicated that patient no. 2 had inherited A215T maternally and R101Q paternally (Fig 2A). His sister had inherited a wild-type allele and R101Q, and her husband was heterozygous for G216R; they had presumably transmitted R101Q and G216R alleles to their ADA− SCID daughter (who was not studied; Fig 2A).
Our initial analysis of cDNA from patient no. 2 suggested an allele-specific skipping of exon 7. To assess this further, we screened a second random group of cDNA clones from his T cells (Fig 2C). Overall, of 32 clones analyzed, 4 of 8 (50%) A215T-derived clones lacked exon 7, compared with 3 of 24 (12.5%) bearing the R101Q mutation (similar to the 10% to 15% frequency of exon 7 skipping in normal cells31 ). In a previous report of a child homozygous for A215T, only four cDNA clones were sequenced, but one lacked exon 7 and another had missplicing of intron 6.13 Our data on a larger number of cDNA clones from a heteroallelic patient more clearly indicate that A215T is associated with both increased loss of exon 7 and a relative decrease in transcript level. Two other exon 7 missense mutations (G216R and R211H) did not appear to have such effects16 27 (and F.X. A.-V., I.S., M.S.H., unpublished data, Duke University).
Activity of Expressed Mutant ADA Alleles
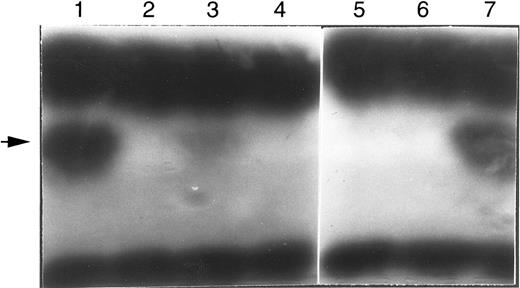
The A215T in vitro translation product was 10% to 20% as active as the wild type; the newly identified P126Q mutant protein was inactive, as were the R101Q and G216R proteins (Fig 3). We have also expressed these cDNAs in Escherichia coli Sø3834 (an ADA deletion strain32 ). A215T had approximately 15% of wild-type ADA activity, compared with approximately 0.1% for P126Q and R101Q and less than 0.01% for G216R (F.X.A., I.S., M.S.H., study in progress). Because exon 7 skipping eliminates activity,33 the loss of ADA activity owing to the A215T mutation may be greater than is evident from the expressed cDNA, which reflects only the effect of the amino acid substitution.
In situ stain for ADA activity of in vitro translation products. Arrow indicates position of human ADA. The dark bands at the bottom and top are rabbit hemoglobin and rabbit ADA, respectively, carried over from the reticulocyte translation reaction. The translation reactions were primed with the normal (wild-type) or mutant human ADA mRNAs (transcribed from cDNA); lane 1, wild-type control; lane 2, blank; lane 3, A215T (patient no. 2); lane 4, R101Q (patient no. 2); lane 5, P126Q (patient no. 1); lane 6, G216R (patient no. 1); lane 7, wild-type control.
In situ stain for ADA activity of in vitro translation products. Arrow indicates position of human ADA. The dark bands at the bottom and top are rabbit hemoglobin and rabbit ADA, respectively, carried over from the reticulocyte translation reaction. The translation reactions were primed with the normal (wild-type) or mutant human ADA mRNAs (transcribed from cDNA); lane 1, wild-type control; lane 2, blank; lane 3, A215T (patient no. 2); lane 4, R101Q (patient no. 2); lane 5, P126Q (patient no. 1); lane 6, G216R (patient no. 1); lane 7, wild-type control.
DISCUSSION
Lymphopenia and immune deficiency were appreciated in patient no. 1 at age 5, but ADA deficiency was then unknown and only aggressive treatment of infections could be offered. Remarkably, she survived and did relatively well from puberty until age 28, when she began to have serious respiratory infections, including tuberculosis. She developed chronic pulmonary and hepatobiliary insufficiency and at 40, fatal leukoencephalopathy caused by JC papovavirus, a disorder prevalent in the acquired immunodeficiency syndrome (AIDS).34 This history suggests that cellular and humoral immune dysfunction were not initially as complete as in SCID, which if uncorrected is fatal by age 1 to 2 years, but decreased to critical levels in the fourth decade, presumably owing to ongoing exposure to toxic metabolites. We wish to speculate that unexplained episodes of hepatitis from childhood in patient no. 1 may also have had a metabolic basis. Thus, hepatocellular degeneration is fatal in newborn ADA knockout mice,35,36 and we have reported a child with ADA− SCID who presented with persistent neonatal hepatitis, apparently without infectious cause, which resolved rapidly with enzyme replacement.37
Both mutations of patient no. 1, G216R and P126Q, greatly reduced ADA activity. However, her RBC dAXP level (28 nmol/mL) was lower than has been reported for immunodeficient patients (350 to >1,800 nmol/mL for SCID; 60 to 300 nmol/mL for those with a later onset16,26,38,39 ). This may reflect a hypoplastic marrow, because RBC dAdo (precursor of dAXP) arises from DNA degraded during normal marrow cell turnover.40,41 However, modest dAXP elevation may reflect greater residual ADA activity in vivo than suggested from cDNA expression. G216R probably eliminates activity irreversibly by interacting with the active site residue glutamate 21742; it is always associated with SCID when homozygous or combined with another severe allele (Hirschhorn et al30 and unpublished data). However, P126Q occurs in a helical segment that has no contact with the active site; this protein might be stabilized or induced to fold properly in lymphoid cells, which normally express very high ADA activity (as discussed elsewhere43 ).
Another phenomenon that should be considered is somatic mosaicism, which was recently found in two ADA-deficient patients who were immunodeficient as children, but then, like patient no. 1, had remissions lasting at least into their second decade.44 45 Selective survival of ADA+ lymphocytes was postulated to explain their low RBC dAXP levels and improved immune function. Tissues from patient no. 1 were not available for study. However, her clinical improvement, although prolonged, was not sustained. If present, mosaicism may not have occurred in a stem cell, and thus might have been unstable.
The A215T allele, discovered in a healthy child with partial ADA deficiency,13 can plausibly explain the benign status of patient no. 2 at age 28. A215T cDNA-encoded protein had 10% to 15% of wild-type activity, 100-fold more than his R101Q cDNA product (the SCID phenotype and high RBC dAXP of the niece of patient no. 2 is consistent with R101Q being a null mutation). Significant in vivo ADA function in patient no. 2 is indicated by activity in lysates of his T cells and by RBC dAXP in a range (≤20 nmol/mL) found in healthy individuals with partial ADA deficiency.20,29,46 47 We plan to monitor dAXP yearly for correlation with immunologic and clinical status.
The mechanism by which A215T alters exon 7 processing (apparently enhancing an effect that occurs with normal ADA pre-mRNA31 ) is unclear. The mutation occurs at nt 37/72 of exon 7, distant from splice junctions, and it does not create a new splice site. Some midexonic nonsense mutations can, through unclear mechanisms, induce exon skipping and reduce mRNA abundance,48,49 eg, the R142X mutation at nt 62/116 of ADA exon 5.29 Skipping of exon 7 maintains reading frame, so A215T does not induce a secondary nonsense codon. Further study of the effects of A215T on splicing are warranted.
ADA deficiency is treatable, but the longer diagnosis is delayed, the less reversible are its chronic consequences. Because of her age and clinical status, patient no. 1 was not a candidate for marrow transplantation. Local regulatory issues prevented access to polyethylene glycol-modified ADA (approved as an “Orphan Drug” in the United States and used in eight other countries for treating ADA deficiency50-52 ). Earlier diagnosis would have enhanced the chance of benefiting from either therapy and might have prompted avoidance of glucocorticoids, which were also used extensively before diagnosis of ADA deficiency in the adult sisters reported previously.38
There are no strict immunological parameters for identifying older ADA-deficient patients. Opportunistic infections may play a less prominent role than in patients with early onset, and older patients have manifested asthma, autoimmunity, and signs of immune dysregulation, eg, IgG subclass deficiency or elevated serum IgE.15,16,38 39 The diagnosis should be considered in adults with persistent lymphopenia and immunodeficiency of unknown cause (eg, some cases of common variable immune deficiency). Once ADA deficiency is diagnosed, metabolite and genotype analysis may help in assessing prognosis and in providing genetic counseling.
ACKNOWLEDGMENT
We gratefully acknowledge the expert technical assistance of Stephan Toutain and Marlis Schmid.
Supported in part by grants from the National Institutes of Health (DK20902) and from Enzon, Inc to M.S.H.
Address reprint requests to Michael S. Hershfield, MD, Box 3049, Duke University Medical Center, Durham, NC 27710.