Abstract
The human blood platelet fibrinogen receptor, integrin αIIbβ3 (glycoprotein IIb-IIIa) is an archetypal member of the integrin family of adhesive molecules and is the only integrin encoded by genes physically linked in the genome. Because studies on the normal and abnormal expression of any gene require a thorough understanding of its organization, the initial goals of the current study were to determine the size and complete the genomic organization for the β3 gene. We now report the isolation of the entire β3 gene in a single P1 plasmid and for the first time have linked the first and second exons on a contiguous fragment of DNA. Using pulsed-field gel analysis, we determined the full size of the β3 gene to be 63 kb and show a large (16.7 kb) first intron; based on this information, we propose a uniform numbering system for the β3 exons. We have completed the 5′ genomic structure and generated a long-range restriction map. The promoter and the 5′ end of the first intron were found to have approximately 50% sequence identity with a region of the avian β3 gene known to possess functional transcriptional activity. Analysis of three different homologous regions led to the identification of a sequence in the 5′-UTR of the human gene, CCGCGGGAGG, which shares 90% identity with the avian gene and which bound nuclear proteins in DNaseI and electrophoretic mobility shift assay studies. Mutating this sequence caused a 2.6-fold reduction in reporter gene activity. In these studies we have (1) determined the full length and 5′ organization of the β3 gene, (2) identified a large region of homology between the 5′ regions of the avian and human genes, and (3) identified a sequence in the 5′-UTR that augments gene expression. Knowing the genomic structure of β3 has permitted the uncovering of new mechanisms of mutagenesis causing Glanzmann thrombasthenia (Jin et al, J Clin Invest 98:1745, 1996), and our findings will be valuable for such genetic analyses as well as for studies on the transcriptional regulation of β3 and other integrin genes.
PLATELET AGGREGATION occurs when fibrinogen binds to its receptor, integrin αIIbβ3 (platelet glycoproteins IIb [GPIIb] and IIIa [GPIIIa]) on the surface of activated platelets.1,2 The αIIbβ3 complex is a member of the integrin family of heterodimeric cellular receptors involved in adhesive interactions.3,4 Alterations in the normal expression and function of the subunits of this important receptor result in a spectrum of hemorrhagic, and perhaps thrombotic, clinical complications. Although αIIb is expressed only in the megakaryocyte/platelet lineage, β3 is nevertheless an early marker for the megakaryocyte lineage. In fact, there is reason to believe the β3 gene may be expressed before αIIb : CD34+ stem cells have been shown to express β3 ,5 but not αIIb .6 Relative to αIIb , β3 expression is less restricted, and in other tissues it pairs with αv to form the vitronectin receptor. In addition to high levels of expression in megakaryocytes and platelets, αvβ3 is expressed in a number of tissues, including endothelial cells,7,8 smooth muscle cells,9 human osteoclasts,10,11 monocyte-derived macrophages,12,13 cultured human embryonic fibroblasts,14,15 human placental syncytiotrophoblast brush border,16 as well as some malignant cell lines. Altered αvβ3 expression has been associated with malignant potential.17,18 The differential pattern of tissue expression for αIIb and β3 indicates that their transcription is regulated, at least in part, by independent factors. But, unlike all other known integrin pairs, the genes for αIIb and β3 are physically linked on 17q21.32,19 20 raising the possibility of shared cis-acting elements coordinating gene expression in megakaryocytes.
Some integrin functions require the ability to upregulate or downregulate different integrin molecules, but relatively little is known about factors that modify this expression. In the case of β3 , for example, expression goes up before menstruation and during pregnancy,21,22 and we have data indicating that this effect is mediated by sex hormones.23 Using nuclear run-on experiments, Zutter et al24 have shown that β3 expression is controlled at the level of transcription. A vitamin D-responsive element has been identified in the avian gene,25 but none of the cis-acting elements necessary for modulating expression of the human gene in response to exogenous stimuli have been identified.
Genetic defects in the β3 gene that result in less than 20% levels of protein cause the inherited bleeding disorder Glanzmann thrombasthenia.26 An increasing number of mutations have been defined,27 but most have been type I patients with nearly absent levels of protein. Type II thrombasthenics have 10% to 20% of the normal level of receptor protein; based on fibrinogen binding and clot retraction studies, this is functionally normal αIIbβ3 protein.26 One type II patient has been shown to have a missense mutation in the αIIb gene.28 Another potential mechanism explaining the type II phenotype would be a mutation in a sequence affecting gene transcription, but to date no such mutations have been identified. In fact, using standard techniques, it has not been possible to identify all mutations in all types of patients with thrombasthenia. Although much of the genomic structure of β3 has been reported,29 30 the first and second exons were never linked, such that neither the size nor the complete β3 genomic structure was known. Perhaps this inability to identify mutations in some patients, particularly in type II patients, may be due in part to this gap in our knowledge of the β3 gene structure. Similarly, the ability to analyze gene transcription has been limited.
We now report the complete size and provide a long-range restriction map of the β3 gene. We also identify a modest degree of homology with the corresponding region of the avian gene. With this information as a guide, we characterized a sequence in the 5′-UTR of the human gene modulating expression. This information should be useful for the study of both the normal (eg, the regulation of tissue-specificity) and abnormal expression of β3 .
MATERIALS AND METHODS
Screening P1 libraries. P1 plasmids are cloning vectors that accommodate very large (typically 75- to 95-kb) fragments of DNA.31 A human genomic DNA P1 plasmid library with inserts of 65 to 110 kb32 was screened by polymerase chain reaction (PCR) analysis using sense and antisense primer pairs from the 5′ region of both the αIIb and β3 genes. This library was contained in 1,500 microtiter plates of 96 wells, with each well holding a single recombinant. The arrayed library was screened as previously described.32 Briefly, the primary screen used template DNA from 24 of the most complex pools, with subsequent screens performed on simpler pools until a single clone was obtained. Host bacteria containing the positive clone were grown by standard techniques, and plasmid DNA was prepared by a modified alkaline lysis method,33 being careful not to shear the DNA.
Southern blotting. Confirmation that D12 contained β3 sequence was accomplished with two sets of Southern bolts. (1) For the first exon, plasmid DNA was digested with restriction endonucleases, separated by electrophoresis through 1.0% agarose gels, transferred to a nylon membrane as previously described,30 and hybridized with a synthetic oligonucleotide 5′ of the first exon, “ex0.S” (Table 1). (2) For downstream exons, P1 plasmids were PCR-amplified with oligonucleotides specific for exons spanning the entire length of the β3 gene, followed by Southern blotting and probing with a 2.6-kb β3 cDNA containing the entire coding region.34 PCR reactions consisted of 50 ng plasmid DNA, 10 pmol of each primer, dNTPs at 200 μmol/L, 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2 , 0.001% gelatin, and 2.5 U Taq polymerase in a 50 μL volume. After 4 minutes at 95°C, 30 cycles of amplification were performed using the following conditions: denaturation for 45 seconds at 94°C, annealing for 30 seconds at 60°C, and extension at 72°C for 60 seconds. All PCR reactions were separated on 3% agarose gels and transferred as described.30
Hybridizations were performed in 2× SSC (300 mmol/L NaCl, 30 mmol/L sodium citrate, pH 7.0), 1% NaDodSO4 , 100 μg/mL salmon sperm DNA, and 10% dextran sulfate with 2 × 106 cpm/mL of 32P-labeled probe at 65°C. Filters were washed in 2× SSC/1% NaDodSO4 at 50°C for 60 minutes with two changes. Filters to be rehybridized were stripped with 0.1× SSC, 1% NaDodSO4 , and 50% formamide at 65°C for 30 minutes and exposed to film for 4 days to ensure complete removal of the first probe.
Pulsed-field gel analyses. Plasmid DNA (7.5 μg) was digested with the indicated restriction enzyme(s) and electrophoresis was performed using two different CHEF pulsed-field gel apparati in 0.5× TBE buffer (45 mmol/L Tris, 45 mmol/L boric acid, 1.25 mmol/L EDTA, pH 8.3). The first set of experiments used an apparatus designed and built according to Chu et al35: a 0.8% agarose gel was electrophoresed at 180 V (10 V/cm) with a switching interval of 5 seconds; total electrophoresis time was 18 hours at 13°C. To obtain better resolution of the DNA fragments, we also used the CHEF Mapper Pulsed Field Electrophoresis System (Bio-Rad Laboratories, Inc, Melville, NY), in which the optimal resolution size was set at 4 to 50 kb; the built-in algorithm determined that the following variables should be used: a 1% agarose gel electrophoresed at 9.0 V/cm at 14°C, forward voltage gradient of 9.0 V/cm, reverse voltage gradient of 6.0 V/cm, initial switching time of 0.08 seconds, and final switching time of 0.92 seconds. The total electrophoresis time was 19 hours and 2 minutes. DNA was transferred to nylon membranes as described above for Southern analysis. Hybridization probes for β3 gene mapping (Table 1) were 5′ to the first exon (“ex0.S”), 3 kb upstream of the second exon (“int0”), and 3′ of the third exon (“ex2.A”). Probe “int0” is in intron 0 and is a 239-bp Acc I-Bsm I genomic DNA fragment that has been previously described.36
DNA sequencing. All sequencing was performed on double-stranded plasmid DNA by the dideoxy method.36 Sequencing primers were SP6, T7, or primers generated from sequence obtained from the P1 inserts.
DNA sequence searches and homology determination. We used the Basic Local Alignment Search Tool (BLAST) software through the National Center for Biotechnology Information (NCBI, Bethesda, MD) to search for nucleic acid homologies to the human β3 gene. We used the blastn program to search all nonredundant sequences in all the GenBank databases as well as those sequences in the eukaryotic promoter database (epd). Nucleotide sequences of the avian (GenBank/EMBL Data Bank, accession no. X75348) and human30 β3 genes were compared and aligned using the GAP program of the Wisconsin Package Version 9.0, Genetic Computer Group (GCG; Madison, WI) and the percentage identities were determined. Dot matrix analysis was performed by the DNASTAR software (DNASTAR Inc, Madison, WI) using the most stringent criteria permissible. This program also provided the Similarity Index, defined as (100 × length of the consensus sequence) divided by (length of the consensus sequence + mispairings + gaps). As a control, eight avian genes were randomly chosen from the GenBank and aligned to the human gene, and the Similarity Index was determined.
Plasmid constructs. The wild-type sequence upstream of the ATG translation start codon to −146 of the transcription start site of the β3 gene was cloned into the luciferase reporter gene in the pGL2 Basic vector (Promega, Madison, WI) and called −146Luc. Mutations were introduced into the −146Luc template using the Site-Directed Mutagenesis kit (Clontech, Palo Alto, CA) according to the manufacturer's recommendations; this construct was called −146mutLuc.
Cell lines and culture conditions. The β3 -expressing cell lines K562,37,38 Dami,39 and HEL40 were cultured as described previously.41 In some experiments, K562 cells were treated with 100 nmol/L phorbol myristate acetate (PMA).41 The human microvascular endothelial cell (HMEC-1) line that expresses β342 was cultured in Endothelial Basal Medium (MCDB 131; Clonetic Corp, San Diego, CA) supplemented with 10% fetal bovine serum (GIBCO BRL, Grand Island, NY), 10 μg/mL hydrocortisone (Sigma, St Louis, MO), and 10 ng/mL epidermal growth factor (EGF; Collaborative Biomedical Products-Becton Dickinson, Bedford, MA). Two additional non–β3 -expressing cell lines were studied. Chinese hamster ovary (CHO) cells, which do not express β3 (data not shown), were cultured in α-modified eagle medium (GIBCO BRL) containing 10% vol/vol fetal bovine serum (GIBCO BRL). The transformed human embryonal kidney cell line 293 (ATCC, Rockville, MD), which expresses little or no β3 ,43 was cultured in Eagle's Minimal essential medium (GIBCO BRL) supplemented with 10% fetal bovine serum (GIBCO BRL).
DNaseI footprint analysis. Crude nuclear extracts were prepared from K562 cells using the method of Andrews and Faller.44 Briefly, cells were lysed in buffer A (10 mmol/L HEPES-KOH, pH 7.9, at 4°C, 1.5 mmol/L MgCl2 , 10 mmol/L KCl, 0.5 mmol/L dithiothreitol, and 0.2 mmol/L phenylmethyl sulfonyl fluoride [PMSF ]) and centrifuged, and the pellets were resuspended in buffer C (20 mmol/L HEPES-KOH, pH 7.9, 25% glycerol, 420 nmol/L NaCl, 1.5 mmol/L MgCl2 , 0.2 mmol/L EDTA, 0.5 mmol/L dithiothreitol, and 0.2 mmol/L PMSF ). Cellular debris was removed by centrifugation, and supernatants were stored at −80°C. A 176-bp fragment containing the −146 to +29 bp portion of the β3 gene was labeled with a 32P-deoxyguanidine triphosphate (dGTP; 3,000 Ci/nmol; Amersham, Arlington Heights, IL) at its 5′ end using Moloney's murine leukemia virus (MMLV) reverse transcriptase (Stratagene, La Jolla, CA). The probe was purified by gel elution after electrophoresis through a 5% native polyacrylamide gel (polyacrylamide; Boehringer Mannheim, Indianapolis, IN). Footprint analysis was performed with 5 μg nuclear extract using the DNaseI HotFoot footprinting kit (Stratagene), and samples were electrophoresed on 12% polyacrylamide gel that was pre-run for 1 hour at 30 W and run for 2.5 to 6 hours at 60 W. Gel was exposed to film (Eastman Kodak, Rochester, NY) for 15 hours at −80°C.
Electrophoretic mobility shift assays (EMSAs). Crude nuclear extracts were prepared as above from K562, Dami, HEL, 293, HMEC-1, and CHO cells. DNA probes were prepared as follows: single-stranded DNA oligonucleotides were slowly annealed to form double-stranded probes by incubating the sense and antisense strands at 95°C followed by slow cooling to room temperature. Double-stranded DNA probes were labeled with α32P-deoxycytidine triphosphate (dCTP; 3,000 Ci/nmol) or α32P-deoxyguanidine triphosphate (dGTP; 3,000 Ci/nmol) using DNA polymerase (Klenow fragment; Boehringer Mannheim). Crude nuclear extracts (5 μg protein/sample) were incubated with double-stranded 32P-labeled oligonucleotide probes in Incubation Buffer (Hotfoot Buffers Kit; Stratagene) for 15 minutes at room temperature. The composition of this latter buffer is proprietary. Competition experiments were performed by the simultaneous incubation with unlabeled, irrelevant DNA probes. Samples were electrophoresed on 6% polyacrylamide gels that were pre-run at 100 V for 1 hour and run for 2.5 hours at 250 V and 4°C. This gel was dried for 1 hour at 80°C and exposed to film (Eastman Kodak) for 12 hours at −80°C.
Luciferase assays. Plasmids (20 μg/sample) were transiently transfected into K562 cells (harvested at a density of 1.5 to 2.0 × 105 cells/mL) by electroporation (1 × 107 cells/sample) using a gene pulser (Bio-Rad, Richmond, CA) set at 500 μF and 400 V. 293 cells were transiently transfected using the CaCl2 method as described previously.45 Cells were incubated on ice for 10 minutes, resuspended in 25 mL of complete media, and incubated at 37°C and 5% CO2 atmosphere. Cells were collected after 24 hours, washed twice with media, lysed, and analyzed using the Luciferase Assay System (Promega). Luciferase activity of the promoterless (pGL2 Basic vector [Promega]), −146Luc, and −146mutLuc constructs was normalized to CAT activity by cotransfection of the pSV40-CAT construct as described previously.30 To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all 3 constructs), the data were normalized and expressed as the fold activation over activity of the promoterless construct, pGL2.
RESULTS
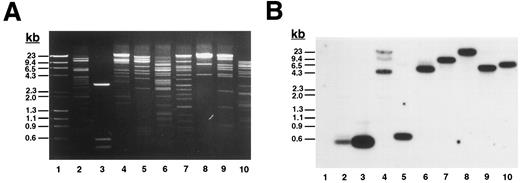
To determine the complete size of the β3 gene and in an initial effort to address the hypothesis that there may be shared cis-acting elements on the long arm of chromosome 17 that affect transcription of the genes for αIIb and β3 , we isolated additional genomic DNA that flanks both genes. Using primers A.S and A.A (Table 1), which flanked exon “i” of the β3 gene identified by Zimrin et al,29 we screened a human genomic DNA P1 library and obtained clone D12. D12 DNA was digested with a series of restriction enzymes (Fig 1A) and probed with an oligonucleotide primer (ex0.S) designed from DNA sequence upstream of the true first β3 exon (Fig 1B). The probe hybridized with a single fragment in each digest (except Xba I, which was only partially digested), and based on the position of this probe, we concluded that D12 contained sequence at least 113 bp upstream of the transcription start site. Subsequent studies showed at least an additional 4 kb of upstream sequence (see below). To determine how much of the downstream portion of the β3 gene was present in D12, we performed additional Southern blot analysis on products derived from PCR amplification of D12 using primers specific for exons 1, 2, 8, 10, 12, and 14 (Fig 1C). Note that exon 14 is the 3′-most exon of the β3 gene.29 Each exon was able to be amplified, and in all cases the major PCR products corresponded to the same fragment size amplified from normal genomic DNA. Each PCR product hybridized to a β3 cDNA probe (Fig 1C), indicating that D12 contained genomic sequence through the 3′ most exon in the gene.
Plasmid D12 contains the entire human β3 gene. (A) Ethidium bromide-stained agarose gel of D12 restriction digests. Lane 1, standard containing λ DNA digested with HindIII plus φx174 DNA digested with HaeIII; lane 2, D12 digested with Xba I + Sma I; lane 3, 1,211-bp Xho I + BamHI fragment of 5′ β3 gene subcloned into pBluescript and digested with Xba I + Sma I; the remaining lanes are D12 cut with Xba I (lane 4), Sma I (lane 5), HindIII (lane 6), Stu I (lane 7), Xho I (lane 8), Spe I (lane 9), and Acc I (lane 10). (B) Southern analysis of gel in (A) probed with 32P-labeled oligonucleotide ex0.S from positions −113 to −95 from the transcription start site of the β3 gene (probe position shown in Fig 3A). Lane 3 contains a positive control plasmid with genomic sequence from which the probe was designed.30 (C) Southern analysis of PCR products using clone D12 as a template, probed with a 32P-labeled β3 cDNA containing the entire coding region. Using primers flanking the indicated exons (Table 1), PCR reactions were performed on no template DNA (lanes 1, 4, 7, 10, 13, and 16), normal genomic DNA (lanes 2, 5, 8, 11, 14, and 17), and clone D12 (lanes 3, 6, 9, 12, 15, and 18). Under the same conditions, using the same PCR primers, no products were obtained using a negative P1 plasmid as a control (not shown). DNA size marker was φx174 digested with HaeIII. Exons are numbered as described in the text.
Plasmid D12 contains the entire human β3 gene. (A) Ethidium bromide-stained agarose gel of D12 restriction digests. Lane 1, standard containing λ DNA digested with HindIII plus φx174 DNA digested with HaeIII; lane 2, D12 digested with Xba I + Sma I; lane 3, 1,211-bp Xho I + BamHI fragment of 5′ β3 gene subcloned into pBluescript and digested with Xba I + Sma I; the remaining lanes are D12 cut with Xba I (lane 4), Sma I (lane 5), HindIII (lane 6), Stu I (lane 7), Xho I (lane 8), Spe I (lane 9), and Acc I (lane 10). (B) Southern analysis of gel in (A) probed with 32P-labeled oligonucleotide ex0.S from positions −113 to −95 from the transcription start site of the β3 gene (probe position shown in Fig 3A). Lane 3 contains a positive control plasmid with genomic sequence from which the probe was designed.30 (C) Southern analysis of PCR products using clone D12 as a template, probed with a 32P-labeled β3 cDNA containing the entire coding region. Using primers flanking the indicated exons (Table 1), PCR reactions were performed on no template DNA (lanes 1, 4, 7, 10, 13, and 16), normal genomic DNA (lanes 2, 5, 8, 11, 14, and 17), and clone D12 (lanes 3, 6, 9, 12, 15, and 18). Under the same conditions, using the same PCR primers, no products were obtained using a negative P1 plasmid as a control (not shown). DNA size marker was φx174 digested with HaeIII. Exons are numbered as described in the text.
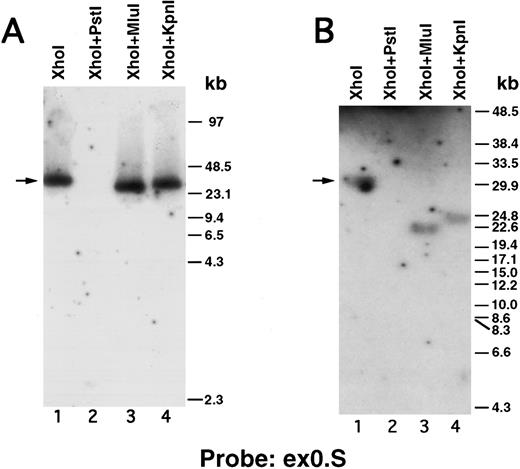
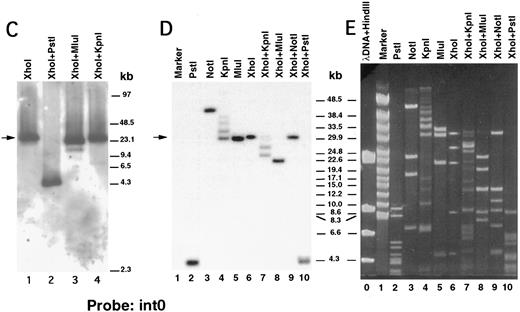
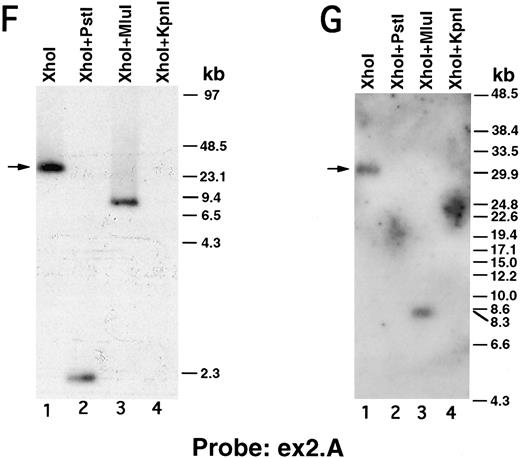
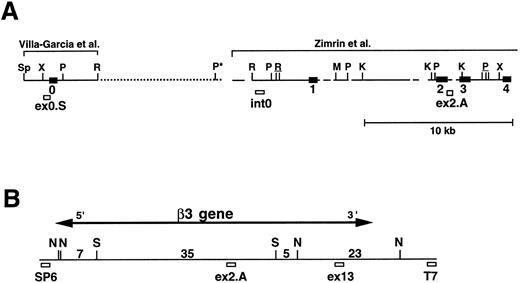
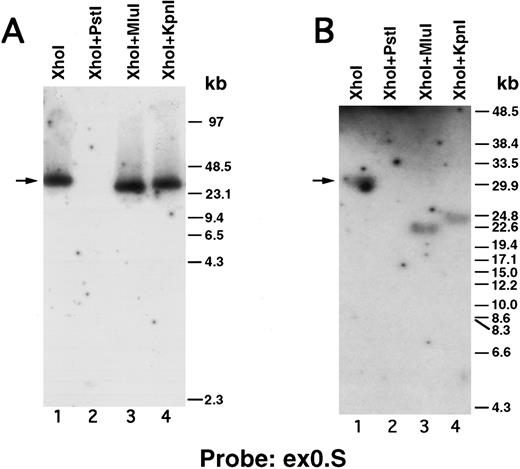
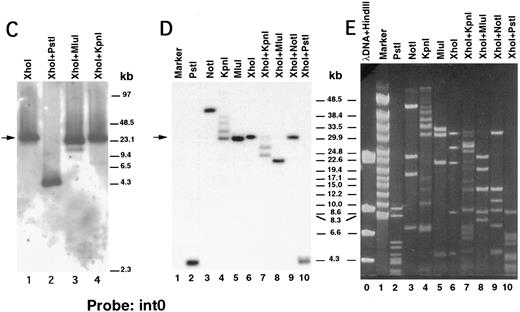
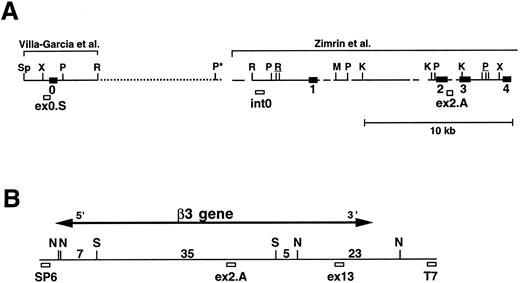
We next performed pulsed-field gel Southern blot analyses on plasmid D12 to construct a restriction map, determine the size of the β3 gene, and link the first two exons. We used probes designed or isolated from the previously published sequences28,29,33 that were located upstream of exon 0, within the first intron, and downstream of exon 2 (probe positions shown in Fig 3A). (To avoid confusion with the numbering system of Zimrin et al,29 in this report we call the true first exon number 0.) Alignment of autoradiograms derived from the sequential hybridization of the same filter with all three probes demonstrated an overlapping approximately 30.5-kb Xho I fragment (indicated by the arrow in Fig 2A-G). Additional experiments using conditions that permitted better resolution of these high molecular weight fragments are shown in Fig 2B, D, E, and G. Prior sequence information had identified the position of one Mlu I and two Kpn I restriction sites located between exons 1 and 2,29 and double digests with these enzymes plus Xho I confirmed that the 3 probes had hybridized to the same 30.5-kb fragment generated by the single Xho I digest in Fig 2A, C, and F. These studies permitted the construction of a map that joins the first two exons (Fig 3A). Note the large 16.7-kb intron between exons 0 and 1. In data not shown, we also sequenced both ends of the insert in plasmid D12, synthesized oligonucleotide probes from these sequences, and used pulsed-field gel analysis to determine a long-range Sfi I and Not I restriction map for the β3 gene (Fig 3B). This mapping indicated that clone D12 had a genomic DNA insert of approximately 80 kb and there was approximately 4 kb of genomic DNA upstream of the first exon. Note that neither of the two Sfi I sites had been previously identified, presumably because not all intronic sequence had been determined. Note also the two 5′ Not I sites that are separated by only 70 bp.30
Pulsed-field gel and Southern blot analysis of the β3 gene contained in clone D12. Two blots, generated using different electrophoretic conditions, are shown for each of the three probes. Panels are presented according to the 5′ to 3′ orientation of the probes (positions shown in Fig 3A). In all panels, the arrow indicates the 30.5-kb Xho I fragment. The same filter was probed, stripped, and reprobed in (A), (C), and (F ). Size markers are λ DNA PFGE Markers (Pharmacia) in (A), (C), and (F ); and 8- to 48-kb (mixed digest of λ) markers (Bio-Rad) in (B), (D), (E), and (G). (A and B) Southern blot analysis with oligonucleotide probe ex0.S, which is 5′ of the first exon. D12 DNA was digested with the indicated restriction enzymes. The Pst I-Pst I fragment (from the Xho I + Pst I digest) containing probe ex0.S is 1.49 kb and has run off the gel. (C, D, and E) Southern blot analysis with probe int0 between the first and second exons. The ethidium bromide-stained gel in (E) was used for the Southern analysis shown in (D) and is shown to demonstrate the ability to resolve the high molecular weight fragments in these studies. D12 DNA was digested with the indicated restriction enzymes. Note that the Kpn I digested completely in (C), but partially in (D) and (E). In (D) and (E), the ”Marker“ in lane 1 contains 8- to 48-kb (mixed digest of λ) markers (Bio-Rad); in (E), lane 0 contains λ DNA digested with HindIII.
(F and G) Southern blot analysis with probe ex2.A, which is 3′ of exon 2. D12 DNA was digested with the indicated restriction enzymes. The Kpn I-Kpn I fragment (from the Xho I + Kpn I digest) containing probe ex2.A is less than 2 kb and ran off the gel.
Pulsed-field gel and Southern blot analysis of the β3 gene contained in clone D12. Two blots, generated using different electrophoretic conditions, are shown for each of the three probes. Panels are presented according to the 5′ to 3′ orientation of the probes (positions shown in Fig 3A). In all panels, the arrow indicates the 30.5-kb Xho I fragment. The same filter was probed, stripped, and reprobed in (A), (C), and (F ). Size markers are λ DNA PFGE Markers (Pharmacia) in (A), (C), and (F ); and 8- to 48-kb (mixed digest of λ) markers (Bio-Rad) in (B), (D), (E), and (G). (A and B) Southern blot analysis with oligonucleotide probe ex0.S, which is 5′ of the first exon. D12 DNA was digested with the indicated restriction enzymes. The Pst I-Pst I fragment (from the Xho I + Pst I digest) containing probe ex0.S is 1.49 kb and has run off the gel. (C, D, and E) Southern blot analysis with probe int0 between the first and second exons. The ethidium bromide-stained gel in (E) was used for the Southern analysis shown in (D) and is shown to demonstrate the ability to resolve the high molecular weight fragments in these studies. D12 DNA was digested with the indicated restriction enzymes. Note that the Kpn I digested completely in (C), but partially in (D) and (E). In (D) and (E), the ”Marker“ in lane 1 contains 8- to 48-kb (mixed digest of λ) markers (Bio-Rad); in (E), lane 0 contains λ DNA digested with HindIII.
(F and G) Southern blot analysis with probe ex2.A, which is 3′ of exon 2. D12 DNA was digested with the indicated restriction enzymes. The Kpn I-Kpn I fragment (from the Xho I + Kpn I digest) containing probe ex2.A is less than 2 kb and ran off the gel.
Map of the 5′ region of the human β3 gene. (A) Southern blot data from Fig 2 were used to generate the map of the 5′ region of the β3 gene. The previously isolated sequences by Villa-Garcia et al30 and Zimrin et al29 are indicated above. Sp, Spe I; X, Xho I; P, Pst I; R, EcoRI; M, Mlu I; and K, Kpn I. The Pst I site with an asterisk is based on the data in Fig 2C. Underlined enzymes indicate clusters of sites. Exons 0 through 4 are indicated as black boxes; probe locations are indicated by small open boxes and labeled ex0.S, int0, and ex2.A. Gaps and dots in horizontal line indicate portions that have not been sequenced; dots represent new information from the current study. It is not known whether additional Pst I, Spe I, or EcoRI sites are contained within the dotted region. (B) Long-range restriction map for the β3 gene. D12 was digested with Sfi I (S) and/or Not I (N), and pulsed-field gel and Southern blot analyses were performed with probes synthesized from the sequences at the SP6 and T7 ends of the P1 plasmid. The entire D12 insert is shown as the horizontal line. Probe ex13 was a 1.5-kb cDNA 3′ to the EcoRI site in exon 13.36 The 5′ ex2.A and 3′ ex13 probes are positioned in the approximate location of the appropriate exon. Numbers are kilobases between indicated restriction sites. The horizontal line with arrow indicates the extent of the β3 gene.
Map of the 5′ region of the human β3 gene. (A) Southern blot data from Fig 2 were used to generate the map of the 5′ region of the β3 gene. The previously isolated sequences by Villa-Garcia et al30 and Zimrin et al29 are indicated above. Sp, Spe I; X, Xho I; P, Pst I; R, EcoRI; M, Mlu I; and K, Kpn I. The Pst I site with an asterisk is based on the data in Fig 2C. Underlined enzymes indicate clusters of sites. Exons 0 through 4 are indicated as black boxes; probe locations are indicated by small open boxes and labeled ex0.S, int0, and ex2.A. Gaps and dots in horizontal line indicate portions that have not been sequenced; dots represent new information from the current study. It is not known whether additional Pst I, Spe I, or EcoRI sites are contained within the dotted region. (B) Long-range restriction map for the β3 gene. D12 was digested with Sfi I (S) and/or Not I (N), and pulsed-field gel and Southern blot analyses were performed with probes synthesized from the sequences at the SP6 and T7 ends of the P1 plasmid. The entire D12 insert is shown as the horizontal line. Probe ex13 was a 1.5-kb cDNA 3′ to the EcoRI site in exon 13.36 The 5′ ex2.A and 3′ ex13 probes are positioned in the approximate location of the appropriate exon. Numbers are kilobases between indicated restriction sites. The horizontal line with arrow indicates the extent of the β3 gene.
We also isolated three different overlapping P1 plasmids that contained the entire αIIb gene (data not shown). Restriction mapping by pulsed-field gel electrophoresis showed that these clones contained a total of approximately 130 kb, including and flanking the αIIb gene. Probing αIIb P1 plasmid DNA on pulsed-field gel Southern blots with oligonucleotide probes from each end of the D12 insert indicated that the genomic DNA contained within the αIIb and β3 P1 plasmids did not overlap. These studies and additional analyses of genomic DNA (not shown) indicated that the genes for αIIb and β3 are separated by a distance of greater than 50 kb.
Because transcriptional regulatory sequences have been identified in other genes with large first introns,46 we used intron 0 sequence to search several databases. The only sequence with substantial homology was the 5′ region of the avian β3 gene, and further comparisons identified three regions that showed 59.2%, 52.2%, and 55.3% sequence identity, respectively (Fig 4A). To begin to assess the significance of the homology of these regions, the human and avian sequences were aligned. The two regions of the human gene that correspond to the vitamin D responsive sequence in the avian gene had relatively little homology and did not match any known consensus vitamin D responsive element (VDRE; Fig 4B and C). Dot matrix analysis was also performed, allowing comparison of these sequences over the entire regions of homology. These alignments and comparisons suggested that only regions H1 and H3 might possess significant similarities (Fig 4D and E). The first region (H1) was studied in more detail because the corresponding region of the avian gene has been shown to regulate transcription.25 To investigate the possibility of a nuclear protein interaction with this sequence, we performed DNaseI footprint analysis with extracts from the β3 -expressing K562 cells (Fig 5A). A protected region was observed corresponding to the sequence CCGCGGGAGG, located at positions +13 to +22 from the transcription start site. This 5′-UTR sequence was identical in 9 of 10 bp to the homologous region of the avian sequence (Fig 5A and the last 10 bp of Fig 4B). Using a DNA probe containing the CCGCGGGAGG sequence in an EMSA (Fig 5B), a DNA-protein interaction was observed. A shifted band was observed in all tissues examined, although nuclear proteins from the megakaryocytic cell lines showed a more prominent interaction with the DNA probe (compare lanes 2 through 7 with lanes 8 through 10). This DNA-protein interaction was specific, because it was not competed by an irrelevant DNA probe (Fig 5B, lane 4), but was competed by an excess of unlabeled probe (lane 3). No other DNA-protein interaction was detected. Functional activity of this sequence was tested with a reporter gene construct containing sequences −146 to +29 of the human β3 gene. This construct had previously been shown to be active in K562 and 293 cells (Wilhide et al47 and manuscript submitted). The footprinted region was mutated at the positions shown in Fig 5A, thus altering 7 of the 10 bp of the 5′-UTR regulatory sequence. As shown in Fig 5C, this disruption of the wild-type sequence caused an approximately 2.6-fold loss of luciferase activity in both K562 and 293 cells, indicating that this sequence was necessary for maximal gene expression. Because an equivalent decrease was also seen in 293 cells, this regulatory sequence is not tissue specific.
Homology between the human and avian β3 genes. (A) Schematic representation showing the homologous regions of the 5′ regions of the human and avian β3 genes. Sequences were compared as described in the Materials and Methods. A1 refers to exon 1 of the avian gene.25 Nucleotide positions are relative to the transcription start sites. The numbers between the horizontal lines refer to the percentage of homology between regions H1, H2, and H3 of each gene. B, Bsm I; H, HindIII; X, Xba I; E, EcoRI; S, Sac I. In (B) through (E), the human and avian sequence numbering is according to the GenBank entry. (B) Alignment of the H1 region of the human and avian β3 genes. The thick underline indicates the VDRE of the avian gene. (C) Alignment between the avian VDRE with the corresponding human sequence from the H2 region. Dot matrix plots of the H1 (D) and H2 (E) regions of the human and avian β3 genes.
Homology between the human and avian β3 genes. (A) Schematic representation showing the homologous regions of the 5′ regions of the human and avian β3 genes. Sequences were compared as described in the Materials and Methods. A1 refers to exon 1 of the avian gene.25 Nucleotide positions are relative to the transcription start sites. The numbers between the horizontal lines refer to the percentage of homology between regions H1, H2, and H3 of each gene. B, Bsm I; H, HindIII; X, Xba I; E, EcoRI; S, Sac I. In (B) through (E), the human and avian sequence numbering is according to the GenBank entry. (B) Alignment of the H1 region of the human and avian β3 genes. The thick underline indicates the VDRE of the avian gene. (C) Alignment between the avian VDRE with the corresponding human sequence from the H2 region. Dot matrix plots of the H1 (D) and H2 (E) regions of the human and avian β3 genes.
Identification of a 5′-UTR sequence modulating β3 gene expression. (A) DNaseI footprint analysis. DNaseI footprint analysis was performed using a 32P-labeled probe of the β3 promoter from position −146 to +29 (shown above), incubated without (lane 1) and with (lane 2) K562 nuclear extracts. The protected region in the human β3 gene is from position +13 to +22 relative to the transcription start site and corresponds to a region of high human-avian sequence homology. The sequence mutated for subsequent functional analysis (C) is indicated. (B) EMSA using the 32P-labeled oligonucleotide probe incubated with nuclear extracts of K562 and K562 cells treated with PMA, Dami, HEL, HMEC-1, 293, or CHO cells. Equivalent amounts of labeled probe were added to each experiment per lane. The amount of probe associated with the shown K562 complex was approximately 20% of the total amount of free probe. The free probe was deliberately run off the gel to maximize resolution of the DNA-protein complex; no other gel shifts were observed. Competitors were used in a 50-fold molar excess and included both an unlabeled EMSA (c.c., cold competition) and an irrelevant (Irrel.) oligonucleotide. A lane with probe but no nuclear extract (lane 1) showed only the fast-migrating radiolabeled probe. (C) Functional assessment of the CCGCGGGAGG sequence. Sequence substitutions in the footprinted region (shown in [A]) were introduced into construct −146mutLuc. K562 and 293 cells were transfected with equivalent amounts of the promoterless (pGL2), −146Luc, and −146mutLuc constructs and assayed for luciferase activity, and all values were normalized for transfection efficiency within each cell line by cotransfection with pSV40-CAT. An approximately 2.6-fold difference between −146Luc and −146mutLuc was observed. To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all three constructs), the data were normalized and expressed as fold activation over activity of the promoterless construct, pGL2. The −146mutLuc construct contained the mutated sequence shown in Fig 5B. Each experiment was performed four times.
Identification of a 5′-UTR sequence modulating β3 gene expression. (A) DNaseI footprint analysis. DNaseI footprint analysis was performed using a 32P-labeled probe of the β3 promoter from position −146 to +29 (shown above), incubated without (lane 1) and with (lane 2) K562 nuclear extracts. The protected region in the human β3 gene is from position +13 to +22 relative to the transcription start site and corresponds to a region of high human-avian sequence homology. The sequence mutated for subsequent functional analysis (C) is indicated. (B) EMSA using the 32P-labeled oligonucleotide probe incubated with nuclear extracts of K562 and K562 cells treated with PMA, Dami, HEL, HMEC-1, 293, or CHO cells. Equivalent amounts of labeled probe were added to each experiment per lane. The amount of probe associated with the shown K562 complex was approximately 20% of the total amount of free probe. The free probe was deliberately run off the gel to maximize resolution of the DNA-protein complex; no other gel shifts were observed. Competitors were used in a 50-fold molar excess and included both an unlabeled EMSA (c.c., cold competition) and an irrelevant (Irrel.) oligonucleotide. A lane with probe but no nuclear extract (lane 1) showed only the fast-migrating radiolabeled probe. (C) Functional assessment of the CCGCGGGAGG sequence. Sequence substitutions in the footprinted region (shown in [A]) were introduced into construct −146mutLuc. K562 and 293 cells were transfected with equivalent amounts of the promoterless (pGL2), −146Luc, and −146mutLuc constructs and assayed for luciferase activity, and all values were normalized for transfection efficiency within each cell line by cotransfection with pSV40-CAT. An approximately 2.6-fold difference between −146Luc and −146mutLuc was observed. To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all three constructs), the data were normalized and expressed as fold activation over activity of the promoterless construct, pGL2. The −146mutLuc construct contained the mutated sequence shown in Fig 5B. Each experiment was performed four times.
DISCUSSION
Previous studies have provided information about the intron-exon structure of the β3 gene, but have not determined the size of the entire gene. Despite the isolation of 22 clones and the DNA sequencing of more than 36 kb of genomic sequence,29,30 no one clone had contained both the first and second exons and no overlapping clones had linked these two exons. In this report, we have used the large capacity of P1 plasmids to isolate the entire β3 gene on a single contiguous fragment of normal human genomic DNA. P1 plasmids are cloning vectors that accommodate very large (typically 75 kb to 95 kb) fragments of DNA.31 These vectors accommodate larger fragments than do cosmid vectors, but do not have the recombination problems associated with yeast artificial chromosomes (YACs). Pulsed-field gel analysis indicated that the full length of the β3 gene is 63 kb. Intron 0 of the β3 gene is quite large (∼16.7 kb), and this may provide a partial explanation for previous difficulties in determining the complete genomic structure. Because of the numerous prior references to the exon numbering of Zimrin et al29 (designated exons i, ii, iii, etc), to maintain consistency, we propose referring to the first exon of the β3 gene as exon 0. We believe that our clone contains the entire gene because (1) Southern blot and PCR mapping show it contains sequence 5′ of the transcription start site and 3′ of the last exon, as well as 5 internal exons that are evenly distributed throughout the gene, and (2) our long-range Sfi I and Not I map is consistent with sequences previously deposited in GenBank.29
Previous estimates on the distance between the αIIb and β3 genes were based on cross-hybridization to an Sfi I fragment ranging in size from 125 kb36 to 260 kb.19 Our studies were not able to refine these previous measurements. Nevertheless, this distance between the two genes remains compatible with the possibility that the genes could share enhancer element(s), considering the large distances involved between other enhancers and the genes they regulate.48
Cao et al25 cloned the first exon and promoter region of the avian β3 gene and determined the sequence of the upstream approximately 800 bp. The avian sequences 702 bp and 161 bp upstream of the translational start site showed the greatest activity, ie, 34-fold and 48-fold, respectively, over the 80-bp minimal promoter. Because birds and humans diverged in evolution 290 to 320 million years ago,49 we were intrigued that computer alignment programs showed considerable sequence identity between the avian promoter region and the promoter and first intron of the human β3 gene. However, the dot matrix plots showed less impressive homologies. Determining whether this degree of homology is significant is not a simple matter. One approach would be to compare this degree of homology to homologies between other regions of the human and avian β3 genes, primarily intronic sequences, because they might be expected to be no better than random. However, the rest of the avian gene sequence is not known. As another approach, we compared the human H1 sequence with 8 avian genes randomly selected from the GenBank. Using the same parameters we used in Fig 4A, the average similarity index was 30.1% ± 1.7%, which is considerably lower than that which we observed for the avian β3 gene. We concluded that this degree of homology may be significant and may prove to be useful in directing further studies on transcriptional regulation — particularly sequences that share the highest homology and that are known transcription factor binding sites. Along these lines, two of the homologous regions with the human gene contained an area corresponding to the avian VDRE, but neither human sequence conforms to any known VDRE. In addition, preliminary transfection studies using the −146 luciferase construct showed no response to treatment with vitamin D (data not shown).
We were able to use the homology information to direct studies that identified a non–tissue-specific sequence in the 5′-UTR of the human β3 gene that augmented gene expression (Fig 5). Regulatory sequences in the 5′-UTR of genes typically affect gene expression by enhancing50 and inhibiting51 translation and possibly by increasing mRNA stability.52 We used two different techniques that showed a nuclear protein interaction with double-stranded DNA (Fig 5A and C), and it is tempting to speculate that the CCGCGGGAGG sequence acts as an enhancer of transcription. The mechanism by which this sequence modulates gene expression is not known, but other genes have been described in which 5′-UTR sequences enhance53 or repress54 transcription. The only matches found from a search of the transcription factor data base were sequences characterized by long strings of G's, and we could not find a consensus similar to CCGCGGGAGG. However, inspection of the 5′-UTR of the αIIb gene shows the sequence CCTGGGAGG at positions +8 to +16. This matches the β3 sequence (starting at +14) in 7 of 9 positions, but has not been studied functionally. In addition, given the high degree of homology between species as divergent as birds and humans, together with the functional activity we have shown for the human β3 gene, it will also be interesting to analyze the corresponding sequence in the avian gene for its ability to enhance gene expression.
It is interesting to note that two regions of the human gene appear homologous to the first 84 bp of the reported avian sequence (Fig 4A). This could represent an artifact of the DNA alignment program or a true duplication of DNA. There is no obvious pseudogene in intron 0 of the human gene and there is not another translation start site corresponding to the avian start site. Perhaps enhancer sequences are buried in the human first intron, but our preliminary attempts to test that region of the human gene (H3) that corresponds to the region of the avian gene showing the greatest functional activity have been inconclusive or negative.
In summary, we have completed and refined the organization of the human β3 gene. Knowledge of the genomic structure of β3 has been essential for the elucidation of novel mechanisms of mutagenesis in Glanzmann thrombasthenia.55 The information provided in this report may permit a greater understanding of the molecular pathogenesis of this disorder, particularly those with type II disease, and also allow more comprehensive gene regulation studies. The observed homology with the avian β3 gene directed studies to the identification of a human sequence that promoted gene expression. Other such homologies may prove useful in identifying transcriptional regulatory elements. Of potential interest, but as yet untested, is the sequence CCACACCC in the 5′-UTR of the avian gene.25 This is a 7 of 8 bp match with the corresponding sequence in the human gene (not shown) and is identical to a sequence in the human β-globin gene promoter and in the GT-I motif of the SV40 enhancer, which have previously been identified as transcription factor binding sites.56 57 Given the structural similarities among the human β integrins, it will be important to consider elements downstream of the transcription start site when studying the expression of these genes. Similarly, the identification of the transcription factor binding the CCGCGGGAGG sequence may help us to understand how β3 expression is regulated and may provide insight into the molecular pathogenesis of thrombasthenia.
ACKNOWLEDGMENT
The authors thank Drs Gary Cutting and Hal Dietz of Johns Hopkins University for the use of their pulsed-field gel apparati.
Supported by Grant No. HL51457 of the National Institutes of Health (Bethesda, MD) and by the Rogers-Wilbur Foundation. Additional 5′ sequence from the β3 gene has been submitted to GenBank (accession no. AF020552).
Address reprint requests to Paul F. Bray, MD, Ross 1015, Johns Hopkins University School of Medicine, 720 Rutland Ave, Baltimore, MD 21205.



![Fig. 5. Identification of a 5′-UTR sequence modulating β3 gene expression. (A) DNaseI footprint analysis. DNaseI footprint analysis was performed using a 32P-labeled probe of the β3 promoter from position −146 to +29 (shown above), incubated without (lane 1) and with (lane 2) K562 nuclear extracts. The protected region in the human β3 gene is from position +13 to +22 relative to the transcription start site and corresponds to a region of high human-avian sequence homology. The sequence mutated for subsequent functional analysis (C) is indicated. (B) EMSA using the 32P-labeled oligonucleotide probe incubated with nuclear extracts of K562 and K562 cells treated with PMA, Dami, HEL, HMEC-1, 293, or CHO cells. Equivalent amounts of labeled probe were added to each experiment per lane. The amount of probe associated with the shown K562 complex was approximately 20% of the total amount of free probe. The free probe was deliberately run off the gel to maximize resolution of the DNA-protein complex; no other gel shifts were observed. Competitors were used in a 50-fold molar excess and included both an unlabeled EMSA (c.c., cold competition) and an irrelevant (Irrel.) oligonucleotide. A lane with probe but no nuclear extract (lane 1) showed only the fast-migrating radiolabeled probe. (C) Functional assessment of the CCGCGGGAGG sequence. Sequence substitutions in the footprinted region (shown in [A]) were introduced into construct −146mutLuc. K562 and 293 cells were transfected with equivalent amounts of the promoterless (pGL2), −146Luc, and −146mutLuc constructs and assayed for luciferase activity, and all values were normalized for transfection efficiency within each cell line by cotransfection with pSV40-CAT. An approximately 2.6-fold difference between −146Luc and −146mutLuc was observed. To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all three constructs), the data were normalized and expressed as fold activation over activity of the promoterless construct, pGL2. The −146mutLuc construct contained the mutated sequence shown in Fig 5B. Each experiment was performed four times.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3951/3/m_bl_0029f5a.jpeg?Expires=1767936249&Signature=LhBSsPZgZcki5BQMCkOkZYeGqlXtt4f8NIUoW4nXhbx6mKIGwV-8neRul0IuB6rRwLFWwsbjgnbfm8-O2t8nwMUjBYZFKU5VCKolbK51LBZegDLtqJ5lG8uEQHOxdjAah9JcMhlHcfu3KagQXItkupIRKyJ1NnX0VoQdItwbAZ3AGFRmrqtjSrS0MyEXzognBRKFCZzQK3oh927yGBBL~Jy3HUQmI~ww9iqzGNnKoShCAoHoDnGdpElc0ZJEVcwRVAdRhqI5KOFETCG0JqFJpHXkT4jZxBMlh6aOFyvfiozj4b88KPppZ7YymMbYPEiK6WhNVB3VjoqpU4PKuOaGHA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Identification of a 5′-UTR sequence modulating β3 gene expression. (A) DNaseI footprint analysis. DNaseI footprint analysis was performed using a 32P-labeled probe of the β3 promoter from position −146 to +29 (shown above), incubated without (lane 1) and with (lane 2) K562 nuclear extracts. The protected region in the human β3 gene is from position +13 to +22 relative to the transcription start site and corresponds to a region of high human-avian sequence homology. The sequence mutated for subsequent functional analysis (C) is indicated. (B) EMSA using the 32P-labeled oligonucleotide probe incubated with nuclear extracts of K562 and K562 cells treated with PMA, Dami, HEL, HMEC-1, 293, or CHO cells. Equivalent amounts of labeled probe were added to each experiment per lane. The amount of probe associated with the shown K562 complex was approximately 20% of the total amount of free probe. The free probe was deliberately run off the gel to maximize resolution of the DNA-protein complex; no other gel shifts were observed. Competitors were used in a 50-fold molar excess and included both an unlabeled EMSA (c.c., cold competition) and an irrelevant (Irrel.) oligonucleotide. A lane with probe but no nuclear extract (lane 1) showed only the fast-migrating radiolabeled probe. (C) Functional assessment of the CCGCGGGAGG sequence. Sequence substitutions in the footprinted region (shown in [A]) were introduced into construct −146mutLuc. K562 and 293 cells were transfected with equivalent amounts of the promoterless (pGL2), −146Luc, and −146mutLuc constructs and assayed for luciferase activity, and all values were normalized for transfection efficiency within each cell line by cotransfection with pSV40-CAT. An approximately 2.6-fold difference between −146Luc and −146mutLuc was observed. To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all three constructs), the data were normalized and expressed as fold activation over activity of the promoterless construct, pGL2. The −146mutLuc construct contained the mutated sequence shown in Fig 5B. Each experiment was performed four times.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3951/3/m_bl_0029f5b.jpeg?Expires=1767936249&Signature=qeCck5b7sv9E90j~qTAGz1YB1UnANsBn9XuObAxd0D8OrHxIZuH5IcoA2PJ4BzZPZ1XoV0DzQZLy9c47lUmVvDHCUORjrjcx5BuzFyy4guoiS111idiPDlOmh23mx7CZyTdxJgx73LqqxfAHq4zvzmwjt-LxvOqwnCJ2XDrGwudVgE-ECSfMQ9gguH2p~~7OjYPoEiRepdQAvgzaeCwLKAFVF77KIuPHplwGnDL-Len~tQaK9dGHkJNKqSAeWAGd-bjp3jtvVZ56U4PZdF7Wf6yRXBBF4NUShcXo~BZe1kjzxLovEH9VKaxzlDI2UyLZWaWAHSAxXpzYabHTcOj5mg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Identification of a 5′-UTR sequence modulating β3 gene expression. (A) DNaseI footprint analysis. DNaseI footprint analysis was performed using a 32P-labeled probe of the β3 promoter from position −146 to +29 (shown above), incubated without (lane 1) and with (lane 2) K562 nuclear extracts. The protected region in the human β3 gene is from position +13 to +22 relative to the transcription start site and corresponds to a region of high human-avian sequence homology. The sequence mutated for subsequent functional analysis (C) is indicated. (B) EMSA using the 32P-labeled oligonucleotide probe incubated with nuclear extracts of K562 and K562 cells treated with PMA, Dami, HEL, HMEC-1, 293, or CHO cells. Equivalent amounts of labeled probe were added to each experiment per lane. The amount of probe associated with the shown K562 complex was approximately 20% of the total amount of free probe. The free probe was deliberately run off the gel to maximize resolution of the DNA-protein complex; no other gel shifts were observed. Competitors were used in a 50-fold molar excess and included both an unlabeled EMSA (c.c., cold competition) and an irrelevant (Irrel.) oligonucleotide. A lane with probe but no nuclear extract (lane 1) showed only the fast-migrating radiolabeled probe. (C) Functional assessment of the CCGCGGGAGG sequence. Sequence substitutions in the footprinted region (shown in [A]) were introduced into construct −146mutLuc. K562 and 293 cells were transfected with equivalent amounts of the promoterless (pGL2), −146Luc, and −146mutLuc constructs and assayed for luciferase activity, and all values were normalized for transfection efficiency within each cell line by cotransfection with pSV40-CAT. An approximately 2.6-fold difference between −146Luc and −146mutLuc was observed. To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all three constructs), the data were normalized and expressed as fold activation over activity of the promoterless construct, pGL2. The −146mutLuc construct contained the mutated sequence shown in Fig 5B. Each experiment was performed four times.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3951/3/m_bl_0029f5c.jpeg?Expires=1767936249&Signature=ovXNCaB8lbFMwALYlkLVQhsJYwzYWyAg7TkAZW0QHu-MCQq-ERzTFA0lTMs-9zZvaFrOPOlu3Bu4oNgipUUpaZ-PA3xj5h~bQ66OXyhdudw3WJjfRTFT0M7-iwouGRsm6yjcfLzgFFafsgtux7983Ka9AZNC09oU1Vzm-lcH4TRzucSfbAQIZGFjJ08QO~~RAbDo5Vn1S4933zxM0v8FtHurjxwsSa98gfZrEVB8qK0eC7xR~vzPBOmyZjCl4RX3Rw3ozZb1LwvCzdawucIFTbjDNg77WvZkRyoVgU2J5vTbwANL0Hm0LhlgGylnjXUiTFaUp0xRUTALSVsVawmTbA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





![Fig. 5. Identification of a 5′-UTR sequence modulating β3 gene expression. (A) DNaseI footprint analysis. DNaseI footprint analysis was performed using a 32P-labeled probe of the β3 promoter from position −146 to +29 (shown above), incubated without (lane 1) and with (lane 2) K562 nuclear extracts. The protected region in the human β3 gene is from position +13 to +22 relative to the transcription start site and corresponds to a region of high human-avian sequence homology. The sequence mutated for subsequent functional analysis (C) is indicated. (B) EMSA using the 32P-labeled oligonucleotide probe incubated with nuclear extracts of K562 and K562 cells treated with PMA, Dami, HEL, HMEC-1, 293, or CHO cells. Equivalent amounts of labeled probe were added to each experiment per lane. The amount of probe associated with the shown K562 complex was approximately 20% of the total amount of free probe. The free probe was deliberately run off the gel to maximize resolution of the DNA-protein complex; no other gel shifts were observed. Competitors were used in a 50-fold molar excess and included both an unlabeled EMSA (c.c., cold competition) and an irrelevant (Irrel.) oligonucleotide. A lane with probe but no nuclear extract (lane 1) showed only the fast-migrating radiolabeled probe. (C) Functional assessment of the CCGCGGGAGG sequence. Sequence substitutions in the footprinted region (shown in [A]) were introduced into construct −146mutLuc. K562 and 293 cells were transfected with equivalent amounts of the promoterless (pGL2), −146Luc, and −146mutLuc constructs and assayed for luciferase activity, and all values were normalized for transfection efficiency within each cell line by cotransfection with pSV40-CAT. An approximately 2.6-fold difference between −146Luc and −146mutLuc was observed. To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all three constructs), the data were normalized and expressed as fold activation over activity of the promoterless construct, pGL2. The −146mutLuc construct contained the mutated sequence shown in Fig 5B. Each experiment was performed four times.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3951/3/m_bl_0029f5a.jpeg?Expires=1767994102&Signature=HjH5mYVoSNoPSC81bdegE5Nr5ps11qfz~V9TtGv08lm7IlJKPMiEXxM8wYUUFixXsurwyYf~agV3Gn~udADxjAie3EaX6rPufxs4Oekw0G-EewU0sMKcNtUQOTiyoq-8cbgj6Gr79gn3JLXvNdFNEPPmN3ScszfDFiDV4FtnFoksYHk4oLVewdjlpDm-yYd46-VIn9IPsC-3J0o0vHGcFCcuBcQ0BOToNfOLqgNUI5rd~nrLvM4fv~x8v~2dMgilEZK2ToLwSTOX0vzitGqvsIelmO6FzF67s1lifNNIUtAbB63BclKL8OI1G~rBUPh9YrIKLP80nBoZDWRsRTGnOw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Identification of a 5′-UTR sequence modulating β3 gene expression. (A) DNaseI footprint analysis. DNaseI footprint analysis was performed using a 32P-labeled probe of the β3 promoter from position −146 to +29 (shown above), incubated without (lane 1) and with (lane 2) K562 nuclear extracts. The protected region in the human β3 gene is from position +13 to +22 relative to the transcription start site and corresponds to a region of high human-avian sequence homology. The sequence mutated for subsequent functional analysis (C) is indicated. (B) EMSA using the 32P-labeled oligonucleotide probe incubated with nuclear extracts of K562 and K562 cells treated with PMA, Dami, HEL, HMEC-1, 293, or CHO cells. Equivalent amounts of labeled probe were added to each experiment per lane. The amount of probe associated with the shown K562 complex was approximately 20% of the total amount of free probe. The free probe was deliberately run off the gel to maximize resolution of the DNA-protein complex; no other gel shifts were observed. Competitors were used in a 50-fold molar excess and included both an unlabeled EMSA (c.c., cold competition) and an irrelevant (Irrel.) oligonucleotide. A lane with probe but no nuclear extract (lane 1) showed only the fast-migrating radiolabeled probe. (C) Functional assessment of the CCGCGGGAGG sequence. Sequence substitutions in the footprinted region (shown in [A]) were introduced into construct −146mutLuc. K562 and 293 cells were transfected with equivalent amounts of the promoterless (pGL2), −146Luc, and −146mutLuc constructs and assayed for luciferase activity, and all values were normalized for transfection efficiency within each cell line by cotransfection with pSV40-CAT. An approximately 2.6-fold difference between −146Luc and −146mutLuc was observed. To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all three constructs), the data were normalized and expressed as fold activation over activity of the promoterless construct, pGL2. The −146mutLuc construct contained the mutated sequence shown in Fig 5B. Each experiment was performed four times.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3951/3/m_bl_0029f5b.jpeg?Expires=1767994102&Signature=c8j0Hf7IItQ6DBJ4Vn7vA70-wUZeTNybS6ywNkbf9iuovDNleRBp6X-QZB7Uuez0rYQhErhc6ohGHKdB0EmJbQk8-vplSG7ZfRUpKzOa6LbFTMMqfuUxDJvAtGvcNcgOLxEtU~OhuR1iTJ2deklYYI4N98gmMpk1AWmgXrX5T1eSh7lKFHun~08XOqjeqViBMlLmrNm8~O~K4MPPrwyspTy9VjDdyAo4FsIkFDKk3jF97E2DfNPniZE7sC-eFsjVgnoelJwdjE5QD6ZDbef~J5SuIE4lANKY1W03e6uT5OvTjoxW5qrLcpLKtgiTg~mGVIfNSj202Ruz~s5W9HjN5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Identification of a 5′-UTR sequence modulating β3 gene expression. (A) DNaseI footprint analysis. DNaseI footprint analysis was performed using a 32P-labeled probe of the β3 promoter from position −146 to +29 (shown above), incubated without (lane 1) and with (lane 2) K562 nuclear extracts. The protected region in the human β3 gene is from position +13 to +22 relative to the transcription start site and corresponds to a region of high human-avian sequence homology. The sequence mutated for subsequent functional analysis (C) is indicated. (B) EMSA using the 32P-labeled oligonucleotide probe incubated with nuclear extracts of K562 and K562 cells treated with PMA, Dami, HEL, HMEC-1, 293, or CHO cells. Equivalent amounts of labeled probe were added to each experiment per lane. The amount of probe associated with the shown K562 complex was approximately 20% of the total amount of free probe. The free probe was deliberately run off the gel to maximize resolution of the DNA-protein complex; no other gel shifts were observed. Competitors were used in a 50-fold molar excess and included both an unlabeled EMSA (c.c., cold competition) and an irrelevant (Irrel.) oligonucleotide. A lane with probe but no nuclear extract (lane 1) showed only the fast-migrating radiolabeled probe. (C) Functional assessment of the CCGCGGGAGG sequence. Sequence substitutions in the footprinted region (shown in [A]) were introduced into construct −146mutLuc. K562 and 293 cells were transfected with equivalent amounts of the promoterless (pGL2), −146Luc, and −146mutLuc constructs and assayed for luciferase activity, and all values were normalized for transfection efficiency within each cell line by cotransfection with pSV40-CAT. An approximately 2.6-fold difference between −146Luc and −146mutLuc was observed. To account for differences in transfection efficiency between K562 and 293 cells (the latter producing much higher activities with all three constructs), the data were normalized and expressed as fold activation over activity of the promoterless construct, pGL2. The −146mutLuc construct contained the mutated sequence shown in Fig 5B. Each experiment was performed four times.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.3951/3/m_bl_0029f5c.jpeg?Expires=1767994102&Signature=p~JZGUzHc0Cek0RfjD4erpNqmu7WI8MXTVqH2ujxREkuvAFAr42cshqc9AKCp4w-tw4dJlUyyTS4vKxb5lmObaF3sEr7sUR2hyjz8MeYC0chW0ZlWspiT0BR5KwRq7HqPgTD4PmsdxNAjTd98yUwzwxNj82kSwj7BGGGFW1Zg1lq3YvWN8GYTEbPuP6XQ8fDbGbqEIjtADJ1-xMQMDrPGhtqVVj1hdC8IsXR3~J~kBZdagDnhZkLOMS5~kOjCUl6TlXV3CC1IDfTEzDSOTp1-V47Qjhw-Q1gcmssF0kz3PGegjPHn0PH3DVvjf4fWo6k7FSTXspFUUmvq1Vnz7tYwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)