Abstract
Previous studies have shown that K562 chronic myelogenous leukemia cells are resistant to induction of apoptosis by a variety of agents, including the topoisomerase II (topo II) poison etoposide, when examined 4 to 24 hours after treatment with an initiating stimulus. In the present study, the responses of K562 cells and apoptosis-proficient HL-60 acute myelomonocytic leukemia cells to etoposide were compared, with particular emphasis on determining the long-term fate of the cells. When cells were treated with varying concentrations of etoposide for 1 hour and subsequently plated in soft agar, the two cell lines displayed similar sensitivities, with a 90% reduction in colony formation at 5 to 10 μmol/L etoposide. After treatment with 17 μmol/L etoposide for 1 hour, cleavage of the caspase substrate poly(ADP-ribose) polymerase (PARP), DNA fragmentation, and apoptotic morphological changes were evident in HL-60 cells in less than 6 hours. After the same treatment, K562 cells arrested in G2 phase of the cell cycle but otherwise appeared normal for 3 to 4 days before developing similar apoptotic changes. When the etoposide dose was increased to 68 μmol/L, apoptotic changes were evident in HL-60 cells after 2 to 3 hours, whereas the same changes were observed in K562 cells after 24 to 48 hours. This delay in the development of apoptotic changes in K562 cells was accompanied by delayed release of cytochrome c to the cytosol and delayed appearance of peptidase activity that cleaved the fluorogenic substrates Asp-Glu-Val-Asp-aminotrifluoromethylcoumarin (DEVD-AFC) and Val-Glu-Ile-Asp-aminomethylcoumarin (VEID-AMC) as well as an altered spectrum of active caspases that were affinity labeled with N-(Nα-benzyloxycarbonylglutamyl-Nε-biotinyllysyl) aspartic acid [(2,6-dimethylbenzoyl)oxy]methyl ketone [z-EK(bio)D-aomk]. On the other hand, the activation of caspase-3 under cell-free conditions occurred with indistinguishable kinetics in cytosol prepared from the two cell lines. Collectively, these results suggest that a delay in the signaling cascade upstream of cytochrome c release and caspase activation leads to a long latent period before the active phase of apoptosis is initiated in etoposide-treated K562 cells. Once the active phase of apoptosis is initiated, the spectrum and subcellular distribution of active caspase species differ between HL-60 and K562 cells, but a similar proportion of cells are ultimately killed in both cell lines.
RECENT STUDIES indicate that a wide variety of chemotherapeutic agents induce apoptotic cell death in susceptible cell lines.1-5 The morphological hallmarks of this process include loss of cell volume, hyperactivity of the plasma membrane, and condensation of peripheral heterochromatin followed by cleavage of the nucleus and cytoplasm into multiple membrane-enclosed bodies containing chromatin fragments.6 Experiments in animals6-9 and studies of circulating blasts from leukemia patients10 have provided evidence that apoptosis also occurs after chemotherapy in vivo. Moreover, it has been suggested that resistance of the apoptotic machinery to activation can lead to resistance to the cytotoxic effects of chemotherapeutic agents.8 11-15 These observations highlight the potential importance of understanding the factors that control the process of apoptosis.
A unique family of cysteine proteases, now called caspases,16 appears to play a critical role in initiating and sustaining the biochemical events that result in apoptotic cell death.4,17-22 This family of proteases, which contains at least 10 human members, cleaves polypeptides on the carboxyl side of aspartate residues.4,17-22 Substrates for these proteases include the nuclear enzyme PARP, which is cleaved at the sequence DEVD-G,23 and the nuclear lamins, which are cleaved by an enzyme that recognizes the sequence VEID-N,24 as well as additional cellular polypeptides.4,25,26 Although it is unclear which of the substrates play a critical role in the apoptotic process, inhibitors of this family of proteases abrogate apoptosis in a variety of cell types.27-30
Human acute leukemia cell lines have proven particularly informative in the study of chemotherapy-associated apoptotic proteolytic events. Early studies indicated that treatment of HL-60 cells with a variety of chemotherapeutic agents was accompanied by proteolytic degradation of a number of nuclear polypeptides, including PARP and lamin B1 .31,32 Immunoblotting and activity assays showed that caspase-3, one of the enzymes that can cleave PARP, is activated in human leukemia lines treated with a number of different agents.29,33,34 This activation of caspase-3 appears to be initiated by release of cytochrome c from mitochondria to cytosol.35-37 More recent studies have indicated that treatment of human leukemia cell lines with the topo II poison etoposide results in the activation of multiple caspases.38,39 In HL-60 cells, for example, two distinguishable activities that cleave the fluorogenic substrates DEVD-AFC and VEID-AMC increase 100-fold and 20-fold, respectively, during the first 3 hours after addition of etoposide.38 Labeling with z-EK(bio)D-aomk, a reagent that reacts with the large subunit of all active caspases tested, showed that active forms of several caspases, including caspase-3 and caspase-6, appear in the cytosol and nuclei of etoposide-treated HL-60 cells over this same time period.38 Affinity labeling techniques have shown a similar pattern of caspase activation in lymphocytic leukemia cell lines undergoing apoptosis after various stimuli.39,40 These observations have led to the suggestion that the same group of effector proteases might be activated in a variety of cell lines after various apoptotic stimuli.39-41
Studies of the K562 cell line, which was derived from a patient during the blast crisis phase of chronic myelogenous leukemia,42 have also provided important insight into the biology of programmed cell death. K562 cells are resistant to the induction of apoptosis by a variety of different agents, including diphtheria toxin, camptothecin, cytarabine, etoposide, paclitaxel, staurosporine, and anti-Fas antibodies.32,43-48 This resistance to apoptosis is specifically reversed by treatment with antisense oligonucleotides that inhibit synthesis of the bcr/abl kinase,44,45,49 50 lending support for the view that the inhibition of apoptosis might be one of the functions of this transforming oncoprotein. On the other hand, studies demonstrating the resistance to apoptosis have generally examined K562 cells only during the first 4 to 24 hours after treatment with the inducing stimulus. In the present study, we have compared the responses of HL-60 and K562 cells to etoposide over a much longer time course. These experiments indicate that K562 cells undergo programmed cell death after etoposide treatment, but the execution phase of this process is delayed compared with HL-60 cells. Consistent with these results, caspase activation is also delayed in K562 cells, suggesting that the bcr/abl kinase exerts its effects upstream of caspase activation. In addition, the species of affinity-labeled caspase molecules detected in cytosol and nuclei during the execution phase of apoptosis differ between the two cell lines, providing evidence that effector caspases can vary in different myeloid cell lines undergoing apoptosis after treatment with the same stimulus.
MATERIALS AND METHODS
Materials.Reagents were obtained from the following suppliers: [14C]-labeled thymidine (57.8 mCi/mmol) and ECL enhanced chemiluminescent reagents from Amersham (Arlington Heights, IL); K562 mRNA from Clontech (Palo Alto, CA); SuperSignal ULTRA chemiluminescent substrate from Pierce (Rockford, IL); horse heart cytochrome c from Sigma (St Louis, MO); and mouse anti-cytochrome c from Pharmingen (San Diego, CA). VEID-AMC and z-EK(bio)D-aomk were synthesized as described previously.38 All other reagents were obtained as previously indicated.38 51
Tissue culture.HL-60 cells were propagated in RPMI 1640 medium containing 100 U/mL penicillin G, 100 μg/mL streptomycin, 2 mmol/L glutamine, and 10% (vol/vol) heat-inactivated fetal bovine serum (medium A). K562 cells, which were kindly provided by Richard J. Jones (Johns Hopkins Oncology Center, Baltimore, MD) or purchased from American Type Culture Collection (Rockville, MD), were cultured in medium A containing 5% (vol/vol) heat-inactivated fetal bovine serum (medium B). Both cell lines were maintained at concentrations of <1 × 106 cells/mL to insure logarithmic growth.
Alkaline elution.Logarithmically growing cells were labeled for 24 hours with 1 μmol/L [14C]-thymidine, sedimented, and incubated for 1 to 2 hours at 37°C in fresh medium A. To measure steady-state levels of topo II–mediated DNA single-strand breaks, aliquots were then incubated for 30 minutes at 37°C with the indicated concentration of etoposide, diluted with ice-cold 75 mmol/L NaCl-2.4 mmol/L EDTA (pH 7.4 at 4°C), and deposited on Nucleopore phosphocellulose filters (1-μm pore size) (VWR Scientific, Minneapolis, MN) by gentle suction. To measure resealing of the same breaks, cells were incubated with 6.8 μmol/L (K562 cells) or 10.5 μmol/L (HL-60 cells) etoposide for 30 minutes at 37°C to stabilize equal numbers of covalent topo II-DNA complexes, diluted 30-fold into prewarmed medium A, and incubated for 2 to 24 minutes at 37°C before being deposited on the phosphocellulose filters. Once cells were deposited on the filters, all further steps were performed as recently described.51 In brief, cells were lysed by allowing 5 mL of buffer consisting of 1% (wt/vol) sodium dodecyl sulfate (SDS), 100 mmol/L glycine, 25 mmol/L EDTA (pH 10), and 0.5 mg/mL proteinase K to drip through the filters. After filters were washed with 20 mmol/L EDTA (pH 10), DNA was eluted with 20 mmol/L EDTA (adjusted to pH 12.1 with tetrapropylammonium hydroxide). The distribution of radiolabel in the eluate, the tubing of the elution apparatus, and the filter was analyzed as described by Covey et al.52 Cells that received 150 to 900 cGy of γ-irradiation from a 137Cs source were included in each experiment as a standard curve.
Induction of apoptosis.At the start of each experiment, nonviable cells were removed by sedimentation at 100 to 150g for 10 to 20 minutes on Ficoll-Hypaque step gradients (density = 1.119 g/cm2 ). Cells obtained from the interface were diluted with RPMI 1640 containing 10 mmol/L N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid (HEPES), sedimented at 150g for 10 minutes, and resuspended in fresh growth medium. Cells were treated with 17 μmol/L etoposide for 1 hour, sedimented at 150g for 10 minutes, and resuspended in drug-free medium A or B, respectively. Alternatively, etoposide was added to a final concentration of 68 μmol/L for continuous exposure experiments. In each case, control samples received an equivalent volume of the diluent dimethylsulfoxide (final concentration 0.1%), which was nontoxic at this concentration.
Immunoblotting.After treatment with etoposide as described in the individual experiments, cells were sedimented at 200g for 10 minutes at 4°C, washed once with ice-cold RPMI containing 10 mmol/L HEPES (pH 7.4 at 4°C); resuspended at a concentration of 3 × 107 cells/mL in SDS sample buffer consisting of 4 mol/L deionized urea, 2% (wt/vol) SDS, 62.5 mmol/L Tris-HCl (pH 6.8 at 21°C), 1 mmol/L EDTA, and 5% (vol/vol) freshly added β-mercaptoethanol; sonicated; and frozen at −70°C. Aliquots were subsequently heated to 65°C for 20 minutes, subjected to electrophoresis on SDS-polyacrylamide gels containing 5% to 15% (wt/vol) acrylamide gradients, transferred to nitrocellulose, and probed with antibodies as described.38
Cell fractionation.Cytosol and nuclei were prepared at 4°C as previously described.38 In brief, cells were washed twice in serum-free RPMI 1640 medium containing 10 mmol/L HEPES (pH 7.4) or in calcium-, magnesium-free phosphate-buffered saline and lysed by homogenization in buffer C [25 mmol/L HEPES (pH 7.5 at 4°C), 5 mmol/L MgCl2 , 1 mmol/L EGTA supplemented immediately before use with 1 mmol/L α-phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL pepstatin A, and 10 μg/mL leupeptin]. Following removal of nuclei by sedimentation at 800g for 10 minutes (see below) or 16,000g for 3 minutes, the supernatant was supplemented with 5 mmol/L EDTA and sedimented at 280,000gmax for 60 minutes. After addition of dithiothreitol (DTT) to a final concentration of 2 mmol/L, the supernatant (cytosol) was frozen in 50-μL aliquots at −70°C. Control experiments confirmed that activities described in the present study were stable at −70°C for at least 3 months, although all experiments were performed within 1 month of extract preparation.
Nuclei were isolated from the cellular homogenates by centrifugation at 800g followed by sedimentation through a layer of 50 mmol/L Tris-HCl (pH 7.4) containing 2.1 mol/L sucrose and 5 mmol/L MgSO4 at 80,000gmax . The nuclei were resuspended in storage buffer53 consisting of 10 mmol/L PIPES (pH 7.4), 80 mmol/L KCl, 20 mmol/L NaCl, 250 mmol/L sucrose, 5 mmol/L EGTA, 1 mmol/L DTT, 0.5 mmol/L spermidine, 0.2 mmol/L spermine, 50% (wt/vol) glycerol, 1 mmol/L PMSF, 10 μg/mL leupeptin, and 10 μg/mL pepstatin and stored at −70°C until use. Specific activity of lactate dehydrogenase, a cytosolic marker, was greater than 20-fold lower in these nuclei than in the corresponding cytosol.38
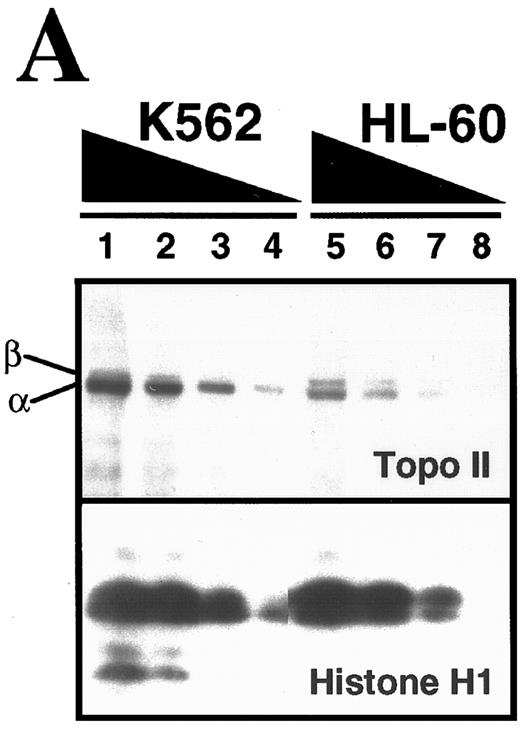
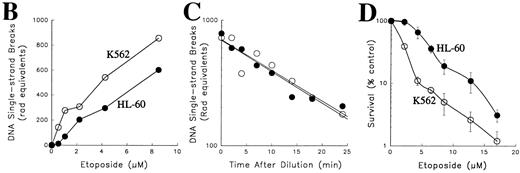
Comparison of topo II–mediated events in HL-60 and K562 cells. (A) Examination of topo II polypeptide content in HL-60 and K562 cells. Whole cell extracts containing polypeptides from 3 × 105 (lanes 1 and 5), 1.5 × 105 (lanes 2 and 6), 0.75 × 105 (lanes 3 and 7) and 0.3 × 105 (lanes 4 and 8) K562 cells (lanes 1 through 4) or HL-60 cells (lanes 5 through 8) were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies that recognize both topo II isoforms (upper panel) or, as a loading control, histone H1 (lower panel). Scanning of the blots (not shown) showed that topo IIβ levels in the two cell lines were equal after correction for differences in loading, whereas levels of topo IIα were twofold higher in K562 cells. (B) Formation of topo II–mediated strand breaks in the presence of etoposide. HL-60 cells (•) and K562 cells (○) were incubated with the indicated concentration of etoposide at 37°C for 30 minutes before application to Nucleopore filters. After elution for 7 hours, the fraction of DNA retained on the filters was compared to the fraction retained after irradiation of HL-60 or K562 cells. (C) Kinetics of resealing of etoposide-induced DNA single-strand breaks. HL-60 cells treated with 10.5 μmol/L etoposide (•) or K562 cells treated with 6.8 μmol/L etoposide (○) for 30 minutes at 37°C were diluted 30-fold and incubated at 37°C for the indicated length of time before deposition on Nucleopore filters. The fraction of DNA retained on the filters after elution under alkaline conditions were compared with the fraction retained after irradiation of HL-60 or K562 cells. (D) Effect of a 1-hour etoposide exposure on clonogenic survival of HL-60 (•) and K562 cells (○). Cells were simultaneously treated for 1 hour with the indicated concentration of etoposide, washed, and plated in 0.3% agar as described in Materials and Methods. Colony formation was assessed 10 to 14 days later. Bars, ± standard deviation of quadruplicate samples. Results in (A), (B), and (D) are representative of four separate experiments. Results in (C) are means of two separate experiments.
Comparison of topo II–mediated events in HL-60 and K562 cells. (A) Examination of topo II polypeptide content in HL-60 and K562 cells. Whole cell extracts containing polypeptides from 3 × 105 (lanes 1 and 5), 1.5 × 105 (lanes 2 and 6), 0.75 × 105 (lanes 3 and 7) and 0.3 × 105 (lanes 4 and 8) K562 cells (lanes 1 through 4) or HL-60 cells (lanes 5 through 8) were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies that recognize both topo II isoforms (upper panel) or, as a loading control, histone H1 (lower panel). Scanning of the blots (not shown) showed that topo IIβ levels in the two cell lines were equal after correction for differences in loading, whereas levels of topo IIα were twofold higher in K562 cells. (B) Formation of topo II–mediated strand breaks in the presence of etoposide. HL-60 cells (•) and K562 cells (○) were incubated with the indicated concentration of etoposide at 37°C for 30 minutes before application to Nucleopore filters. After elution for 7 hours, the fraction of DNA retained on the filters was compared to the fraction retained after irradiation of HL-60 or K562 cells. (C) Kinetics of resealing of etoposide-induced DNA single-strand breaks. HL-60 cells treated with 10.5 μmol/L etoposide (•) or K562 cells treated with 6.8 μmol/L etoposide (○) for 30 minutes at 37°C were diluted 30-fold and incubated at 37°C for the indicated length of time before deposition on Nucleopore filters. The fraction of DNA retained on the filters after elution under alkaline conditions were compared with the fraction retained after irradiation of HL-60 or K562 cells. (D) Effect of a 1-hour etoposide exposure on clonogenic survival of HL-60 (•) and K562 cells (○). Cells were simultaneously treated for 1 hour with the indicated concentration of etoposide, washed, and plated in 0.3% agar as described in Materials and Methods. Colony formation was assessed 10 to 14 days later. Bars, ± standard deviation of quadruplicate samples. Results in (A), (B), and (D) are representative of four separate experiments. Results in (C) are means of two separate experiments.
Fluorogenic assays.Aliquots containing 50 μg of cytosolic or nuclear protein (estimated by the bicinchoninic acid method54 ) in 50 μL buffer C were diluted with 225 μL of freshly prepared buffer D [25 mmol/L HEPES (pH 7.5), 0.1% (wt/vol) 3-[(cholamidopropyl)dimethylammonio]-1-propane sulfonate, 10 mmol/L DTT, 100 U/mL aprotinin, 1 mmol/L PMSF] containing 100 μmol/L substrate and incubated for 2 hours at 37°C. Reactions were terminated by addition of 1.225 mL ice cold buffer D. Fluorescence was measured using an excitation wavelength of 360 nm and emission wavelength of 475 nm. Reagent blanks containing 50 μL of buffer C and 225 μL of buffer D were incubated at 37°C for 2 hours, then diluted with 1.225 mL ice cold buffer D. Standards containing 0 to 3,000 pmol of aminomethylcoumarin or 0 to 1,500 pmol of aminotrifluoromethylcoumarin were used to determine the amount of fluorochrome released. Control experiments (not shown) confirmed that the release of substrate was linear with time and with protein concentration.
Affinity labeling.Aliquots containing the indicated amounts of nuclear or cytosolic protein were incubated for 1 hour at room temperature with 1 μmol/L z-EK(bio)D-aomk, diluted with 1/2 vol of 3X concentrated SDS sample buffer, heated to 95°C for 3 minutes, subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 16% (wt/vol) acrylamide gels, transferred to nitrocellulose, probed with peroxidase-labeled streptavidin, and visualized using ECL enhanced chemiluminescent reagents. Control experiments revealed that P20 subunits of active caspases-1, -2, -3, -4, and -6 labeled with this reagent.38
Two-dimensional analysis was performed using isoelectric focusing for the first dimension and SDS-PAGE for the second dimension as described.38 Labeled polypeptides were visualized using peroxidase-coupled streptavidin followed by SuperSignal ULTRA chemiluminescent substrate. Caspases expressed in Sf9 cells38 were subjected to this analysis in parallel to permit identification of the labeled polypeptide species.
Activation of caspases under cell-free conditions.Cytosol was prepared from untreated HL-60 or K562 cells as described above with the following changes: (1) 2 mmol/L DTT was added directly to homogenization buffer; and (2) the postnuclear supernatant was sedimented at 100,000gmax . In the wells of a 96-well microtiter plate, the following components were assembled on ice: 50 μg cytosolic protein, 1.2 mmol/L dATP, and 0.5 mg/mL horse heart cytochrome c.26 After the volume was brought to 40 μL with ice-cold buffer C, the reaction was initiated by shifting the plate to 37°C. At the indicated times, samples were removed, mixed with 4X SDS sample buffer, heated to 70°C for 10 to 15 minutes, subjected to SDS-PAGE on gels with 5% to 15% acrylamide gradients, and transferred to PVDF. Blots were probed with commercially available anti-caspase-2 or with affinity-purified rabbit antisera raised against recombinant large subunit of caspase-3.
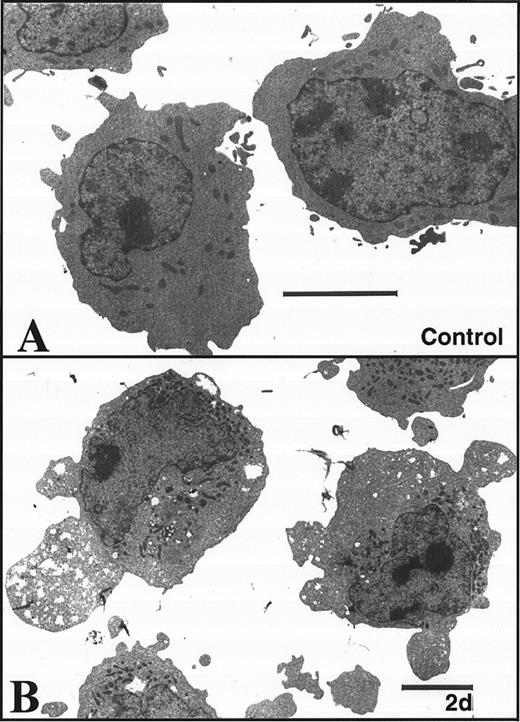
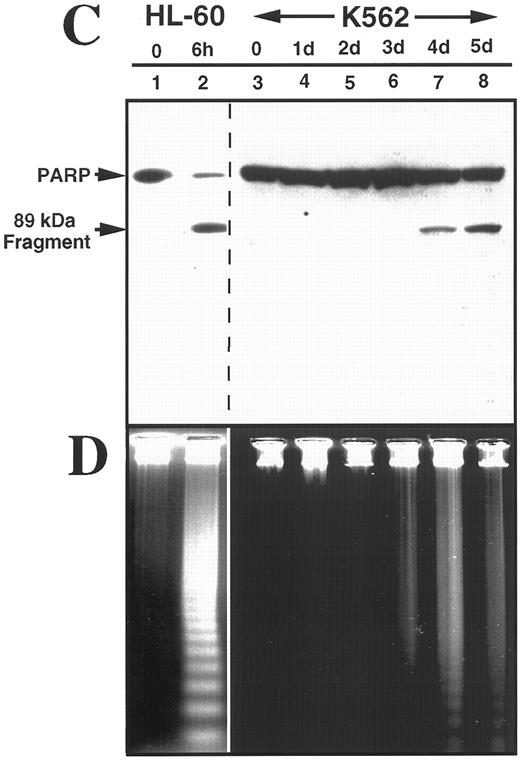
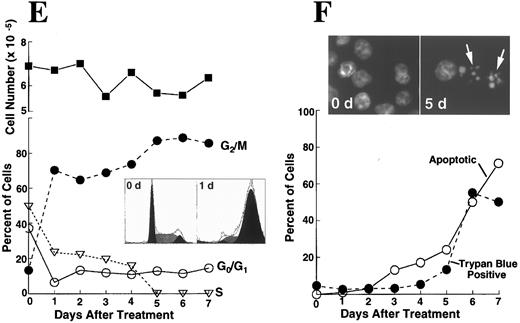
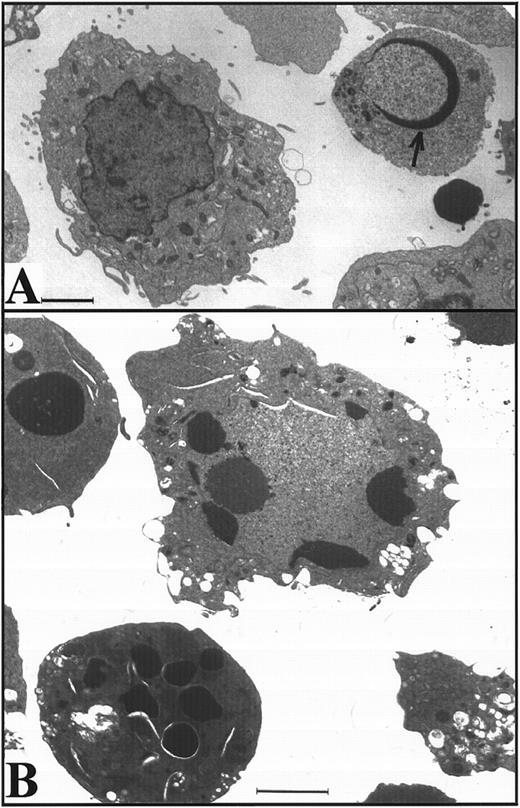
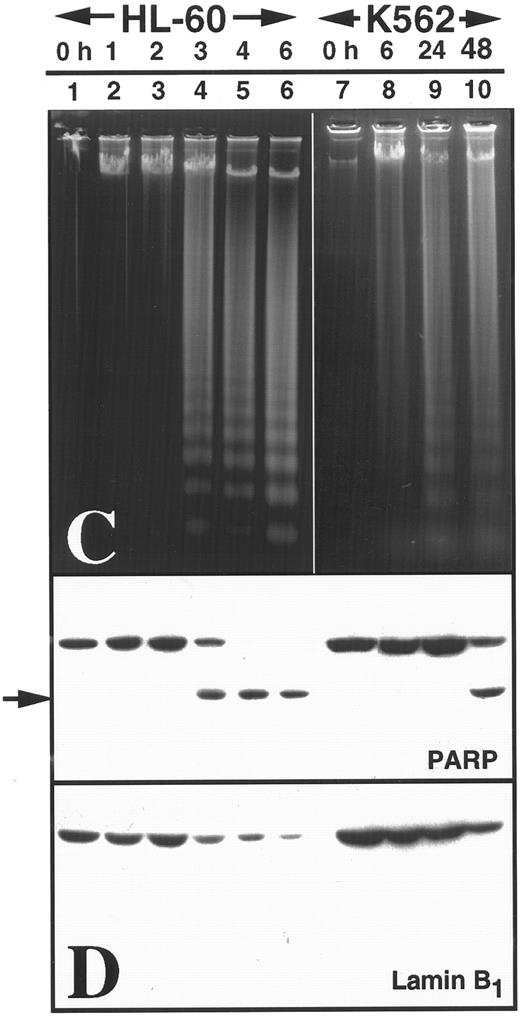
K562 cells treated with 17 μmol/L etoposide for 1 hour undergo delayed apoptosis. (A and B) Electron micrographs of K562 cells before (A) and 48 hours after (B) a 1-hour treatment with 17 μmol/L etoposide. Bar, 2 μm. (C and D) Samples of HL-60 cells (lanes 1, 2) and K562 cells (lanes 3 through 8) were treated with 17 μmol/L etoposide for 0 hours (lanes 1 and 3) or 1 hour (lanes 2 and 4 through 8) and incubated in drug-free medium thereafter. At the indicated times, samples were obtained and subjected to SDS-PAGE followed by immunoblotting with monoclonal anti-PARP (C) or agarose gel electrophoresis (D). (E) Total cell number of K562 cells following a 1-hour exposure to 17 μmol/L etoposide and cell-cycle distribution of the resulting nonapoptotic cells. (Inset) DNA histograms obtained before and 1 day after a 1-hour exposure of K562 cells to 17 μmol/L etoposide. (F) Percentage of K562 cells that exhibit apoptotic morphology and trypan blue uptake after a 1-hour exposure to 17 μmol/L etoposide. (Inset) Photograph of K562 cells stained with 1 μg/mL Hoechst 33342 at the indicated time after a 1-hour etoposide treatment. Arrows, cells with chromatin condensation and nuclear fragmentation indicative of apoptosis. Each panel is representative of at least three experiments.
K562 cells treated with 17 μmol/L etoposide for 1 hour undergo delayed apoptosis. (A and B) Electron micrographs of K562 cells before (A) and 48 hours after (B) a 1-hour treatment with 17 μmol/L etoposide. Bar, 2 μm. (C and D) Samples of HL-60 cells (lanes 1, 2) and K562 cells (lanes 3 through 8) were treated with 17 μmol/L etoposide for 0 hours (lanes 1 and 3) or 1 hour (lanes 2 and 4 through 8) and incubated in drug-free medium thereafter. At the indicated times, samples were obtained and subjected to SDS-PAGE followed by immunoblotting with monoclonal anti-PARP (C) or agarose gel electrophoresis (D). (E) Total cell number of K562 cells following a 1-hour exposure to 17 μmol/L etoposide and cell-cycle distribution of the resulting nonapoptotic cells. (Inset) DNA histograms obtained before and 1 day after a 1-hour exposure of K562 cells to 17 μmol/L etoposide. (F) Percentage of K562 cells that exhibit apoptotic morphology and trypan blue uptake after a 1-hour exposure to 17 μmol/L etoposide. (Inset) Photograph of K562 cells stained with 1 μg/mL Hoechst 33342 at the indicated time after a 1-hour etoposide treatment. Arrows, cells with chromatin condensation and nuclear fragmentation indicative of apoptosis. Each panel is representative of at least three experiments.
Reverse transcriptase-polymerase chain reaction (RT-PCR).Procaspase transcripts were analyzed as previously described.38 In brief, 1 μg of poly A+ RNA from K562 cells was reverse transcribed with a combination of random hexamer and oligo-dT primers using Superscript RT (Life Sciences, Gaithersburg, MD) under standard conditions.55 One twentieth of the cDNA product was amplified using AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, CT) and the sets of procaspase primers previously described38 or the following primers for procaspase-7 (Accession No. U37449): forward primer, TCAAGTGCTTCCGAAGCCTGG; reverse primer, TGACCCATTGCTTCTCAGCTAGAATGTAC. Except as indicated, reactions were performed on a Perkin Elmer PE-480 DNA thermal cycler for 30 cycles at 92°C denaturation, 60°C annealing, and 72°C extension, 45 seconds per step. Procaspase-5 cDNA was amplified for 35 cycles using an annealing temperature of 55°C. Control PCR reactions included (1) K562 cDNA template and human β-actin primers (forward-GTGGGGCGCCCCAGGCACCA, reverse-CTCCTTAATGTCACGCACGATTTC) to validate the cDNA, and (2) .05 μg K562 poly (A+ ) RNA template to confirm that the products observed were derived from cDNA rather than contaminating genomic DNA. After amplification, 10% of each PCR reaction was applied to a 3.5% composite agarose gel (2.5% NuSieve, 1% Seakem-GTG; FMC Bioproducts, Rockland, ME) containing 0.5 μg/mL ethidium bromide in 1X TBE buffer. After electrophoresis for 160 volt-hours, reaction products were visualized on a UV transilluminator (256 nm), photographed, and excised for sequencing.
Assessment of clonogenic survival.Colony-forming assays were performed as described.56 In brief, 1-mL aliquots containing 4 to 8 × 105 log phase cells were treated for 1 hour at 37°C with the indicated concentration of etoposide or diluent, sedimented at 200g for 10 minutes, and washed. Aliquots containing 1 to 5 × 103 cells were plated in gridded 35-mm plates in the medium of Pike and Robinson57 containing 0.3% agar and incubated for 10 to 14 days at 37°C before colonies containing ≥50 cells were counted. Untreated K562 and HL-60 cells had cloning efficiencies of 40% and 20%, respectively, under these conditions.
RESULTS
Comparison of etoposide action in HL-60 and K562 cells.The present studies focused on HL-60 cells, a human leukemia cell line that readily undergoes apoptosis in response to a variety of chemotherapeutic agents,31,32,34,47,58,59 and K562 cells, a bcr-abl–expressing human chronic myelogenous leukemia line that has been reported to resist induction of apoptosis by many of the same stimuli.32,43,44,46-49 These two cell lines have similar population doubling times (20 to 24 hours) and similar pretreatment cell cycle distributions (data not shown). In addition, both of these cell lines lack the oncoprotein p53.60 Initial comparison of these two cell lines indicated that K562 cells contain approximately twofold higher levels of topo IIα (Fig 1A) and form more protein-linked DNA single-strand breaks (indicative of covalent topo II-DNA cleavage complexes) during etoposide treatment (Fig 1B). When the two cell lines were treated with drug concentrations that resulted in approximately equal numbers of topo II-DNA complexes, drug dilution was followed by topo II–mediated resealing of the protein-linked DNA single-strand breaks, a process that occurred with indistinguishable first-order kinetics in the two cell lines (Fig 1C). In further experiments, soft agar cloning assays indicated that K562 cells were at least as sensitive as HL-60 cells when exposed to etoposide for 1 hour and washed to remove drug (Fig 1D). These last results, which were somewhat surprising in view of previous reports that K562 are resistant to etoposide-induced apoptosis, prompted us to examine the response of these two cell lines in greater detail.
Brief etoposide exposure triggers delayed apoptosis in K562 cells.In subsequent experiments, HL-60 and K562 cells were treated with 17 μmol/L etoposide for 1 hour, washed, incubated in drug-free medium for up to 7 days, and examined by various techniques. Within 6 hours after this treatment, HL-60 cells displayed multiple features of apoptosis, including morphological changes (not shown), PARP cleavage (Fig 2C), and internucleosomal DNA degradation (Fig 2D). In contrast, K562 cells displayed no morphological signs of apoptosis (Fig 2B), no evidence of PARP cleavage (Fig 2C), and no internucleosomal DNA degradation 2 days after etoposide treatment (Fig 2D). These results were consistent with previous descriptions of the resistance of K562 cells to apoptosis.
Although the preceding results suggested that K562 cells might be unaffected by the etoposide treatment, further examination of the cells showed an entirely different picture. Cell counts revealed that the etoposide-treated K562 cells ceased increasing in cell number after etoposide treatment (Fig 2E). Flow cytometry (inset, Fig 2E) combined with microscopic examination showed that the cells arrested in G2 and remained in this phase of the cell cycle for several days. Thereafter there was evidence that the cells began dying, as indicated by accumulation of increasing numbers of particles with subdiploid DNA content and by accumulation of cells that failed to exclude trypan blue (Fig 2F). Examination of the dying cells by light (inset, Fig 2F) and electron microscopy (not shown) showed that overt apoptotic morphological changes accompanied the loss of membrane integrity. Consistent with these observations, PARP cleavage and internucleosomal DNA degradation were detectable 4 to 5 days after etoposide treatment (Fig 2C and D, respectively). These observations suggest that etoposide-treated K562 cells are dying an apoptotic cell death.
Apoptosis is delayed in K562 cells even after treatment with higher doses of etoposide.The long latent phase and lack of synchrony after treatment of K562 cells with 17 μmol/L etoposide made it difficult to perform additional biochemical studies under these conditions. Previous studies have indicated that increasing the etoposide concentration to 68 μmol/L, a concentration that is readily achievable in the bone marrow transplant setting,61,62 results in more synchronous induction of apoptosis in HL-60 cells, with greater than 85% of cells exhibiting apoptotic morphology within 6 hours.32 When this same strategy was applied to K562 cells, a population of cells with condensed peripheral chromatin was visible at 24 hours (arrow, Fig 3A). At 48 hours, 80% of cells displayed increased electron density (indicating loss of H2O) and peripheral chromatin condensation or nuclear fragmentation when examined by light (not shown) or electron microscopy (Fig 3B). Oligonucleosomal DNA degradation (Fig 3C) as well as proteolytic cleavage of PARP and lamin B1 (the latter detected as loss of intact 67-kD polypeptide in Fig 3D) were readily evident 48 hours after addition of etoposide. Collectively, these observations again indicated that K562 cells demonstrate typical apoptotic changes, although their development was markedly delayed relative to HL-60 cells.
Apoptosis in K562 cells treated continuously with 68 μmol/L etoposide. (A and B) Morphology of K562 cells after 24 hours (A) and 48 hours (B) continuous exposure to etoposide. Arrow in (A), peripheral chromatin condensation in K562 cell after 24-hour etoposide exposure. Bar, 2 μm. (C and D) HL-60 cells treated with etoposide for 0 to 6 hours (lanes 1 through 6) and K562 cells treated with etoposide for 0 to 48 hours (lanes 7 through 10) were harvested for agarose gel electrophoresis (C) and SDS-PAGE followed by blotting with C-2-10 anti-PARP or polyclonal anti-lamin B1 antibodies (D). Arrow indicates 89-kD cleavage product of PARP.
Apoptosis in K562 cells treated continuously with 68 μmol/L etoposide. (A and B) Morphology of K562 cells after 24 hours (A) and 48 hours (B) continuous exposure to etoposide. Arrow in (A), peripheral chromatin condensation in K562 cell after 24-hour etoposide exposure. Bar, 2 μm. (C and D) HL-60 cells treated with etoposide for 0 to 6 hours (lanes 1 through 6) and K562 cells treated with etoposide for 0 to 48 hours (lanes 7 through 10) were harvested for agarose gel electrophoresis (C) and SDS-PAGE followed by blotting with C-2-10 anti-PARP or polyclonal anti-lamin B1 antibodies (D). Arrow indicates 89-kD cleavage product of PARP.
Expression and activation of caspases in HL-60 and K562 cells.Recent studies have indicated that caspase activation plays a critical role in initiation of the active phase of apoptosis.4,17,19-21,26,29,30 In addition, it has been suggested that the apoptotic regulator Bcl-2 exerts its anti-apoptotic effects upstream of events that result in activation of effector caspases.29,36 37 These observations prompted us to compare the expression and activation of caspase precursors in the HL-60 and K562 cell lines.
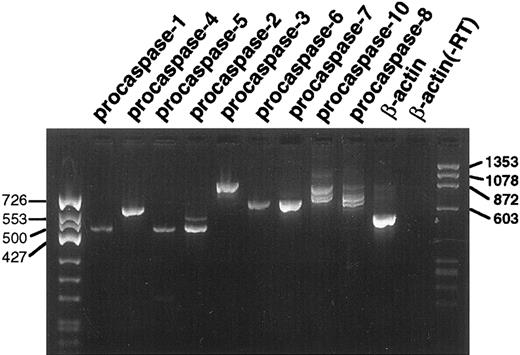
Previous experiments indicated that transcripts for all procaspases examined, including procaspases-1, -2, -3, -4, -5, -6, -7, -8, and -10 were detectable in untreated HL-60 cells.38 RT-PCR likewise demonstrated transcripts for the nine procaspases in untreated K562 cells (Fig 4). The identity of the observed gel bands was confirmed by DNA sequencing. This sequence analysis also showed that the multiple PCR products observed using primers for procaspases-8 and -10 were derived from alternatively spliced transcripts as described by Boldin et al.63
RT-PCR analysis of procaspase transcripts in untreated K562 cells. Each lane contains 10% of the product from the PCR reaction for the indicated target mRNA. The lane marked β-actin (−RT) is a control PCR reaction using poly A+ RNA as the template. The lack of a product in this lane confirms that the products observed in the other lanes are not derived from contaminating genomic DNA.
RT-PCR analysis of procaspase transcripts in untreated K562 cells. Each lane contains 10% of the product from the PCR reaction for the indicated target mRNA. The lane marked β-actin (−RT) is a control PCR reaction using poly A+ RNA as the template. The lack of a product in this lane confirms that the products observed in the other lanes are not derived from contaminating genomic DNA.
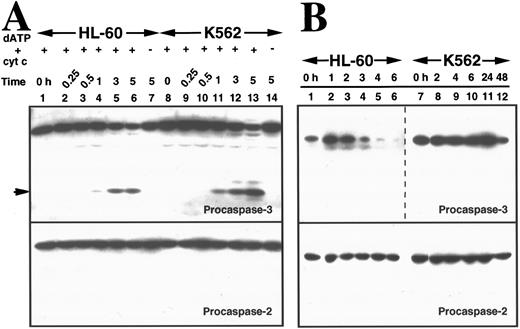
To compare the ability of cytosol from the two cell lines to support caspase activation, cytosol samples prepared from untreated aliquots of the two cell lines were incubated with cytochrome c and dATP. This treatment has recently been shown to promote cleavage of procaspase-3 to the processed, catalytically active enzyme in HeLa cytosol in vitro.35,36 Immunoblotting showed that procaspase-2 and procaspase-3 were readily detectable in cytosol from HL-60 and K562 cells (Fig 5A). Treatment with dATP and cytochrome c resulted in generation of active caspase-3, a critical downstream effector of apoptosis,26 35 in cytosol from both cell lines. This activation process was indicated by a decrease in levels of procaspase-3, detection of the 17-kD large subunit of the active enzyme with an antibody raised against that subunit, and appearance of peptidase activity that cleaved DEVD-AFC (Fig 5A and data not shown). The time course for activation of procaspase-3 in vitro was similar in cytosol from the two cell lines (Fig 5A, lanes 4 through 6 and 11 through 13, top panel). In contrast, procaspase-2 was not activated under these conditions in either cytosol (Fig 5A, bottom panel).
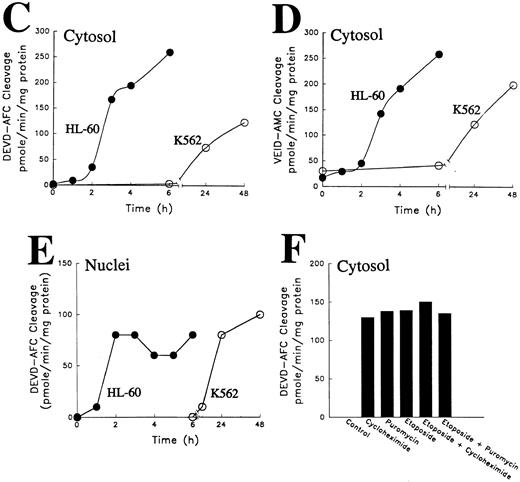
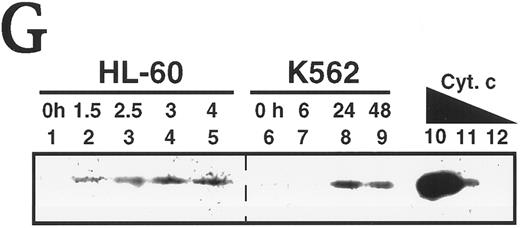
Activation of caspases in vitro and in intact cells. (A) Activation of caspases in vitro. Aliquots containing 50 μg of cytosolic protein from untreated HL-60 or K562 cells were incubated with or without dATP and cytochrome C for the indicated length of time, subjected to SDS-PAGE, transferred to PVDF membrane, and probed with the indicated antibody. Arrow indicates 17-kD large subunit of active caspase-3. (B) Activation of caspases in vivo. HL-60 and K562 cells were treated with 68 μmol/L etoposide for the indicated length of time. Samples containing total cellular polypeptides from 3 × 105 cells were subjected to SDS-PAGE followed by immunoblotting with antibodies to the indicated caspase precursor. The lower panel was derived from a single x-ray film, permitting direct comparison of polypeptide levels. Because procaspase-3 levels were roughly fivefold higher in K562 cells than in HL-60 cells, the exposure of the blot shown in lanes 7 through 12 was adjusted to give a signal comparable to that shown in lanes 1 through 6 of the top panel. (C and D) Detection of peptidase activity in cytosol from etoposide-treated HL-60 or K562 cells. Cytosol was simultaneously prepared from HL-60 (•) or K562 cells (○) treated with 68 μmol/L etoposide for the indicated length of time. Aliquots (50 μg protein) from the same set of extracts were incubated with DEVD-AFC (C) or VEID-AMC (D). The amount of fluorochrome released was determined by comparison to an AFC or AMC standard curve. (E) Detection of DEVD-AFC cleavage activity in nuclei prepared from HL-60 or K562 cells after treatment with 68 μmol/L etoposide for the indicated length of time. (F) Effect of protein synthesis inhibitors on etoposide-induced activation of DEVD-AFC cleavage activity. K562 cells were treated for 24 hours with 70 μmol/L cycloheximide or 100 μmol/L puromycin alone or in combination with 68 μmol/L etoposide. At the completion of the incubation, cytosolic extracts were prepared and assayed for DEVD-AFC cleavage activity. (G) Time course of appearance of cytochrome c in cytosol. Aliquots containing 50 μg of cytosol protein prepared from HL-60 or K562 cells treated with 68 μmol/L etoposide for the indicated length of time were subjected to SDS-PAGE and blotting with anti-cytochrome c. Lanes 10 through 12, 270, 27, and 2.7 ng of cytochrome c, respectively. In contrast to HL-60 cells, where cytochrome c release to cytosol is evident within 1.5 hours (lane 2 and Yang et al36 ), cytochrome c release to cytosol of K562 cells was not evident until 24 hours. Results are representative of three to six independent experiments.
Activation of caspases in vitro and in intact cells. (A) Activation of caspases in vitro. Aliquots containing 50 μg of cytosolic protein from untreated HL-60 or K562 cells were incubated with or without dATP and cytochrome C for the indicated length of time, subjected to SDS-PAGE, transferred to PVDF membrane, and probed with the indicated antibody. Arrow indicates 17-kD large subunit of active caspase-3. (B) Activation of caspases in vivo. HL-60 and K562 cells were treated with 68 μmol/L etoposide for the indicated length of time. Samples containing total cellular polypeptides from 3 × 105 cells were subjected to SDS-PAGE followed by immunoblotting with antibodies to the indicated caspase precursor. The lower panel was derived from a single x-ray film, permitting direct comparison of polypeptide levels. Because procaspase-3 levels were roughly fivefold higher in K562 cells than in HL-60 cells, the exposure of the blot shown in lanes 7 through 12 was adjusted to give a signal comparable to that shown in lanes 1 through 6 of the top panel. (C and D) Detection of peptidase activity in cytosol from etoposide-treated HL-60 or K562 cells. Cytosol was simultaneously prepared from HL-60 (•) or K562 cells (○) treated with 68 μmol/L etoposide for the indicated length of time. Aliquots (50 μg protein) from the same set of extracts were incubated with DEVD-AFC (C) or VEID-AMC (D). The amount of fluorochrome released was determined by comparison to an AFC or AMC standard curve. (E) Detection of DEVD-AFC cleavage activity in nuclei prepared from HL-60 or K562 cells after treatment with 68 μmol/L etoposide for the indicated length of time. (F) Effect of protein synthesis inhibitors on etoposide-induced activation of DEVD-AFC cleavage activity. K562 cells were treated for 24 hours with 70 μmol/L cycloheximide or 100 μmol/L puromycin alone or in combination with 68 μmol/L etoposide. At the completion of the incubation, cytosolic extracts were prepared and assayed for DEVD-AFC cleavage activity. (G) Time course of appearance of cytochrome c in cytosol. Aliquots containing 50 μg of cytosol protein prepared from HL-60 or K562 cells treated with 68 μmol/L etoposide for the indicated length of time were subjected to SDS-PAGE and blotting with anti-cytochrome c. Lanes 10 through 12, 270, 27, and 2.7 ng of cytochrome c, respectively. In contrast to HL-60 cells, where cytochrome c release to cytosol is evident within 1.5 hours (lane 2 and Yang et al36 ), cytochrome c release to cytosol of K562 cells was not evident until 24 hours. Results are representative of three to six independent experiments.
Delayed activation of caspases in etoposide-treated K562 cells.Despite the similar kinetics of caspase-3 activation observed in cytosol in vitro, caspase activation was markedly delayed in intact K562 cells compared with HL-60 cells. Three hours after addition of etoposide to HL-60 cells, the signal for full-length procaspase-3 diminished markedly (Fig 5B, top panel, lanes 4 through 6). In contrast, there was no detectable decrease in the signal for procaspase-3 in K562 cells during the first 24 hours after addition of etoposide, although the signal did eventually diminish somewhat (Fig 5B, top panel, lane 12).
This delay in activation of caspases was also evident when cytosol from etoposide-treated cells was directly assayed using fluorogenic substrates. Aliquots of cytosol prepared after various lengths of etoposide treatment were incubated with DEVD-AFC or VEID-AMC, which mimic the apoptotic cleavage sites of PARP23 and lamin A,24 respectively. Previous experiments have indicated that the VEID-AMC cleavage activity was at least 10-fold more sensitive than the DEVD-AFC cleavage activity to DEVD-fluoromethylketone (IC50 < 1 nmol/L v 10 nmol/L) and threefold more sensitive to YVAD-chloromethylketone (IC50 ∼ 3 μmol/L v 10 μmol/L),38 supporting the suggestion that DEVD-AFC and VEID-AMC cleavages are mediated by different caspases. When HL-60 and K562 cells were treated with etoposide, a marked increase in DEVD-AFC and VEID-AMC cleavage activities was observed in the cytosol fractions of both cell lines, but the kinetics of peptidase activation differed markedly (Fig 5C and D). DEVD-AFC and VEID-AMC cleavage activity increased 100-fold and 20-fold, respectively, within 3 hours of addition of etoposide to the HL-60 cells. In contrast, DEVD-AFC and VEID-AMC activity in cytosol remained at baseline levels 6 hours after addition of etoposide to K562 cells and increased more gradually over the ensuing 2 days.
The delay in appearance of peptidase activity was observed in nuclei as well as cytosol from K562 cells. When highly purified nuclei from etoposide-treated HL-60 cells were assayed, DEVD-AFC cleavage activity was low or undetectable in nuclei from control cells (0 hours) but increased to ∼100 pmol product released/min/mg protein 2 hours after addition of etoposide to the cells (Fig 5E). In contrast, DEVD-AFC cleavage activity was not detectable in K562 nuclei until 24 hours after addition of etoposide to the cells.
To assess the possibility that the delayed caspase activation in K562 cells reflected a need to synthesize new polypeptides, K562 cells were treated with 68 μmol/L etoposide in the absence or presence of cycloheximide or puromycin at concentrations that inhibited [35S]-methionine incorporation into protein by greater than 90%. Results of this experiment (Fig 5F) showed that neither cycloheximide nor puromycin inhibited the etoposide-induced increase in DEVD-AFC cleavage activity. On the contrary, treatment with cycloheximide alone or puromycin alone was associated with increased DEVD-AFC cleavage activity. These results argue against the possibility that the delay in caspase activation in K562 cells reflects a need for de novo protein synthesis. Instead, examination of the cytosol by immunoblotting showed that the delay in caspase activation reflected a delay in release of mitochondrial cytochrome c to the cytosol of K562 cells (Fig 5G).
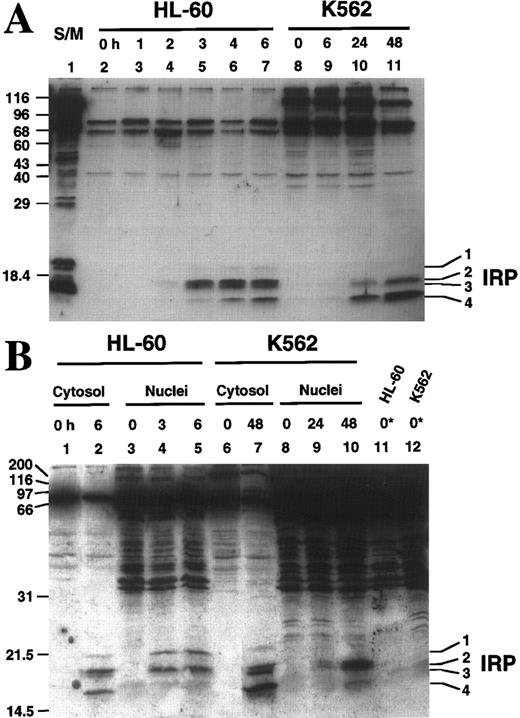
The spectrum of active caspases differs between HL-60 and K562 cells.To more completely delineate the spectrum of caspases activated during etoposide-induced apoptosis, aliquots of cytosol and nuclei were labeled with z-EK(bio)D-aomk, a recently described affinity labeling reagent designed to mimic the EVD motif preferred by the apoptotic proteases detected in Fig 5.38 Six hours after addition of etoposide, four discrete z-EK(bio)D-aomk-reactive bands termed IRP1 (Mr 21.5 kD), IRP2 (Mr 19.1 kD), IRP3 (Mr 18.4 kD), IRP4 (Mr 16.6 kD) were detected in HL-60 cytosol (Fig 6A, lane 7). None of these bands were detected in cytosol from control cells (Fig 6A, lane 2). Comparison to affinity-labeled recombinant caspases suggested that IRP2 and IRP4 correspond to active forms of caspase-3, whereas IRP3 and possibly IRP1 correspond to active forms of caspase-6.38 As previously reported, each active caspase appeared at a characteristic time after etoposide addition (Fig 6A, lanes 4 through 7). IRP2 and IRP3 were faintly visible 2 hours after addition of etoposide, increased dramatically by 3 hours and remained relatively constant thereafter, a pattern that parallels the activation of DEVD-AFC and VEID-AMC cleavage activities (Fig 5). In contrast, IRP4 was first evident at 3 hours and increased progressively during the remainder of the incubation; and IRP1 did not become detectable until 4 hours after the start of etoposide treatment.
Affinity labeling of etoposide-treated HL-60 and K562 cytosol and nuclei. (A) Aliquots containing 37 μg of cytosolic protein prepared from HL-60 (lanes 2 through 7) or K562 (lanes 8 through 11) cells treated with 68 μmol/L etoposide for the indicated length of time were reacted with z-EK(bio)D-aomk, subjected to SDS-PAGE, and reacted with peroxidase-coupled streptavidin. Additional experiments (see B) indicate that bands detected in 0-hour samples bind streptavidin in the absence of reaction with z-EK(bio)D-aomk. Lane 1, apoptosis-inducing S/M extract prepared from DU249 chicken hepatoma cells.24 53 (B) z-EK(bio)D-aomk labeling of cytosol and nuclei from HL-60 cells and K562 cells treated with 68 μmol/L etoposide for the indicated length of time. Samples containing cytosol or nuclei from 3 × 106 cells were incubated with 1 μmol/L z-EK(bio)D-aomk for 1 hour, then subjected to SDS-PAGE and blotting as described in the legend for (A). Lanes marked 0* contain nuclei that were subjected to SDS-PAGE without z-EK(bio)D-aomk treatment, thereby revealing the streptavidin-binding polypeptides that are normal constituents of these nuclei.
Affinity labeling of etoposide-treated HL-60 and K562 cytosol and nuclei. (A) Aliquots containing 37 μg of cytosolic protein prepared from HL-60 (lanes 2 through 7) or K562 (lanes 8 through 11) cells treated with 68 μmol/L etoposide for the indicated length of time were reacted with z-EK(bio)D-aomk, subjected to SDS-PAGE, and reacted with peroxidase-coupled streptavidin. Additional experiments (see B) indicate that bands detected in 0-hour samples bind streptavidin in the absence of reaction with z-EK(bio)D-aomk. Lane 1, apoptosis-inducing S/M extract prepared from DU249 chicken hepatoma cells.24 53 (B) z-EK(bio)D-aomk labeling of cytosol and nuclei from HL-60 cells and K562 cells treated with 68 μmol/L etoposide for the indicated length of time. Samples containing cytosol or nuclei from 3 × 106 cells were incubated with 1 μmol/L z-EK(bio)D-aomk for 1 hour, then subjected to SDS-PAGE and blotting as described in the legend for (A). Lanes marked 0* contain nuclei that were subjected to SDS-PAGE without z-EK(bio)D-aomk treatment, thereby revealing the streptavidin-binding polypeptides that are normal constituents of these nuclei.
When cytosol from K562 cells was labeled using the same procedure, a different pattern emerged. As was the case with HL-60 cells, there was no detectable labeling of active caspase subunits in cytosol from control cells (Fig 6A, lane 8). After 6 hours of continuous etoposide treatment, labeled caspases were difficult to discern (Fig 6A, lane 9), although faint labeling of IRP4 was evident. At 24 hours, IRP4 labeling increased dramatically, and IRP2 was detectable (Fig 6A, lane 10). Although these polypeptides comigrated with the polypeptides labeled in extracts from apoptotic HL-60 cells, the relative intensities of the labeled species differed substantially between the two cell lines (see Fig 6A, lanes 7 and 11).
Labeling of nuclei with z-EK(bio)D-aomk (Fig 6B) likewise revealed differences between HL-60 and K562 cells after etoposide treatment. Nuclei that were not treated with z-EK(bio)D-aomk contained a number of streptavidin-reactive bands (Fig 6B, lane 11) that did not change during the course of etoposide treatment (Fig 6B, lanes 3 through 5; and data not shown). These included an Mr ∼17,000 polypeptide that migrated just below the expected position of IRP3 . Treatment of HL-60 cells with etoposide resulted in the appearance of additional z-EK(bio)D-aomk-reactive bands that comigrated with IRP1 , IRP2 , and IRP4 (Fig 6B, lanes 3 through 5). Treatment of K562 cells likewise resulted in the appearance of three nuclear polypeptides that labeled with z-EK(bio)D-aomk. However, these bands did not appear until 24 hours after addition of etoposide, and they comigrated with IRP2 , IRP3 , and IRP4 rather than IRP1 , IRP2 , and IRP4 (see Fig 6B, lanes 5 and 10).
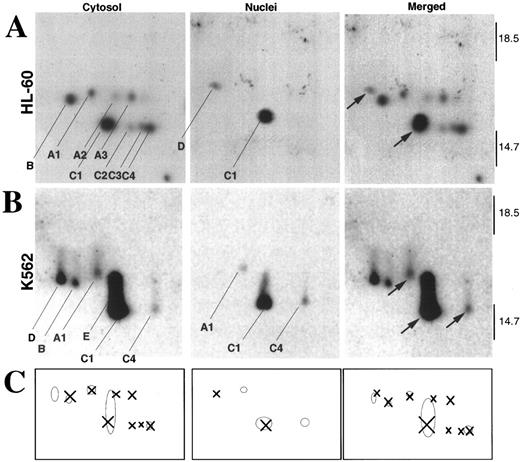
Differences in both the pattern and subcellular localization of active caspases were even more striking when the z-EK(bio)D-aomk–labeled samples from the two cell lines were analyzed by two-dimensional isoelectric focusing/SDS-PAGE (Fig 7). HL-60 cells contained a total of nine active caspase species (indicated by X in Fig 7C). In contrast, K562 cells contained six active caspase species that could be resolved in this gel system (indicated by O in Fig 7C). Thus, several of the active caspase species detected in HL-60 cells were not detected in K562 cell fractions. Strikingly, when corresponding species were detected, they were not always found in the same subcellular compartments in the two cell lines. For example, a new species (spot D) that was not resolved in our previous study38 was located in nuclei of HL-60 cells (Fig 7A, nuclei) and in the cytosol of K562 cells (Fig 7B, cytosol). Conversely, spots A1 and C4, which were previously shown to comigrate with isoforms of active caspase-3,38 were detected in K562 nuclei but not in HL-60 nuclei. Finally, K562 cytosol also contained an abundant active caspase (labeled E in Fig 7B) that ran with an isoelectric point similar to spot C1 (an active form of caspase-338 ) but was larger. These results clearly indicate that the populations of active caspase species detected in the two cell lines during etoposide-induced apoptosis were distinguishable.
Analysis of z-EK(bio)D-aomk-labeled caspases by two-dimensional gel electrophoresis. (A) Analysis of active caspases in HL-60 cytosol (left) and nuclei (middle). The spots are indexed as recently described.38 In these gels, the pH decreases from left to right. This analysis showed a novel active species (D) in HL-60 nuclei. The merged picture (right) was obtained by co-electophoresing mixtures of, respectively, HL-60 cytoplasm + active caspase-2 and HL-60 nuclei + active caspase-2 (data not shown). Caspase-2, which was previously shown not to comigrate with any of the caspases detected in HL-60 cytosol,38 provided a reference point for alignment of the cytosolic and nuclear images. Arrows in the merged image point to species observed in the nuclear fractions. (B) Analysis of active caspases in K562 cytosol (left) and nuclei (middle). Images were merged (right) as described for (A). (C) Comparison of the pattern of active caspases in cytosol and nuclei of HL-60 and K562 cells. Crosses correspond to the exact positions of the active species in HL-60, and circles correspond to the exact positions of the active species in K562.
Analysis of z-EK(bio)D-aomk-labeled caspases by two-dimensional gel electrophoresis. (A) Analysis of active caspases in HL-60 cytosol (left) and nuclei (middle). The spots are indexed as recently described.38 In these gels, the pH decreases from left to right. This analysis showed a novel active species (D) in HL-60 nuclei. The merged picture (right) was obtained by co-electophoresing mixtures of, respectively, HL-60 cytoplasm + active caspase-2 and HL-60 nuclei + active caspase-2 (data not shown). Caspase-2, which was previously shown not to comigrate with any of the caspases detected in HL-60 cytosol,38 provided a reference point for alignment of the cytosolic and nuclear images. Arrows in the merged image point to species observed in the nuclear fractions. (B) Analysis of active caspases in K562 cytosol (left) and nuclei (middle). Images were merged (right) as described for (A). (C) Comparison of the pattern of active caspases in cytosol and nuclei of HL-60 and K562 cells. Crosses correspond to the exact positions of the active species in HL-60, and circles correspond to the exact positions of the active species in K562.
DISCUSSION
Previous studies have suggested that expression of the bcr/abl kinase renders chronic myelogenous leukemia cell lines such as K562 cells resistant to the induction of apoptosis by a variety of treatments.32 43-49 These previous studies generally involved the assessment of cell survival 4 to 24 hours after treatment with an inducing stimulus. In the present study, the effect of the topo II poison etoposide on HL-60 and K562 cells was assessed by various techniques over a period of up to 14 days. These studies have led to several novel observations. First, K562 cells that appear normal 24 hours after brief etoposide treatment subsequently die an apoptotic cell death. Second, activation of apoptotic proteases in K562 cells is delayed, suggesting that the anti-apoptotic effect of bcr/abl is a result of action upstream from the effector caspases. Third, once the execution phase of apoptosis occurs in K562 cells, the pattern of active proteases differs from that observed in HL-60 cells even though the same inducing stimulus has been applied. Each of these observations has potential implications for understanding the process of programmed cell death.
It is important to emphasize that the present observations fully confirm previous reports that K562 cells fail to undergo apoptosis during the first 4 to 24 hours after treatment with a variety of stimuli (Fig 2). However, additional experiments showed that the resistance of K562 cells to etoposide-induced apoptosis reported by this laboratory32 and others was not observed when drug sensitivity was assessed by colony-forming assays (Fig 1C). There were two potential explanations for this discrepancy. On the one hand, it was possible that etoposide treatment was causing prolonged cell-cycle arrest in K562 cells without producing subsequent loss of viability. This would result in the failure of K562 cells to form colonies even though they resisted the killing effects of the drug. On the other hand, it was possible that etoposide was killing the K562 cells, but the terminal events were not occurring during the time period previously examined. The present results clearly distinguish between these explanations. Flow cytometry and light microscopy showed that etoposide treatment induced prolonged arrest in G2 (Fig 2E) followed by loss of membrane integrity (Fig 2F). Examination of the dying cells showed typical apoptotic changes in morphology (inset, Fig 2F) as well as PARP cleavage and internucleosomal DNA degradation (Fig 2C and D). Aside from the G2 arrest, all of these changes were undetectable at time points up to 3 days in this cell line.
These results were not limited to the use of etoposide as an apoptosis-inducing stimulus. Instead, K562 cells were observed to undergo apoptotic cell death, albeit delayed, after treatment with other stimuli, including protein synthesis inhibitors (Fig 5F), staurosporine, cisplatin, and ionizing radiation (S.H.K., unpublished observations). The observation of delayed cell death in K562 cells after these various treatments raises the possibility that rapid, high throughput measures currently used to assess chemotherapeutic sensitivity in many drug-screening programs might be underestimating the true effectiveness of agents in killing the K562 cell line and other cell lines that express the bcr/abl kinase. This could potentially result in failure to identify compounds that have unique or promising activity against bcr/abl-dependent chronic myleogenous leukemia cells. To the extent that cell lines derived from other malignancies also show a delay in induction of apoptosis, the same screening methods might underestimate the effectiveness of agents in those lines as well.
Although previous observations have made it clear that the ability of K562 cells to undergo apoptosis in response to various stimuli can be restored by treatment with antisense oligonucleotides that inhibit synthesis of the bcr/abl kinase,44,45,49,50 the apoptotic step that is inhibited by the bcr/abl kinase has remained unclear. Our results suggest that bcr/abl delays apoptosis rather than preventing it altogether. A similar phenomenon was recently reported in Bcl-2–expressing cells treated with a variety of different agents.64-66 Furthermore, our results suggest that bcr/abl kinase does not appear to be activating a soluble caspase inhibitor in K562 cells. Caspase activation induced by dATP and cytochrome c follows a similar time course in aliquots of HL-60 and K562 cytosol (Fig 5A). Additional experiments have shown that cytosol from untreated K562 cells cannot inhibit active caspases in cytosol from apoptotic HL-60 cells when the two are mixed (T.J.K. and S.H.K., unpublished observations). Collectively, these observations rule out the possibility that the bcr/abl kinase is acting downstream of caspase-3 activation. Instead, bcr/abl appears to act upstream of events that result in cytochrome c release and caspase activation (Fig 5), presumably by slowing the poorly understood biochemical events that occur between the generation of topo II–mediated DNA damage and the execution phase of programmed cell death. Consistent with this conclusion, it has been observed that K562 cells readily undergo an apoptotic cell death when treated with cytotoxic lymphocytes,67,68 which bypass the signaling aspects of the death cascade and directly activate the effector caspases.69
Once the execution phase of apoptosis was triggered in HL-60 and K562 cells, there were surprising differences in the caspase species that were activated. The observation that a different spectrum of active species was detected in the cytoplasm of apoptotic HL-60 and K562 cells (Figs 6 and 7) agrees with results of Faleiro et al,39 who observed a similar difference when comparing active caspases in cytosol of three etoposide-treated lymphoid cell lines. In our study, there were also differences in the subcellular distributions of the active caspases. Two-dimensional gel electrophoresis (Fig 7) showed that K562 nuclei contained three species previously shown to comigrate with three active forms of caspase-3,38 whereas HL-60 nuclei contained one active form of caspase-3 and another caspase species (D) that was not detected in the previous study of HL-60 cytosolic caspases.38 These observations, which indicate that preferences of active caspase molecules for various subcellular compartments can vary between different cell types, raise questions about the mechanisms of subcellular targeting of these important enzymes. Our results suggest that caspase targeting is unlikely to be determined solely by sequences of the caspases themselves (unless such signals are “read” differently in different cell types), but instead might reflect the presence of distinct targeting factors in different cell lines. Further study of the individual caspase species and the factors that control their subcellular distribution is required to determine the biochemical basis for these differences in caspase localization.
ACKNOWLEDGMENT
We thank Dr Ivan Lieberburg for making this collaboration possible; Michael Power for DNA sequence confirmation of clones and constructs used in this work; Richard J. Jones, Pierre Henkart, and Guy Poirier for stimulating conversations; and Deb Strauss for secretarial assistance.
Supported in part by Public Health Service Grant No. CA69008 (S.H.K. and W.C.E.), a studentship from Programa Gulbenkian de Doutoramento em Biologia e Medicina (L.M.M.), and a Leukemia Society of America Scholar Award (S.H.K.).
L.M.M., P.W.M., and T.J.K. contributed equally to this work.
Address reprint requests to Scott H. Kaufmann, MD, PhD, Division of Oncology Research, Guggenheim 1342C, Mayo Clinic, 200 First St, SW, Rochester, MN 55905.