Abstract
Many diseases might be treated by gene therapy targeted to the hematopoietic system, but low rates of gene transfer achieved in humans and large animals have limited the application of this technique. We have developed a competitive hematopoietic repopulation assay in baboons to evaluate methods for improving gene transfer and have used this method to compare gene transfer rates for retroviral vectors having an envelope protein (pseudotype) from amphotropic murine retrovirus with similar vectors having an envelope protein derived from gibbon ape leukemia virus (GALV). We hypothesized that vectors with a GALV pseudotype might perform better based on our previous work with cultured human hematopoietic cells. CD34+ marrow cells from each of four untreated baboons were divided into two equal portions that were cocultivated for 48 hours with packaging cells producing equivalent titers of either amphotropic or GALV pseudotyped vectors containing the neo gene. The vectors contained small sequence differences to allow differentiation of cells genetically marked by the different vectors. Nonadherent and adherent cells from the cultures were infused into animals after they received a myeloablative dose of total body irradiation. Polymerase chain reaction (PCR) analysis for neo gene-specific sequences in colony-forming unit–granulocyte-macrophage from cell populations used for transplant showed gene transfer rates of 2.7%, 7.1%, <15%, and 3.9% with the amphotropic vectors and 7.1%, 11.3%, <15%, and 26.4% with the GALV pseudotyped vector. PCR analysis of peripheral blood and marrow cells after engraftment showed the neo gene to be present in all four animals analyzed at levels between 0.1% and 5%. Overall gene transfer efficiency was higher with the GALVpseudotyped vector than with the amphotropic vectors. Southern blot analysis in one animal confirmed a gene transfer efficiency of between 1% and 5%. The higher gene transfer efficiency with the GALV-pseudotyped vector correlated with higher levels of GALV receptor RNA compared with the amphotropic receptor in CD34+ hematopoietic cells. These results show that GALV-pseudotyped vectors are capable of transducing baboon marrow repopulating cells and may allow more efficient gene transfer rates for human gene therapy directed at hematopoietic cells. In addition, our data show considerable differences in gene transfer efficiency between individual baboons, suggesting that a competitive repopulation assay will be critical for evaluation of methods designed to improve gene transfer into hematopoietic stem cells.
HEMATOPOIETIC repopulating cells (stem cells) are appealing targets for gene transfer. Although efficient and reproducible gene transfer has been shown in hematopoietic progenitor cells in in vitro studies for multiple species and also in in vivo studies in mice, retroviral gene transfer in hematopoietic repopulating cells in large animal models and in human gene transfer trials has remained low.1-5 Several reasons may account for this low gene transfer efficiency. Commonly used vectors based on murine leukemia viruses require cell division to integrate into the host genome, and primitive hematopoietic stem cells are quiescent and rarely divide in culture.6,7 Additionally, retroviral vectors can be made with different viral envelope (Env) proteins that recognize different receptors for cell entry, and low levels of receptors may impede vector entry into stem cells.8
Several studies have reported a correlation between receptor RNA expression and retroviral gene transfer efficiency.9-11 Hematopoietic cells express the amphotropic retrovirus receptor Ram1 only at low levels, which may explain the low gene transfer efficiency observed in vivo using amphotropic vectors.10,11 Most hematopoietic cells analyzed express higher levels of the gibbon ape leukemia virus (GALV) receptor Glvr1,10,12 and we have recently reported improved gene transfer in vitro into human progenitor cells using a retroviral vector that carried the GALV envelope protein.13 Based on these observations, we investigated whether GALV-pseudotyped vectors would be able to genetically mark baboon hematopoietic repopulating cells more efficiently than amphotropic vectors. To date, studies directly comparing gene transfer into marrow repopulating cells by amphotropic and GALV-pseudotyped vectors have not been reported.
Our results show that GALV-pseudotyped vectors were able to transduce baboon hematopoietic repopulating cells. Overall, gene transfer efficiency was higher with the GALV-pseudotyped vector than with the amphotropic vectors, which correlated with the higher level of GALV receptor RNA compared to amphotropic receptor RNA in baboon hematopoietic CD34+ cells. Because of interanimal variation, our findings also emphasize the importance of using a competitive repopulation assay when studying methods to improve gene transfer into hematopoietic stem cells.
MATERIALS AND METHODS
Animals. Four healthy juvenile baboons (Papio cynocephalus cynocephalus or Papio cynocephalus anubis ) were housed at the University of Washington Regional Primate Research Center under conditions approved by the American Association for Accreditation of Laboratory Animal Care. Studies were conducted under protocols approved by the Institutional Review Board and Animal Care and Use Committee. All animals were provided with water, biscuits, and fruit ad libitum. All procedures and blood draws were performed after animals had been anesthetized with a combination of ketamine-HCL (Aveco, Fort Dodge, IA) and Xylazine (Haver, Shawnee, KS). Before transplantation, all animals had central venous catheters placed for use with a tether system. Animals were treated with prophylactic broad spectrum antibiotics until they engrafted and were transfusion independent. All animals were treated with a single dose of 1,020 cGy total body irradiation (TBI) administered from opposing 60Cobalt sources at 7 cGy per minute as preparation for transplant.14
Retrovirus vectors. The LN, LNL6, and LNX vectors carry the bacterial neomycin phosphotransferase gene (neo gene) conveying G418 resistance and are identical with the exception of different length sequences between the neo gene and the 3′ vector long terminal repeat (LTR). This makes it possible to use a single pair of primers to amplify different length sequences with polymerase chain reaction (PCR) that can distinguish cells genetically marked by the different vectors. Both LN and LNL6 vectors have been described.15 LNX was made from LNSX15 by blunt ligation of BstI and StuI to remove the SV40 promoter. The packaging cell lines PA317 and PG13 have been described.16 17 Vector titers for all three vectors (LN, LNX, and LNL6) were 5 × 105 colony-forming unit (CFU)/mL on D17 canine and HeLa human cells. Individual producer cell clones with equal titers were selected for this study.
Gene transfer into baboon CD34 enriched cells. Bone marrow (BM) buffy coat cells from untreated baboons were labeled with IgM monoclonal antibody (MoAb) 12-8 (CD34) at 4°C for 30 minutes, washed, bound to rat monoclonal antimouse IgM microbeads (Miltenyi Biotec, Auburn, CA ) for 30 minutes at 4°C, washed, and then separated using an immunomagnetic column technique (Miltenyi Biotec) according to manufacturer's instructions. The purity of the CD34-enriched cells was between 57% and 91%.
Equal numbers of enriched CD34+ cells were placed into 75 cm2 canted-neck flasks (Corning, Corning, NY) that had been seeded 6 to 8 hours earlier with 5 × 106 nonirradiated packaging cells producing either LNL6 (PA317), LNX (PA317), or LN (PG13). Cells were cultured for 48 hours in 30 mL Iscove's modified Dulbecco's medium (GIBCO-BRL, Gaithersburg, MD) supplemented with 12.5% horse serum (GIBCO-BRL), 12.5% fetal calf serum (FCS) (GIBCO-BRL), 10-6 mol/L hydrocortisone (Sigma Chemical Co, St Louis, MO), 0.1 mmol/L 2-mercaptoethanol (Sigma), 1% glutamine, and penicillin/streptomycin (GIBCO-BRL) in the presence of 50 ng/mL of recombinant human interleukin-3 (IL-3), IL-6, and stem cell factor (SCF ) and 4 μg/mL protamine sulfate. Supernatants collected at the end of cocultivation had vector titers of 3 to 4 × 105 for all three vectors (LN, LNX, and LNL6) as determined using HeLa and D17 cells as targets for infection. Nonadherent and adherent cells collected from the cultures were infused into the same animals from which the cells were obtained after the animals had received a myeloablative dose of 1,020 cGy TBI. Both PA317 and PG13 packaging cells are rapidly lysed by human and baboon serum, thus preventing persistence of the packaging cells in animals.
Colony-forming unit–granulocyte-macrophage (CFU-GM) assays and PCR. CD34-enriched cells were cultured in a double layer agar culture system as previously described.18 Briefly, isolated cells were cultured in alpha minimal essential medium supplemented with 25% fetal bovine serum (FBS; Hyclone, Logan, UT), 0.1% bovine serum albumin (BSA; fraction V; Sigma), 0.3% (wt/vol) agar (DIFCO, Detroit, MI) overlaid on medium with 0.5% agar (wt/vol) containing 100 ng/mL recombinant human SCF, IL-3, IL-6, granulocyte-macrophage colony-stimulating factor (GM-CSF ), G-CSF, and 4 U/mL Epo (provided by Dr Ian McNiece, Amgen, Thousand Oaks, CA). Cells were plated at 1,000 to 5,000 cells per dish. Cultures were incubated at 37°C in 5% O2 , 5% CO2 , 90% air in a humidified incubator. All cultures were performed in triplicate. Colonies were enumerated at day 14 of culture.
PCR analysis of CFU-GM colonies. CFU-GM colonies were picked after 10 to 14 days and DNA was prepared from pools of 2, 5, and 15 colonies. Colonies were individually plucked from agar cultures and pooled in 90 μL sterile water, heated to 100°C for 10 minutes, cooled to 4°C for 10 minutes, treated with Proteinase K at 55°C for 2 hours; 60 μL supernatant was removed and 20 μL supernatant was lyophilized and resuspended in 30 μL PCR reaction buffer. PCR was used to amplify a 500-bp sequence of the neo gene using primers designated CANEO2 5′ ATG CTC TTC GTC CAG ATC AT 3′19 and NEO1 5′ CAA GAT GGA TTG CAC GCA GG 3′.20 Negative controls were run without DNA (water) or with DNA extracted from normal baboon CFU-GM. As positive controls, we used samples containing 2 μg DNA purified from K562/LXSN, a K562 subclone marked by the retroviral vector LXSN,15 DNA from a single K562/LXSN colony grown in agar and DNA from a single K562/LXSN colony pooled with 14 CFU-GM grown from normal baboon marrow. PCR reactions were scored + or −. Samples with −PCR for the neo gene were assayed by PCR for the β-2 microglobulin sequence to confirm the integrity of the DNA. The estimated frequency of gene transfer or the probability that any given colony was genetically marked P, was determined as P = 1 − (1 − r/n)1/m, where r = the number of PCR positive pools, n = the number of pools tested, and m = the number of colonies per pool. In the first three animals, only pools with 15 colonies were tested allowing the determination of gene transfer efficiency in pooled CFU-GM colonies up to an estimated efficiency of 15%. In the most recent animal, M94142, smaller pools with two and five colonies, as well as pools of 15, were tested allowing a more accurate determination of gene transfer efficiency.
PCR analysis posttransplant. Genomic DNA was prepared using DNAzol (MRC Inc, Cincinnati, OH). For the amplification of the LN and LNL6 vectors, the following oligonucleotide primers were used: LN/LNL6 2229 5′ TGC TTG CCG AAT ATC ATG GTG G 3′ and LN/LNL6 3210 5′ TTC ATT CCC CCC TTT TTC TGG 3′, which yielded a 982-bp product for LNL6 and a 301-bp PCR product for the LN vector. The neo gene was amplified with neo 350 5′ AAG AGA CAG GAT GAA GGA TCG 3′ and neo 1150 5′ CAG AAG AAC TCG TCA AGA 3′, which resulted in an 800-bp product and neo 450 5′ ACA AAC AGA CAA TCG GCT GCT 3′ and neo 1027 5′ GCC AAC GCT ATG TCC TGA TA 3′ to make a hybridization probe. One hundred (for samples with signal intensity between 1% and 10%) or 500 ng (for samples with signal intensities between 0.1% and 1%) of genomic DNA were amplified with either LN/LNL6 2229 and LN/LNL6 3210 or with neo 350 and neo 1150 using 2.5 U Taq polymerase (Perkin Elmer Cetus, Norwalk, CT). Conditions were optimized to ensure linear amplification in the range of the intensity of the positive PCR samples: denaturation at 95°C, followed by 31 cycles of 62°C annealing (1 minute), and 95°C denaturation (1 minute) with a final extension at 72°C for 7 minutes. For the detection of LN and LNL6, reaction products were separated on 1.5% agarose gels and transferred onto zeta-probe blotting membrane (Bio-Rad Laboratories, Hercules, CA). Hybridization was performed overnight at 50°C using a 32P-labeled 577-bp internal neo gene fragment amplified with primers neo 450 and neo 1027 as described.3 For the detection of LN and LNX, PCR was run in the presence of 10 μCi/mL 32P deoxycytidine triphosphate, and PCR products were separated on a 10% polyacrylamide gel. To correct for differences in actual DNA amount between samples, all samples were analyzed for the β-actin gene using 100 ng of genomic DNA and the following primers: actin-1 5′ TCC TGT GGC ATC CAC GAA ACT 3′ and actin-2 5′ GAA GCA TTT GCG GTG GAC GAT 3′. Phosphorimage analysis was used to identify the linear range of β-actin amplification. Conditions for β-actin PCR were the same as for the neo gene, except that only 16 cycles were run.
Retrovirus receptor analysis. Reverse transcriptase (RT)-PCR for receptor amplification has been described.10 Briefly, RNA samples from hematopoietic cells were reverse transcribed using a GeneAmp kit from Perkin Elmer, Branchburg, NJ, and subjected to PCR analysis using forward primer 5′ TTC CAG TTC CTG CAG GTC CT 3′ and reverse primer 5′ TCC TTC CCC ATG GTC TGG AT 3′. The reverse primer is identical to both Glvr1 and Ram1, while the forward primer differs by one base from each of the Glvr1 and Ram1 cDNAs. PCR conditions included initial denaturation at 94°C for 1 minute; five cycles of 94°C for 15 seconds, 55°C for 15 seconds, and 72°C for 15 seconds; 30 cycles of 94°C for 15 seconds and 60°C for 30 seconds; and a final extension of 60°C for 15 minutes. Reaction products were precipitated with ethanol and end-labeled with 32P using T4 Polynucleotide kinase (Promega, Madison, WI). The labeled products were digested with HaeIII and separated on a 10% nondenaturing acrylamide gel at 200 V 50 W for 6 to 8 hours.
Northern blots. RNA from hematopoietic cells and cell lines was isolated using RNAzol (MRC Inc, Cincinnati, OH) according to the manufacturer's instructions. Northern blots were hybridized with the human cDNAs for Glvr1 and Ram1. An 800-bp Kpn I-Xba I fragment was used for Glvr1 and a 488-bp Dra I-Xho I fragment was used for Ram1.
Assay for helper virus. After transplantation, peripheral blood (PB) mononuclear cell DNA was screened by PCR for the presence of amphotropic and GALV envelope sequences. Positive control log dilutions of DNA from PG13/LN and PA317/LNL6 packaging cells into normal baboon DNA were run concurrently. The primers used to amplify amphotropic envelope gene have been described.3 The sequence of the primers used to amplify the GALV envelope gene were as follows: GLVEnv2 5′ AAC CTG ATG TAT GTG CCT TGG 3′; GLVEnv3 5′ TTT CCC GTT ATC CAG TCC TTG G 3′. PCR conditions were as described above for the neo gene. PB and serum from the longest surviving animal was also assayed by S+ L− assay as described,21 22 and no helper virus was detected.
RESULTS
Engraftment and gene transfer efficiency. Four baboons engrafted after autologous transplantation of enriched CD34 marrow cells genetically marked by cocultivation with PA317/LNL6 or PG13/LN vector producing cells for three animals and PA317/LNX or PG13/LN for one animal (Table 1). Gene transfer efficiency in CFU-GM from animals J94090, M94264, and M94142 was significantly higher with LN (PG13) (P = .01) and for one animal, we estimated the gene transfer efficiency at >15% for both LN and LNL6 marked cells, given that all 12 pools of colonies tested (15 colonies/pool) were neo-positive by PCR, indicating at least one genetically marked colony was present in each pool (see Materials and Methods). All four animals engrafted with granulocyte counts >500/μL on days 32, 28, 21, and 14, respectively. Two animals died of infectious complications 20 and 7 weeks after transplantation and two animals are alive and well >7 and >50 weeks after transplantation. All animals were followed for the presence of the neo gene in unseparated PB and marrow cells. Neo positive cells were detected at levels between 0.1% and 1% in animals J94090 and M94264, and between 1% and 5% of PB cells and bone marrow in animals F93449 and M94142 (Figs 1 4). Southern blot analysis of DNA samples from PB and BM from animal F93449 showed faint bands with the expected sizes for LN and LNL6 at levels between 1% and 5% confirming the PCR results (Fig 5). We have studied the longest surviving animal, J94090, in greater detail to determine if the neo gene was present in mature and immature myeloid cells, as well as in lymphoid cells (Fig 1). Granulocytes and CD2+ T lymphocytes purified to >98% purity from PB at week 34 both contained between 0.1% to 1% neo-positive cells as determined by PCR. Similarly, between 0.1% and 1% of CD34+ cells purified from marrow at week 11 contained the neo gene. To further confirm that myeloid progenitors were marked, we used PCR to assay pools of 15 CFU-GM colonies grown in the absence of selection from the marrow at week 36. Of 36 pools assayed, three were neo-positive, giving an estimated marking frequency of 0.6% ± 0.3% of marrow-derived CFU-GM containing the neo gene. Thus, the neo gene was present at similar frequencies in mature cells of both myeloid and lymphoid lineages, and it was present in a portion of granulocytic and erythrocytic progenitors in marrow.
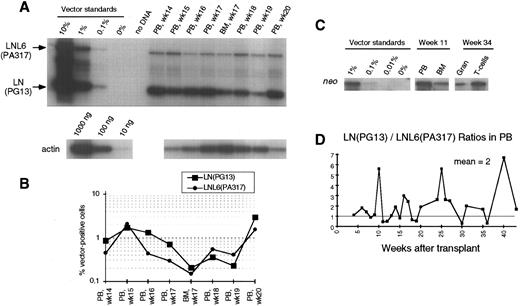
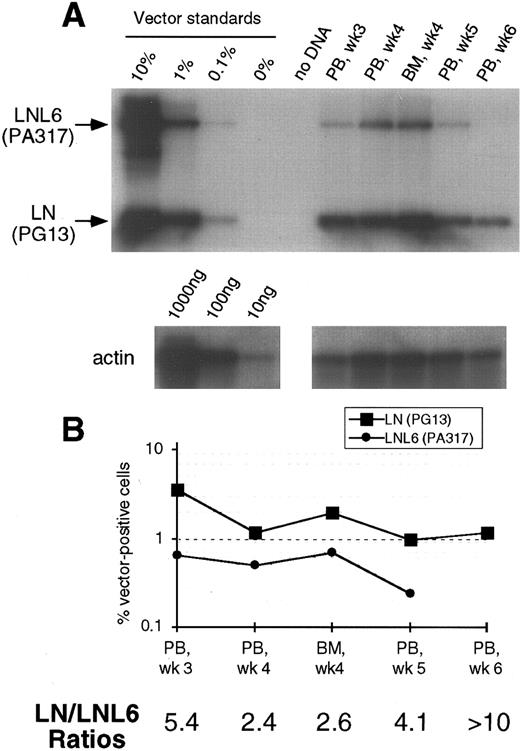
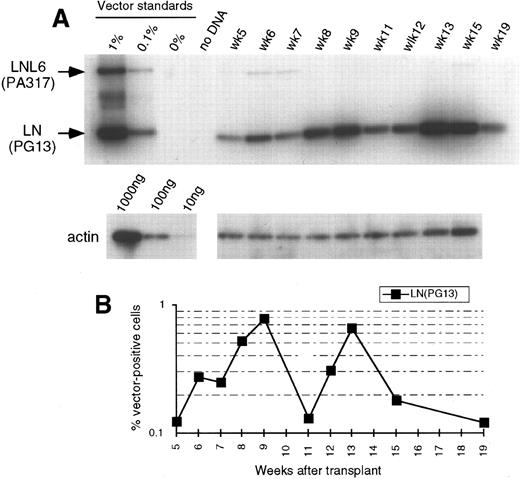
(A) Southern blot analysis of amplified vector sequences from baboon, J94090. Primers LN/LNL6 2229 and LN/LNL6 3210 were used to distinguish signals derived from LN (301 bp) versus LNL6 (982 bp). PCR for actin DNA was performed on the same samples with 100 ng DNA. Standards consist of single copy HT1080 DNA, HT1080/LN, and HT1080/LNL6 at a 1:1 ratio, mixed with normal baboon DNA in a log dilution series. PB, peripheral blood; BM, bone marrow. (B) Percentage vector-positive cells measured by phosphorimage analysis of signal intensities for LN and LNL6 corrected for the LN/LNL6 ratio in the standards (see Results) and for the amount of DNA as determined by actin PCR. (C) Southern blot analysis of neo-amplified sequences from PB, BM, peripheral blood granulocytes (Gran), and CD2+ T lymphocytes (using neo primers neo 350 and neo 1150). (D) LN/LNL6 ratios over time in PB cells of the long-term surviving animal, J94090.
(A) Southern blot analysis of amplified vector sequences from baboon, J94090. Primers LN/LNL6 2229 and LN/LNL6 3210 were used to distinguish signals derived from LN (301 bp) versus LNL6 (982 bp). PCR for actin DNA was performed on the same samples with 100 ng DNA. Standards consist of single copy HT1080 DNA, HT1080/LN, and HT1080/LNL6 at a 1:1 ratio, mixed with normal baboon DNA in a log dilution series. PB, peripheral blood; BM, bone marrow. (B) Percentage vector-positive cells measured by phosphorimage analysis of signal intensities for LN and LNL6 corrected for the LN/LNL6 ratio in the standards (see Results) and for the amount of DNA as determined by actin PCR. (C) Southern blot analysis of neo-amplified sequences from PB, BM, peripheral blood granulocytes (Gran), and CD2+ T lymphocytes (using neo primers neo 350 and neo 1150). (D) LN/LNL6 ratios over time in PB cells of the long-term surviving animal, J94090.
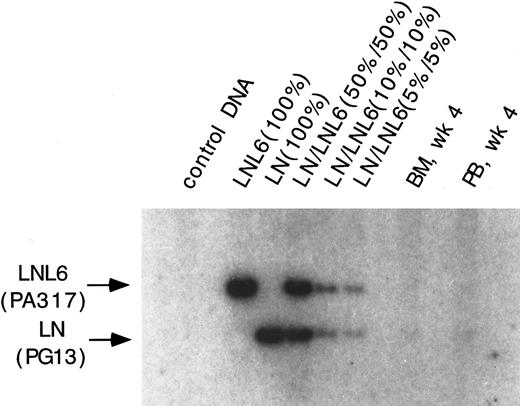
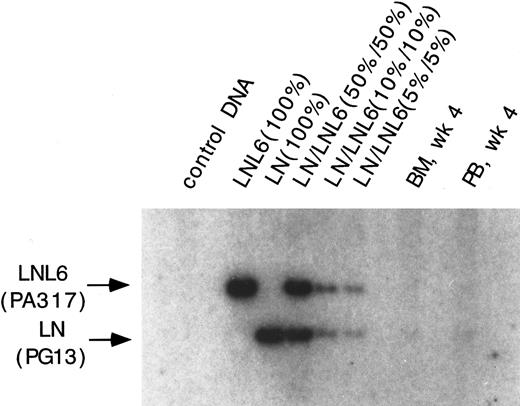
Southern blot analysis for the presence of the LN and LNL6 vector in DNA from PB and marrow in baboon, F93449, on week 4 after transplant. DNA was restricted with Sac I, which cuts both vectors outside of the neo gene. The expected size for LNL6 is 3052 bp and for LN, 2371 bp. Positive control standards and dilutions consist of the indicated ratios of DNA from HT1080 cells marked with a single copy of either the LN or the LNL6 vector mixed with normal DNA.
Southern blot analysis for the presence of the LN and LNL6 vector in DNA from PB and marrow in baboon, F93449, on week 4 after transplant. DNA was restricted with Sac I, which cuts both vectors outside of the neo gene. The expected size for LNL6 is 3052 bp and for LN, 2371 bp. Positive control standards and dilutions consist of the indicated ratios of DNA from HT1080 cells marked with a single copy of either the LN or the LNL6 vector mixed with normal DNA.
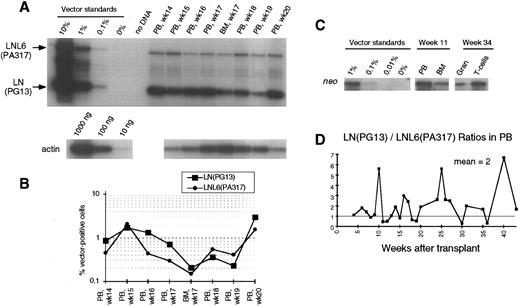
Contribution of LNL6 (PA317), LNX (PA317), and LN (PG13) marked cells to hematopoietic repopulation. To study the contribution of cells marked with vectors having different retroviral pseudotypes, LNL6/LNX and LN amplification products were separated and the intensities of the radioactive bands were quantitated by phosphorimaging. The PCR conditions used resulted in linear amplification for the LN, LNX, and LNL6 sequences in the range of 0.1% to 10%, allowing accurate correction for the differential amplification intensities of the different sized vector sequences. All four animals showed reconstitution with hematopoietic cells carrying either both the LN and LNL6 vector or the LN and LNX vectors. In animal J94090, the contribution of cells marked by the different vectors as measured by LN/LNL6 ratios fluctuated over time, although the overall mean LN/LNL6 ratio was 2.0 (Fig 1). In the three other animals, all blood and marrow samples tested contained more LN marked cells. In two animals, M94142 and F93449, LN/LNX or LN/LNL6 ratios ranged from 2.4 to >10 (Fig 3 and 4). In M94264, only three PB samples had LNL6 marked cells at around 0.1%; all the other PB samples had LNL6 marked cells <0.1% or nondetectable (Fig 2). In contrast, all PB samples contained LN marked cells at frequencies between 0.1% to 1% (Fig 2). Southern blot analysis of DNA from PB and BM in animal F93449 showed a slightly stronger band for the LN vector, confirming the PCR data (Fig 5).
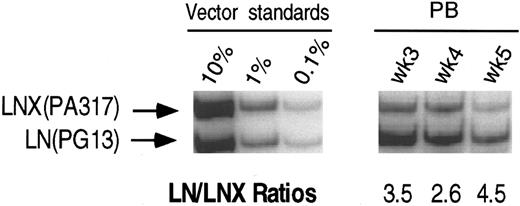
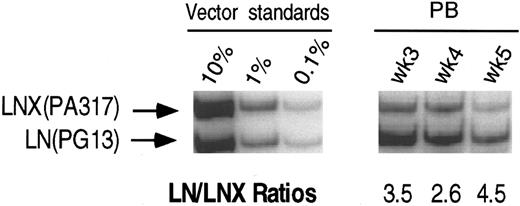
PCR analysis of vector sequences from baboon, M94142. Primers LN/LNL6 2229 and LN/LNL6 3210 were used to distinguish signals from LN (301 bp) versus LNX (326 bp) derived sequences. Amplification products were separated on a 10% polyacrylamide gel. LN/LNX ratios.
PCR analysis of vector sequences from baboon, M94142. Primers LN/LNL6 2229 and LN/LNL6 3210 were used to distinguish signals from LN (301 bp) versus LNX (326 bp) derived sequences. Amplification products were separated on a 10% polyacrylamide gel. LN/LNX ratios.
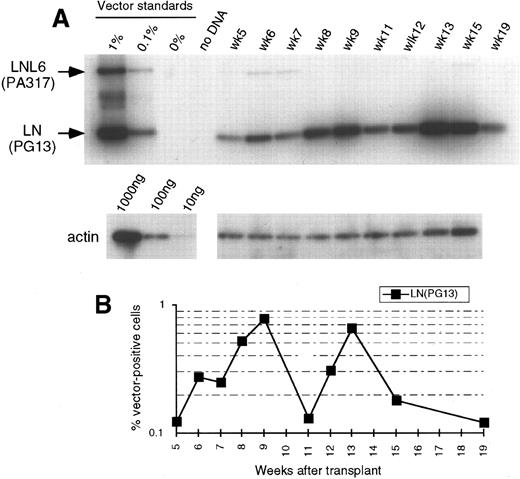
(A) Southern blot analysis of amplified vector sequences from baboon, M94264, as described for Fig 1. (B) Phosphorimage analysis of signal intensities for LN adjusted for the amount of DNA as determined by actin PCR. Numbers for LNL6 were either below 0.1% or bands were undetectable.
(A) Southern blot analysis of amplified vector sequences from baboon, M94264, as described for Fig 1. (B) Phosphorimage analysis of signal intensities for LN adjusted for the amount of DNA as determined by actin PCR. Numbers for LNL6 were either below 0.1% or bands were undetectable.
Autopsy and helper virus analysis. Autopsy of the expired animals showed trilineage engraftment. Both animals died of infectious complications. PCR analysis of PB samples for the presence of amphotropic and GALV envelope sequences were negative in all four animals indicating that there was no helper-virus infection. S+ L− assay in the longest surviving animal, J94090, was negative, as well.
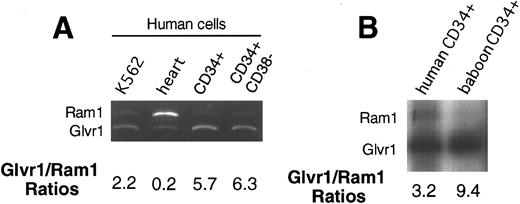
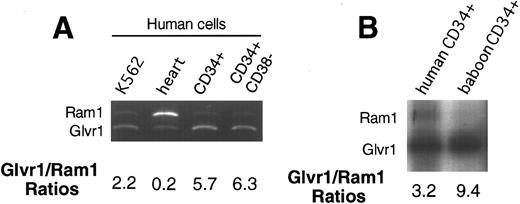
Retroviral receptor expression. Retroviral receptor expression was determined by Northern analysis in different human hematopoietic cell lines (KG1, HL60, KMT2, and K562) and in different human tissues (data not shown). All hematopoietic cell types analyzed showed higher levels of Glvr1 than Ram1 RNA. This was also the case for unseparated and CD34-enriched marrow from baboons. To more directly compare the relative levels of the Glvr1 and Ram1 mRNAs, we used an RT-PCR technique10 involving a pair of primers that recognize both receptor sequences followed by restriction digestion of the products with HaeIII, which yields different size fragments for Glvr1 or Ram1. This technique showed more abundant Glvr1 RNA expression in the human hematopoietic cell line K562, human and baboon CD34+ cells, and human CD34+CD38− cells (Fig 6). In human heart tissue, this ratio was reversed (Fig 6), as has previously been observed for rat heart.12
Analysis of relative retroviral receptor expression. RT-PCR was performed on RNA from heart, different hematopoietic cell lines, and hematopoietic CD34 cells and CD34+ CD38− cells in humans and CD34+ cells in baboons. Samples were electrophoresed on nondenaturing gels and either stained with ethidium bromide (A) or exposed to x-ray film (B) and analyzed by densitometry. The predicted DNA fragment for the amphotropic receptor is 182 bp and for GALV receptor, it is 149 bp.
Analysis of relative retroviral receptor expression. RT-PCR was performed on RNA from heart, different hematopoietic cell lines, and hematopoietic CD34 cells and CD34+ CD38− cells in humans and CD34+ cells in baboons. Samples were electrophoresed on nondenaturing gels and either stained with ethidium bromide (A) or exposed to x-ray film (B) and analyzed by densitometry. The predicted DNA fragment for the amphotropic receptor is 182 bp and for GALV receptor, it is 149 bp.
DISCUSSION
GALV and amphotropic pseudotyped retroviral vectors have been used for gene transfer into human hematopoietic cells and are being used in clinical trials. We have previously shown increased gene transfer efficiency into hematopoietic progenitor cells in vitro using a GALV-pseudotyped vector compared with an amphotropic vector.13 In this report, we used a competitive repopulation assay to compare gene transfer efficiency in baboon hematopoietic repopulating cells using GALV and amphotropic-pseudotyped vectors. We transplanted four animals and found that the GALV-pseudotyped vector was able to mark short-term and long-term repopulating cells. Our results also suggest that gene transfer into baboon repopulating cells is more efficient using GALV-pseudotyped vectors than amphotropic vectors, as indicated by the higher frequency of cells marked with the GALV-pseudotyped vector in the blood and marrow of all animals. Although factors affecting hematopoietic stem/progenitor cell survival and proliferation could conceivably vary between the packaging cell lines used to transduce baboon CD34-enriched marrow cells, the recovery of colony-forming cells from the different cocultures in our study was similar.
We and others have shown that mRNAs encoding the receptors for these two viruses are expressed at different levels on hematopoietic cells.10,11,13 23 Most hematopoietic cells express more Glvr1 than amphotropic receptor mRNA. In this study, we have analyzed receptor RNA expression of Ram1 and Glvr1 in human and baboon hematopoietic progenitor by Northern blot analysis and RT-PCR. The ratio of Glvr1 to Ram1 expression in samples showed that human and baboon marrow-derived CD34+ cells expressed more Glvr1 than Ram1; a similar ratio was also seen for human CD34+ CD38− cells, suggesting that even more primitive hematopoietic progenitor cells express more Glvr1 than Ram1. Northern analysis confirmed that baboon unseparated marrow and CD34+ cells expressed more Glvr1.
Gene transfer efficiency was between 0.1% and 1% in two animals and between 1% and 5% in the other two animals. We have not seen significant decreases in the proportion of marked cells in PB and BM in the longest surviving animal 1 year posttransplant, suggesting that there has not been a significant immune response to genetically marked cells in these animals. Although follow-up has been short in three animals, our data also suggest that there are considerable differences in gene transfer efficiency between animals. The only differences between our animals were the variable number of genetically modified cells infused and the use of G-CSF and megakaryocyte growth and development factor (MGDF ) in one animal (M94142). The animals transplanted with a higher number of genetically marked CD34+ cells had a higher gene transfer efficiency in vivo, suggesting a possible correlation between cell dose and gene transfer efficiency. On the other hand, this observation could simply reflect a stochastic event as suggested by others.24 We also saw higher in vivo gene transfer in animals that had a higher in vitro gene transfer rate in CFU-GM. The fact that the differences in in vivo gene transfer efficiencies between animals were in the 10-fold range, although the same gene transfer protocol was used, emphasizes the importance of testing variables that may change gene transfer efficiency in one animal by using a competitive repopulation assay.4
To establish a baseline gene transfer efficiency in our baboon model, we have used a 2-day cocultivation gene transfer protocol. We hypothesized that this would give us the best gene transfer results using unstimulated marrow CD34+ cells. Growth factor stimulation is known to alter receptor expression,9 and it is conceivable that receptor expression of certain retroviral receptors can be affected by administration of specific growth factors. Dunbar et al25 have recently shown that marrow or PB obtained from animals after treatment with hematopoietic growth factors were more efficiently marked compared with cells obtained from untreated animals. We would, therefore, expect that the gene transfer efficiency seen in our animals could be further improved by using growth factor-stimulated stem cells for the gene transfer.
Two animals died of pulmonary infections 7 and 20 weeks after transplant. Marrow examination at the time of autopsy confirmed that both animals had engrafted. There was no evidence of helper virus in these animals. PCR analysis for amphotropic and GALV envelope sequences was negative in these animals, as well as in the surviving animals.
In summary, we have shown that a GALV-pseudotyped retroviral vector is capable of transducing baboon marrow repopulating cells, and the rate of gene transfer was higher than that of similar vectors with an amphotrophic pseudotype. The competitive repopulation assay used here will be useful to evaluate additional variables that affect gene transfer to hematopoietic stem cells and should lead to improved gene transfer conditions in human gene therapy trials.
NOTE ADDED IN PROOF
Recent modifications of the tether system used for intravenous therapy in our animals from a Single Channel Swivel (Spalding Medical Products, PA) to a Rotating Infusion Stand III-BB (Washington Regional Primate Center, WA) has virtually eliminated posttransplant mortality. In the last 5 animals transplanted with transduced CD34+ blood and marrow cells there have been no deaths, indicating that deaths in the current study were unrelated to the use of retroviral vectors.
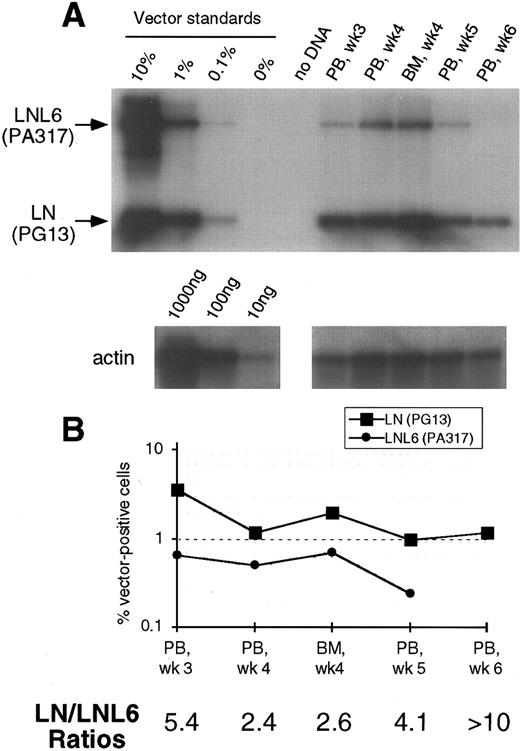
(A) Southern blot analysis of amplified vector sequences from baboon, F93449, as described for Fig 1. (B) Phosphorimage analysis of signal intensities for LN and LNL6 adjusted for the amount of DNA loaded as determined by actin PCR.
(A) Southern blot analysis of amplified vector sequences from baboon, F93449, as described for Fig 1. (B) Phosphorimage analysis of signal intensities for LN and LNL6 adjusted for the amount of DNA loaded as determined by actin PCR.
ACKNOWLEDGMENT
We thank Mike Gough, Gary Millen, and the staff of the University of Washington Regional Primate Research Center and the staff of the Fred Hutchinson Cancer Research Center Clinical Hematology laboratory for their assistance. We also acknowledge the assistance of Bonnie Larson and Harriet Childs in preparing the manuscript.
Supported by National Institutes of Health (Bethesda, MD) Grants and Contracts No. HL54881, CA15704, AI35191, NIHRR00166, and DK47754. H.P.K. was supported by the Lucille P. Markey Charitable Foundation.
Address reprint requests to Hans-Peter Kiem, MD, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, Seattle, WA 98109-1024.