TRANSCRIPTION FACTORS play a major role in differentiation in a number of cell types, including the various hematopoietic lineages.1-4 In the hematopoietic system, stem cells undergo a process of commitment to multipotential progenitors, which in turn give rise to mature blood cells. Although a number of transcription factors have been identified that play a role in the development of erythroid or lymphoid lineages,5-9 only in the past few years have those factors that influence development of myeloid cells been identified and studied. In particular, recent studies of the regulation of normal myeloid genes, as well as the study of leukemias, have suggested that transcription factors play a major role in both myeloid differentiation and leukemogenesis.10-12 To understand the process of normal myeloid differentiation, it is important to identify and characterize the transcription factors that specifically activate important genes in the myeloid lineage. These factors may play a role in acute myeloid leukemia (AML), in which this normal differentiation program is blocked. Finally, some of the transcription factor genes identified at the sites of consistent chromosome rearrangements in AML have now been shown to play a role in normal myeloid gene regulation.
The purpose of this review will be to summarize these recent data regarding myeloid gene regulation and the transcription factors mediating this process to understand normal myeloid development and leukemia. Emphasis will be placed on the identified or potential role of these factors in leukemia; we will focus on how alteration of myeloid transcription factors (changes in expression and structure) could lead to alterations in normal function in myelopoiesis, leading to a block in differentiation. In addition, we will discuss those factors that are involved in normal human myeloid maturation or human leukemia. Although this review will of necessity discuss different classes of factors, it does not aim to be an exhaustive compilation or listing of different factors and does not intend to be historical. Rather, it will attempt to highlight how recent findings can help to explain normal myeloid development and myeloid leukemia and will focus on certain examples to make important general points. In addition, this review will focus on early events in myeloid development and only briefly mention transcription factors involved in activation of mature myeloid cells. Finally, because there have been several recent reviews on hematopoietic factors in general, GATA, AML1, and the retinoic acid receptors, we will attempt as much as possible not to repeat what has been presented in these reviews.3,4 10-19 Some of the key issues that we will attempt to address in this review are the following: (1) How is gene expression restricted to myeloid cells? (2) How is the temporal expression of myeloid genes controlled? (3) Which transcription factors are thought to be necessary for myeloid development based on knockout and/or inactivation studies and which are thought to be necessary based on their redundancy? (4) If certain transcription factors are expressed in myeloid and nonmyeloid cells, why are certain target genes expressed in myeloid cells specifically? (5) How does a common progenitor cell become one type (myeloid) rather than another?
Currently, models have been advanced proposing that hematopoietic lineage determination is driven extrinsically (through growth factors, stroma, or other external influences),20,21 intrinsically (as described in stochastic models),22 or both.23 Our working hypothesis is that, regardless of whether the environmental or stochastic models are invoked, transcription factors are the final common pathway driving differentiation and that hematopoietic commitment to different lineages is driven by alternative expression of specific combinations of transcription factors, which often induce expression of growth factor receptors or growth factors. The transcription factors are often activated in positive autoregulatory loops, helping to explain the irreversible differentiation process found in human hematopoiesis. Experimental support of the hypothesis that transcription factors are the nodal decision points for myeloid differentiation include the observations that many genes cloned at the site of leukemic translocation breakpoints are transcription factors,11,12,24 genes isolated by their regulation of a phenotype (such as differentiation) are transcription factors,25 and, finally, knockouts of these transcription factors show gross defects in myeloid development.3 26-28
METHODS USED TO IDENTIFY AND STUDY MYELOID TRANSCRIPTION FACTORS
Our understanding of myeloid factors has been brought about by a number of different methods. Initially, myeloid factors were identified by investigations of known transcription factors, particularly oncogenes; examples include myb, myc, and the homeobox genes.12a Similarly, a number of factors have been isolated using hybridization and/or polymerase chain reaction (PCR) methods based on similarity to known genes, such as MZF-1, with subsequent studies investigating the normal and forced expression of these factors in myeloid cells.29-36 A second approach is subtraction hybridization or differential screening, which identified Egr-1 as a potential mediator of monocytic development.25 A third method has been identification of factors through the analysis of myeloid-specific promoters. Examples are PU.1 and C/EBPα, and these studies will be described in further detail below. Finally, a number of myeloid transcription factors have been identified through their involvement in leukemias, either as a result of abnormal expression, such as PU.1,37,38 or through their involvement at the site of a consistent chromosome translocation (examples include AML139 and PLZF40; see Table 1).
Once identified, the role of these factors has been ascertained by a number of inhibitory studies. These include the use of antisense and competitor oligonucleotide techniques, which have shown the role of myc, myb, MZF-1, and PU.1 in myeloid development.41-44 A limited number of studies have used a trans-dominant mutant approach and have been used to demonstrate the role of the retinoic acid receptor α in myeloid development and of PU.1 in activation of specific gene targets.45-47 Finally, the technique of gene disruption (knockout) has recently been used to investigate the role of specific factors in myeloid development.3,7,26-29 48-50 A recent review has focused on the effects of these knockout studies on hematopoiesis in general3; therefore, we will discuss them only as they relate to myeloid transcription factors.
HOW DO MYELOID PROMOTERS WORK? WHAT ARE GENERAL RULES SO FAR?
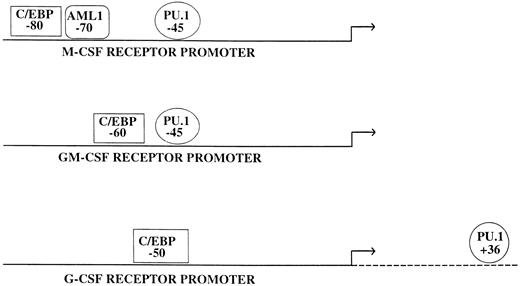
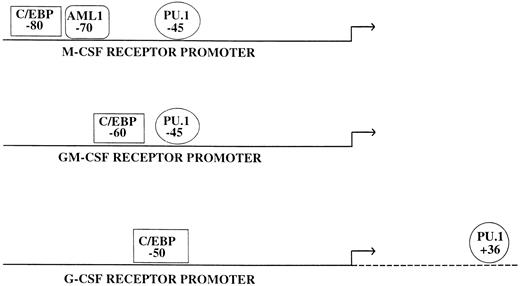
Many myeloid promoters appear to differ from what has been described previously for tissue-specific promoters.51 In general, a relatively small upstream region (usually a few hundred basepairs) is capable of directing cell-type–specific expression in tissue culture studies, although it appears that additional elements are necessary for position independent, high-level expression in a chromosomal locus (Fig 1). For example, almost all of the specificity and activity detected in transfection of tissue culture cells by the macrophage colony-stimulating factor (M-CSF) receptor, granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor α, and granulocyte colony-stimulating factor (G-CSF) receptor promoters are contained within 80, 70, and 74 bases of the major transcription start sites, respectively,52-54 and similar results have been obtained with the promoters for myeloid transcription factors such as PU.1.55,56 Although most of these promoters direct lineage and developmental specific expression, in general they lack a TATA box or defined initiator sequence. Interestingly, many myeloid promoters have a functional PU.1 binding site upstream of the transcription start site at a location corresponding to one similar to that of a TATA box. Because PU.1 can bind to the TATA binding protein (TBP) in vitro,57 one mechanism by which PU.1 could activate a whole class of myeloid promoters is by recruiting TBP, the primary component of the basal transcription factor TFIID, and the rest of the transcriptional apparatus. Consistent with this mechanism are studies showing that mutation of a PU.1 site in the FcγR1 promoter can abolish myeloid-specific expression, and replacement of this mutant site with a TATA box can restore myeloid expression.58 Other factors involved in myeloid gene regulation, including Sp1, C/EBPα, and Oct-1, have all been shown to interact with TBP,59-61 and these interactions could also help mediate recruitment of the basal transcription machinery. Finally, whereas some of these TATA-less and initiator-less myeloid promoters have multiple transcriptional start sites,55,62 others appear to have a single strong start site.63 64 The mechanism for how the start sites are determined in these promoters is unknown.
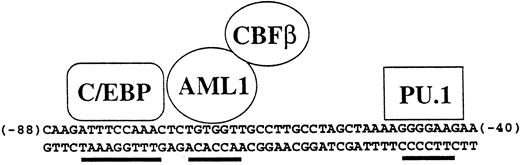
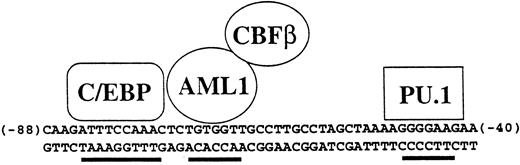
Specificity of the human M-CSF receptor promoter is mediated by combinatorial activity of factors in myeloid cells. The activity and specificity of the human M-CSF receptor promoter in transient transfection studies is mediated by the 50-bp region lying between bp −88 and −40 relative to the major transcription start site in monocytic cells.52,117,215 Shown are the binding sites for C/EBP, AML1, and PU.1, all of which are important for M-CSF receptor promoter activity. In the hematopoietic system, C/EBPα is myeloid specific, AML1 is expressed in all white blood cells, and PU.1 is B-cell– and myeloid-specific, so therefore the major cell type in which all three are expressed is myeloid cells. In addition, C/EBPα and AML1 can physically interact and synergize to activate this promoter.215 Others have shown using other promoters that C/EBP proteins can synergize with PU.1.88 Also shown is CBFβ, which forms a heterodimer with AML1 and augments its ability to bind to DNA.
Specificity of the human M-CSF receptor promoter is mediated by combinatorial activity of factors in myeloid cells. The activity and specificity of the human M-CSF receptor promoter in transient transfection studies is mediated by the 50-bp region lying between bp −88 and −40 relative to the major transcription start site in monocytic cells.52,117,215 Shown are the binding sites for C/EBP, AML1, and PU.1, all of which are important for M-CSF receptor promoter activity. In the hematopoietic system, C/EBPα is myeloid specific, AML1 is expressed in all white blood cells, and PU.1 is B-cell– and myeloid-specific, so therefore the major cell type in which all three are expressed is myeloid cells. In addition, C/EBPα and AML1 can physically interact and synergize to activate this promoter.215 Others have shown using other promoters that C/EBP proteins can synergize with PU.1.88 Also shown is CBFβ, which forms a heterodimer with AML1 and augments its ability to bind to DNA.
Consistent with their lack of a TATA box, these promoters often are dependent on a functional Sp1 site, which can interact not only with the TATA binding protein, TBP, as well as a TBP-associated factor, TAF110.59 These Sp1 sites not only mediate activity, but also specificity and inducibility with differentiation.65-67 How the Sp1 factor mediates this specificity is not clear, but in one case Sp1 binds to its myeloid target in vivo in myeloid cells only,65 and in another, increased relative expression in myeloid cells of a phosphorylated form of Sp1 that shows markedly increased binding to its DNA target site is observed.66 Although Sp1 is ubiquitous, it is preferentially expressed in hematopoietic cells.68 Differential expression in distinct hematopoietic lineages has not been explored. In addition, Sp1 can mediate responses to retinoic acid, thyroid hormone, and retinoblastoma protein, all of which may influence myeloid differentiation.69-72 Finally, it has been recently shown that Sp1 belongs to a family of related factors and that at least one of these, Sp3, may mediate repression of target genes.73,74 Sp1 can also cooperate in repression with GATA-1,75 and this interaction could potentially repress myeloid targets in erythroid cells (see below).
Several of the myeloid promoters appear to have GATA sites that can bind GATA proteins specifically in gelshift assays. However, mutation of GATA binding sites in the CD11b76 or PU.155 promoters does not lead to significant loss of promoter function. Coexpression of either GATA-1 or GATA-2 leads to a slight downregulation (2-fold) of these promoters. Interestingly, there is a functional GATA-1 site in the promoter for the platelet-specific integrin IIb that is located at a position almost identical to that of the nonfunctional GATA site found in the myeloid integrin CD11b,77 suggesting evolutionary conservation of binding sites in related integrin promoters, but not function. The differences between the ability of GATA-1 to activate promoters such as IIb in erythroid or megakaryocytic cells, in which GATA-1 is thought to play an important role in lineage development and not in myeloid cells, may be due to cell-type–specific modifications of GATA and/or interactions with other transcription factors. In addition, GATA-1 is not expressed at significant levels in myeloid cells, and both GATA-144 and GATA-278 are downregulated in myeloid development. These results are consistent with the preservation of myeloid cell development in mice with targeted mutations of GATA-1.5 In contrast to monocytic and neutrophilic cells, high levels of GATA-1 are detected in eosinophils,79 but to date functional GATA sites have not been detected in eosinophil promoters. In summary, it appears that GATA proteins do not play an important positive role in myeloid development; indeed, downregulation may be critical for myeloid maturation.44,80 81
As noted above, almost all myeloid promoters have a functional PU.1 site close to the transcription start site. There are a number of interesting exceptions to this rule. The CD14 promoter has a TATAA-like sequence and does not have a functional PU.1 site in its promoter.66 This promoter region is highly active and cell-type–specific in transient assays66 but not in transgenic mice.82,83 This scenario is somewhat similar to that of chicken lysozyme, which lacks a PU.1 site in its promoter but contains one in an enhancer located 2.7 kb upstream and which is part of a genomic fragment that is capable of high-level specific expression in mice.84 85 Whether other myeloid promoters that lack PU.1 sites will turn out to have PU.1-containing enhancer elements located several kilobases from the promoter remains to be determined.
Another set of myeloid genes that appear to have TATAA boxes are the primary granule protein promoters, including myeloperoxidase,86,87 neutrophil elastase,87,88 proteinase 3,89 and cathepsin G.90 At least three of these have been noted to have functional PU.1 sites, and in this class of genes PU.1 plays an important role, but perhaps has a different mechanism than other myeloid genes in which there is no TATAA box. At this point it is not clear what other common factors account for the distinct pattern of expression of these genes, in which mRNA is dramatically upregulated at the promyelocytic stage and then rapidly downregulated with further myeloid development. It has also been observed that a number of these same primary granule myeloid promoters (cathepsin G, myeloperoxidase, neutrophil elastase, aminopeptidase N, c-fes, and MRP8) contain a common sequence CCCCnCCC.91 Recently, a protein binding to this site has been described, and mutations of this site in the proteinase-3 promoter that abrogate binding of this factor lead to a significant loss in promoter function.89 Therefore, identification of this protein may lead to important understanding of the regulation of this whole family of myeloid genes.
In contrast to the primary granule protein genes, most other myeloid-specific genes are upregulated during the course of myeloid development and are expressed at highest levels in mature myeloid cells. Examples include key receptors, such as the G-CSF receptor92 and CD11b,93,94 as well as critical transcription factors, such as PU.1.44,95 In some cases, there are emerging clues into the mechanisms of upregulation during differentiation. For example, the upregulation of the CD14 promoter during vitamin D3-induced monocytic differentiation is mediated by an Sp1 site,67 and recently a novel transcription factor has been described that mediates upregulation of CD11b promoter activity during 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced differentiation of myeloid cell lines.96 The binding site for this factor is shared by the three leukocyte integrin promoters. The factor identified, MS-2, like Sp1, is not specific for myeloid cells. In other cases, the promoter elements and transcription factors mediating upregulation during myeloid differentiation have not been well defined. It should be noted that the transcription factors critical for upregulation during myeloid development versus those invoked during activation of mature myeloid cells may well be distinct. For example, the macrophage scavenger receptor has motifs mediating lineage specificity (including a PU.1 site) and a distinct element mediating upregulation of stimulated macrophages (through an AP-1/ets site).97 Similarly, distinct elements mediating lineage upregulation of the interleukin-1β (IL-1β) promoter in macrophage lines have been described.47,98 99
Functionally important transcriptional regulatory elements have been described in the 5′ untranslated regions of a number of genes,100,101 and several myeloid promoters also contain important functional sites in their 5′ untranslated region. The human and murine PU.1 promoters are almost totally conserved in their 5′ untranslated region, and both contain a functional PU.1 site.55 In the case of the human and murine G-CSF receptor promoters, conservation in this region is mainly limited to the PU.1 site found there.53
TRANSGENIC STUDIES
Often myeloid regulatory factors have been identified through the investigation of myeloid-specific promoter elements in studies involving transient transfection studies in tissue culture cell lines. In transgenic mice, other regulatory elements, such as locus control elements, are very important.102 A number of myeloid promoters have worked to some extent in transgenic studies involving the expression of heterologous genes in myeloid cells. In some instances, there is variability of levels of expression and significant position dependence. For example, the proximal 1.5 kb of the gp91-phox gene, normally expressed at high levels in granulocytes, targeted reporter expression to a small subset of monocytic cells only, and most monocytes and neutrophils did not express the transgene.103 In other examples, the responsible regulatory elements have not yet been defined in detail. These studies include human c-fes/fps,104 chicken lysozyme,85,105 or human cathepsin G,90 in which the investigators reported high-level, properly regulated transgene expression. In these studies, relatively large pieces of DNA (6 to 21.5 kb) were used that included the gene itself as the reporter. In some instances, the expression patterns of the transgenes were not described in detail, but were stated to be specific for macrophages and neutrophils, respectively. Examples include those involving a 5-kb portion of the monocytic M-CSF (CSF-1) receptor promoter, which was reported to target macrophages,106,107 and the MRP8 promoter, which was used to express bcl-2 in neutrophils.108 The MRP8 promoter has also been used to induce expression of a PML/RARα transgene in myeloid cells, resulting in a high percentage of expressing transgenic lines, altered granulocytic differentiation, and, in some animals, an acute leukemia resembling human acute promyelocytic leukemia.109 A recent study has shown that the human lysozyme promoter can direct reporter gene expression in macrophages.110
A number of studies have looked at the ability of myeloid integrin promoters to direct the expression of reporter genes in transgenic studies.111,112 The CD11b promoter is capable of driving high-level expression of a reporter gene in myeloid cells at levels comparable to the endogenous CD11b promoter. This promoter construct is position-dependent, perhaps explaining why another group did not observe similar levels of expression.112 Although in these studies the activity of the promoter was highest in mature myeloid cells, this promoter construct has also been used to direct expression of the t(15:17) PML/RARα fusion protein in early myeloid cells.113 The reason for these differences is not clear, but may reflect a significant amount of position dependence on promoter activity and specificity. Attempts to direct expression of reporter genes into early myeloid cells using promoters such as CD3483 and c-kit114 have not been successful. However, recent reports suggest that upstream regions of the cathepsin G115 and MRP8108,109 genes may be useful in directing expression of an inserted cDNA into promyelocytes. Many of the promoters used in these studies, like most myeloid-specific genes, lack a TATAA box,63,76 and previous studies using transient transfections in tissue culture cells have shown the role of PU.1 in the myeloid specificity of the promoters (Table 2). Of the myeloid genes that have been shown to function in transgenic studies to date, several of them, including CD11b,116 the M-CSF receptor,117 the chicken lysozyme enhancer,84 and c-fes,118,119 have important functional PU.1 sites. A recent study has confirmed the importance of the PU.1 site in macrophage expression of the macrophage scavenger receptor.120 These studies strongly suggest that PU.1 will in general be a critical factor in directing myeloid expression in transgenic studies.
In summary, it appears from the transgenic studies that myeloid promoters can drive expression in monocytic and neutrophilic cells in transgenic mice and that the PU.1 site is likely to be critical. However, the details of the other critical elements necessary for myeloid-specific and position-independent expression are not defined yet, and much work needs to be performed in this area in the future.
GENE THERAPY APPLICATIONS OF MYELOID PROMOTERS
An exciting prospect would be to use the myeloid regulatory elements to target given genes to proper hematopoietic compartments for gene therapy applications, particularly in terms of directing macrophage expression of enzymes involved in glycogen storage diseases.121 Myeloid promoter elements may be useful for viral vectors, especially adeno-associated virus (AAV), because, at least in transient assays, very small DNA fragments of a few hundred basepairs direct activity that is as high and as specific as much larger fragments. A note of caution is that larger elements or a combination of promoter and upstream elements (locus control regions) may be necessary, as noted above from the description of transgenic studies. Very few reports on the use of myeloid elements in gene therapy vectors have been reported. In one study, the CD11b promoter failed to direct significant reporter expression when incorporated into a retroviral vector,122 but a second study showed the same promoter directing significant amounts of glucocerebrosidase mRNA and enzymatic activity in myeloid cell lines and in bone marrow.123 Again, the reason for the discrepancies observed between these two studies using a similar promoter is not clear, and much more work needs to be performed in this area of research.
PU.1 (Spi-1) AND OTHER ets FACTORS
PU.1: A master regulator of myeloid genes.Recent studies have focused on PU.1 as a major regulator of myeloid development.26,44,161 PU.1 is the product of the Spi-1 oncogene identified as a consequence of its transcriptional activation in Friend virus-induced erythroleukemias.14,37,38,124 The role of PU.1 in the malignant transformation of the proerythroblast was confirmed by subsequent studies. Overexpression of PU.1 from a retroviral construct in long-term bone marrow cultures is able to stimulate the proliferation of proerythroblast-like cells that differentiate at a low frequency into hemoglobinized cells,125 and overexpression in transgenic mice can also induce erythroleukemia.126 Moreover, PU.1 antisense oligonucleotides reduce the capacity of Friend tumor cells to proliferate.127 Taken together, these data suggest that deregulation of PU.1 increases the self-renewal capacity of erythroid precursor cells and blocks their differentiation. Furthermore, it shows the importance of proper regulation of PU.1 in the hematopoietic system.
PU.1 is a member of the Ets transcription family.124 Ets factors all contain a characteristic DNA binding domain of approximately 80 amino acids.14,128,129 PU.1 and the related Ets family member Spi-B130 form a distinct subfamily within the larger group of Ets proteins, with very distinct structure, patterns of expression, binding specificity, and functions that are nonoverlapping with other members of the Ets family. The PU.1 protein consists of 272 amino acids, with the DNA binding domain located in the carboxyl terminal part of the protein, whereas the amino terminus contains an activation domain that has been implicated in interactions with other regulatory proteins.57,124,131,132 The structure of the DNA binding domain has been recently determined and demonstrates a winged helix-turn-helix motif.133
PU.1 shows specific patterns of hematopoietic expression. PU.1 is expressed at highest levels in myeloid and B cells, but not in T cells.37,95,124 During hematopoietic development, PU.1 mRNA is expressed at low levels in murine ES cells and human CD34+ stem cells and is specifically upregulated with myeloid differentiation.44 The timing of this upregulation coincides with the first detection of early myeloid maturation, suggesting that this increase in expression may be an important process in myeloid development, and inhibition of PU.1 function at this juncture can block myeloid progenitor formation.44 These findings were confirmed by another report showing that PU.1 is expressed in the earliest Go CD34+ stem cells and upregulated as single CD34+/CD38− cells developed into myeloid colonies.134 Studies in both primary CD34+ cells and human leukemic cell line models suggest that PU.1 mRNA and DNA binding activity do not increase further with subsequent myeloid maturation from the promyelocytic to more mature stages,44,95 but these studies do not preclude the possibility that the high levels of mRNA observed in human monocytes and neutrophils95 are a result of subsequent final maturation and/or activation. Although initially thought to be B-cell and monocyte specific,124,135 high levels of both PU.1 mRNA and DNA binding activity have been subsequently found in neutrophils.95 PU.1 mRNA has also been detected in human eosinophils (S.J. Ackerman and D.G. Tenen, unpublished observations). In murine Friend erythroleukemia cells, PU.1 is downregulated during chemically induced erythroid differentiation.127,136 137
These expression studies suggest that regulation of PU.1 mRNA may play a significant role in the commitment of early multipotential progenitors to the myeloid lineages, as well as in the further differentiation and maturation of these cells. Characterization of the murine and human PU.1 promoters shows that a highly conserved region surrounding the transcription start sites can direct myeloid-specific activity in transient transfection studies. The important functional sites identified thus far in the promoter, an octamer site at bp −54, an Sp1 site at bp −39, and a site for PU.1 itself at bp +20, are conserved in the two species. In B cells, the octamer site plays a major role in PU.1 expression,56,138 whereas in myeloid cells the PU.1 site is the most important for function of the promoter.55 This study suggests the presence of a positive feedback mechanism in the regulation of the PU.1 promoter similar to that shown for another Ets family member, Ets-1.139 Such autoregulation of transcription factors could play a major role in commitment and differentiation of hematopoietic multipotential progenitor cells (see below). The PU.1 promoter also contains a site that can bind GATA proteins, but no binding could be detected using nuclear extracts from myeloid cell lines, and cotransfection of either a GATA-1 or GATA-2 expression construct repressed the PU.1 promoter twofold. Important unanswered questions are whether GATA proteins affect PU.1 expression in multipotential progenitors44 or in early erythroid cells, in which PU.1 is repressed in the course of differentiation.136,137,140 Finally, although the elements described above direct expression of PU.1 in tissue culture cells, as much as 2 kb of upstream murine DNA sequence failed to direct expression of a reporter cDNA in spleen or peritoneal macrophages in transgenic studies, so other elements are required for high level expression in the chromosomal context.83
PU.1, like other Ets factors,128 was first noted to bind to a sequence characterized by a purine-rich core (GGAA), hence its name.124 However, the DNA binding specificity of PU.1 appears to be quite distinct from that of other Ets factors. For example, PU.1 and Spi-B bind to the CD11b PU.1 site, but not the other Ets family members Ets-1, Ets-2, Elf-1, or Fli-1.44 In addition, it is not possible to predict PU.1 binding by observation of a 5′-GAGGAA-3′ sequence. For example, there are three such sequences in the M-CSF receptor promoter, and PU.1 does not bind to any of them with high affinity. Instead, PU.1 binds to the nearby sequence 5′-AAAGAGGGGGAGAG-3′.117 This example is consistent with previous work indicating that binding of PU.1 and other Ets factors is dependent on sequences outside the GGAA core.141 In addition, many functional PU.1 binding sites in myeloid promoters have a string of adenosine residues immediately 5′ of the GA core.54,116,117,137 These results are consistent with a recently described consensus sequence for PU.1 binding.118 The availability of an x-ray crystallographic structure for PU.1133 will assist in further studies of PU.1 binding specificity. In summary, PU.1 is distinct from other Ets factors in DNA binding specificity. Identification of the transactivation domain of PU.1 has been relatively difficult because PU.1 acts as a weak transactivator; indeed, most transactivation studies have required multiple PU.1 binding sites and used artificial promoter constructs.53,55,124,142 Studies using myeloid promoters have in general shown weak transactivations, on the order of twofold to fivefold.116,117 Recently, using multimerized reporter constructs, the transactivation domain of PU.1 was identified in the amino terminus.131 These studies defined multiple relatively weak transactivation domains; whether this indicates that each interacts with different proteins (see below) remains to be determined.
PU.1, like other Ets proteins, interacts with other transcriptional regulators. In B cells, PU.1 recruits a second, B-cell –specific DNA binding factor, NF-EM5 or Pip, to a site important for Ig κ 3′ enhancer function.143-146 For this interaction, phosphorylation of a serine residue at position 148 is necessary.144 However, such interactions have not been shown previously to play a functional role in previously described myeloid targets of PU.1, such as CD11b and the M-CSF receptor, and mutation of Ser148 does not affect the ability of PU.1 to activate these myeloid promoters.116 117
As noted above, PU.1 can interact with TBP in vitro.57,132 An attractive hypothesis is that PU.1 may recruit the basal transcription machinery to myeloid promoters, which in general lack TATAA boxes, but the functional significance of this interaction is not known at this time. Similarly, these same studies showed that PU.1 can interact physically with RB. It has been shown that hypophosphorylated RB can negatively regulate the activity of another Ets factor, Elf-1, in the course of T-cell activation.147 Because RB becomes hypophosphorylated during myeloid differentiation,148,149 it possibly could regulate PU.1 function in these cells. Experiments to date have failed to show an effect of RB on PU.1 function on myeloid promoters,83 although it has been shown that RB can repress PU.1 function on an artificial promoter.132 A recent study using a protein interaction screen has identified a number of proteins that can bind to PU.1, although the functional consequences of these interactions remains to be determined.142 One of these is HMG(I)Y, which has been shown to augment the ability of the Ets factor Elf-1 to activate the IL-2 receptor β chain.150 To date, similar experiments with HMG(I)Y have failed to show binding to myeloid targets such as the M-CSF receptor or GM-CSF receptor α promoters or to augment the ability of PU.1 to activate these promoters. Another factor cloned in this study and shown to bind to PU.1 is NF-IL6β (same as C/EBPδ); in this case, it was shown that PU.1 and C/EBPδ could cooperatively activate an artificial reporter plasmid. These findings are interesting because they suggest that PU.1 might also interact with C/EBPα (see below). Similarly, in another study, PU.1 has been shown to interact with another basic leucine zipper (BZIP) transcription factor, c-Jun.151 Finally, PU.1 has been shown to functionally interfere with steroid hormone gene activation, and steroid hormone receptors can inhibit PU.1 transactivation function.152 Of particular relevance to myeloid development is the fact that these interfering effects were seen with the retinoic acid receptor.152 However, the functional significance with respect to myeloid development of all of these interactions is not known.
Ets family members have been shown to play an important role in several signal transduction pathways.153-156 PU.1 is phosphorylated in vitro by casein kinase II and JNK kinase, but not by ERK1 (MAP) kinase.157 A recent study has shown that lipopolysaccharide (LPS) can activate casein kinase II and phosphorylation of PU.1 in LPS-stimulated macrophage lines.158 The LPS stimulation of PU.1 transactivation of a reporter construct was abolished by a serine to alanine mutation of residue 148, suggesting that stimulation of macrophages with LPS leads to increased phosphorylation and activity of PU.1. The same PU.1 phosphorylation site has been shown to be important for B-cell gene activation (see above). A recent study implicated PU.1 in M-CSF–induced proliferation of murine macrophages, and this effect was abrogated by mutating two potential phosphorylation sites (Ser41 and Ser45) in the transactivation domain of PU.1 that are distinct from the postulated site of LPS stimulation at Ser148.159 Finally, a recent study suggested that there may be differences in PU.1 phosphorylation between myeloid cells and B cells, but not during myeloid commitment of multipotential progenitors.160
A number of studies have addressed PU.1 function in myeloid cells by inhibiting its function. Addition of competitor oligonucleotides to human CD34+ cells specifically blocked myeloid CFU formation.44 The inhibition was only observed if the competitors were added very early during GM-CSF–induced myeloid maturation, before the upregulation of PU.1 expression. Several groups have addressed the function of PU.1 on murine development using targeted disruption of the PU.1 gene.26,161 One group reported that PU.1 −/− embryos died in utero, usually at day E16.26 These animals demonstrated a variable anemia, but no production of any type of white blood cells, including monocytes, neutrophils, and B cells, as might be expected based on the expression of PU.1 and its known target genes. The concomitant failure to produce T cells in the −/− animals was surprising, given that PU.1 has not been known to be expressed in this lineage (see above). These studies suggested that the defect was either due to a block of a very early multilineage progenitor cell or that T-cell development is dependent on the presence of macrophages and/or B cells.26
A second PU.1 knockout animal yielded a phenotype with some distinct differences.161 The PU.1 −/− animals in this case were viable at birth and could be kept alive for days by housing them in a sterile environment with the administration of antibiotics. These animals also lacked monocytes and mature B cells, but were capable of producing B-cell progenitors. Several days after birth, T cells and cells resembling neutrophils in the peripheral blood were detected. In this case, the findings suggested a role for PU.1 in B-cell and monocytic development, which is more in keeping with its known pattern of expression and its known gene targets. Although neutrophilic cells were observed, they were Mac-1–negative and reduced in number, suggesting that targeting PU.1 results in a partial defect in neutrophil development. The reason for these marked differences between the two knockout phenotypes is not known at this time, but will be important to determine in that the former knockout model suggests that PU.1 plays an important role in multipotential cells,26 whereas the latter suggest a more limited role in B-cell and myeloid development.161 In vitro differentiation of PU.1 −/− ES cells does not produce macrophages and points to the M-CSF receptor as a major target for PU.1 in understanding its effect on myeloid differentiation.162 163
Consistent with the finding that PU.1 has a major role in myeloid development has been the identification in the past three years of multiple myeloid genes regulated by PU.1 (Table 2). In transfection studies, PU.1 regulates a number of genes that appear as late markers of myeloid cells (Table 2). In addition, PU.1 sites are critical for the activity of a number of myeloid CSF receptor promoters, including the M-CSF receptor,117,164 the GM-CSF receptor α,54 and the G-CSF receptor promoter.53 Quantitative studies will be needed to establish whether hematopoietic cells in PU.1 knockout animals have decreased or absent expression of these three myeloid CSF receptors. If so, the role of expression of these receptors in myeloid development can be tested by restoring their expression singly or in combination in these PU.1 −/− animals. As noted above, studies in PU.1 −/− ES cells have confirmed that CD11b and the M-CSF receptor are major targets for PU.1.162,163 In addition to transactivating the promoters of the genes cited above, it is possible that PU.1 has other mechanisms of action. Recently, it was noted that PU.1 (but not Spi-B or other Ets factors) can bind RNA, suggesting that it could possibly play a role in posttranscriptional regulation.165 The biologic significance of these observations remains to be determined.
Although PU.1 was first isolated as a gene activated in murine erythroleukemia,37 so far it has not been implicated in the pathogenesis of human leukemias. The PU.1 gene has been mapped to 11p11.22, not a frequent site of chromosomal translocations found in human leukemia. PU.1 expression in leukemic myeloid lines is not higher than that observed in primary myeloid cells,95 and a recent survey of primary human leukemias found no significant discordance between PU.1 expression in leukemic samples and that predicted by the apparent differentiation stage of the leukemic cells (M.T. Voso, unpublished observations). However, no studies to date have addressed whether PU.1 is mutated in human leukemias.
Other Ets factors and myeloid development.Spi-B was isolated by hybridization with a PU.1 cDNA probe and, together with PU.1, forms a distinct subfamily within the Ets factors.130 In contrast to other Ets proteins,44 Spi-B can bind to many of the targets for PU.1 and can transactivate many of these myeloid genes.55,95 However, the observation that PU.1 knockout mice failed to produce certain myeloid lineages suggested that Spi-B might be expressed quite differently from PU.1. This was confirmed by studies showing that Spi-B is not expressed at significant levels in myeloid cells, suggesting that its role in myeloid differentiation and function is likely to be limited.55,95 Recently, it has been shown that the binding specificity of Spi-B is very similar but not identical to that of PU.1,118,166 and perhaps this can also contribute to the differences in function of these two factors. In addition, Spi-B and PU.1 are phosphorylated by different kinases.157
To date the role of other Ets factors in myeloid development has not been well defined. A number of other Ets factors are expressed in myeloid cells, including Ets-2, Fli-1, and Elf-1167,168 (D.E.Z. and D.G.T, unpublished observations). Ets-2 is capable of transactivating the murine M-CSF receptor promoter,169 but targeted disruption of the Ets-2 gene did not affect the ability of ES cells to form macrophages in vitro,163 suggesting that Ets-2 does not play a necessary role in myeloid development. Ets-2 may play an important role in activation or signaling in macrophages.107,156,164 The ubiquitous Ets protein GABP is important for the activity of the CD18 protein and can functionally cooperate with PU.1.170 GABP from myeloid cells can also bind to the neutrophil elastase promoter and cooperates with c-myb and C/EBP to transactivate this promoter in nonmyeloid cells (A. Rosmarin, unpublished observations). Recently, another ubiquitously expressed Ets protein, TEL, was isolated from a patient with chronic myelomonocytic leukemia as a fusion protein with the platelet-derived growth factor (PDGF) receptor.171 TEL can also form a fusion protein with AML1 in a large percentage of cases of pre-B–cell acute lymphoblastic leukemia (ALL).172 At this time, the precise role of TEL in myeloid development and in the pathogenesis of myeloid leukemia is not known.173
C/EBP, A REGULATOR OF GRANULOCYTIC DEVELOPMENT
C/EBP (CCAAT/enhancer binding protein; now known as C/EBPα) was isolated as a rat liver protein that bound to viral enhancer sequences174,175 and is a member of one of several leucine zipper transcription factor families, including the fos/Jun family and the ATF/CREB family.176-178 C/EBPα binds as a homodimer or heterodimer with other C/EBP proteins or other transcription factors and has been shown to regulate a number of hepatic and adipocyte genes. C/EBPα is upregulated and plays an important role in adipocyte differentiation and in this cell type functions in fully differentiated, nonproliferative cells; inhibition of C/EBPα blocks differentiation; and expression of C/EBPα induces differentiation.179,180 Indeed, several studies have indicated that C/EBPα can act as a general inhibitor of cell proliferation and in this respect resembles a tumor suppressor.181-183 C/EBPα is part of a family of leucine zipper transcription factors, including C/EBPβ and C/EBPδ, that heterodimerize and are differentially regulated during adipocyte differentiation.179,184,185 Another very interesting C/EBP gene, C/EBPε, was very recently isolated and shown to be preferentially expressed in myeloid cells.186,187 Both C/EBPα and C/EBPβ have single mRNAs that can encode transcriptionally active and repressive forms, depending on the usage of alternative start AUG codons in the same reading frame.188,189 In addition, another C/EBP family member, CHOP-10, can dimerize with C/EBP proteins to inhibit transcriptional activation and is found at a translocation breakpoint in liposarcomas.190,191 Of note is that CHOP can specifically block C/EBPα activity and induce increased apoptosis of myeloid 32D cells.191
In murine adipocyte cell lines, C/EBPα is transcriptionally upregulated (and C/EBPβ is downregulated) with adipocyte differentiation and binds to its own promoter.192 In addition, c-myc overexpression can block differentiation of adipocytes, possibly by repressing the C/EBPα promoter.193 Interestingly, although both murine and human C/EBPα promoters are autoregulated, the mechanisms of autoregulation are different. The murine C/EBPα promoter has a binding site for C/EBPα.192 The human C/EBPα promoter does not conserve the C/EBPα binding site found in the murine promoter, and the autoregulation is indirect and mediated by the action of C/EBPα on a helix-loop-helix protein, upstream stimulatory factor (USF).194 Studies of C/EBPα promoter analysis in myeloid cells have not been reported.
Whereas the C/EBP proteins are expressed in a number of different tissues, their expression in the hematopoietic system may be limited to myeloid cells. It has shown that C/EBPα is specifically expressed in human myelomonocytic cell lines and not in human erythroid, B-cell, and T-cell lines,195 consistent with the myeloid-specific expression previously noted for avian C/EBP.196-199 In studies of murine 32D cells and human leukemic lines, such as HL-60 and U937 cells, the C/EBP proteins showed a pattern of expression quite different from that observed in adipocytes.195,200 In these studies, C/EBPα was observed to be highly expressed in proliferating myelomonocytic cells upon induction of differentiation and downregulated with maturation, whereas C/EBPβ and C/EBPδ were upregulated. However, recent Northern blot analysis of human primary CD34+ cells shows that C/EBPα expression is maintained during granulocytic differentiation but is markedly downregulated with monocytic or erythroid differentiation. In addition, Northern blot analysis of mature peripheral blood neutrophils shows very high levels of C/EBPα mRNA, which was completely undetectable in adherent peripheral blood monocytes (H. Radomska and D.G.T., manuscript submitted). In addition, C/EBPα was noted to be upregulated as single CD34+/CD38− cells were differentiated into granulocytic colonies; no such upregulation was observed during colony-forming unit-macrophage (CFU-M) colony formation.134 These studies point to a role of C/EBPα in neutrophilic but not monocytic lineage development.
As noted above, C/EBPα was the first protein noted to have the leucine zipper motif.176,201 The leucine zipper consists of repetitive leucine residues spaced every 7 amino acids that results in an α helical amphipathic structure with hydrophobic and hydrophilic faces on each side of the helix. Just amino terminal to the zipper is a basic region that is highly positively charged and directly interacts with DNA. The leucine zipper is directly involved in homodimerization and heterodimerization. C/EBPα, C/EBPβ, and C/EBPδ are strongly similar in their C terminal basic region and leucine zipper domains and diverge in the N-terminal transactivation domain.61,179,184,202,203 Adding to the complexity, other DNA binding proteins, including CTF/NF-1204 and NF-Y (also known as CBF and CP1),205-207 can bind to CAAT containing sequences. C/EBP binding sites consist of two half sites, but even the sequence CCAAT was not always required for binding. Despite the high degree of similarity of the carboxyl terminal basic-zipper DNA binding domain, C/EBPα and C/EBPβ can bind with vastly different affinities to the same promoter site.208 The consensus binding site for C/EBPα has been recently determined using PCR binding site selection.209
In addition to other members of the C/EBP family, the C/EBP proteins can interact with a number of transcription factors, including NF-κB and Rel proteins,210,211 members of the CREB/ATF family,98,178 Sp1,212 RB,213 and members of the fos/jun zipper family.214 The amino terminal region of C/EBPα has been shown to physically interact with TBP.61 As noted above, C/EBPδ has been shown to interact with PU.1,142 and PU.1 can physically interact with C/EBPα83 and C/EBPβ as well (P. Auron, unpublished observations). As noted above, C/EBPα and C/EBPβ are most similar in their carboxyl terminal basic-zipper DNA binding domain and less so in the amino terminus, perhaps explaining why they can also show very different abilities to interact with other proteins.212 Another functionally important interaction of relevance to myeloid gene regulation is that between C/EBPα and AML1, which is important for M-CSF receptor promoter function in transfection studies (see below).215 Recently, it has been shown that the underphosphorylated form of the retinoblastoma (RB) protein can interact physically with C/EBP proteins to increase DNA binding and transactivation of target genes during adipocyte differentiation.216 As RB becomes underphosphorylated with myeloid differentiation,148 149 it could positively regulate C/EBPα-induced granulocytic maturation.
Recently, lines of mice harboring a knockout of the C/EBPα gene were generated.217,218 Heterozygous mice are outwardly normal, but homozygous mice die within the first few hours after birth of impaired glucose metabolism; their viability can be extended to about 1 day with injections of glucose. The mice are unable to properly synthesize and mobilize glycogen and fat. Analysis of the hematopoietic system in embryonic and newborn mice has demonstrated a significant defect in production of granulocytic cells.28 Newborn knockout (−/−) animals do not produce any mature neutrophils, which comprise 90% of the peripheral blood white blood cells of newborn heterozygote (+/−) or wild-type (+/+) mice. Cells that appear to be immature myeloid cells are observed in the peripheral blood. These cells stain with Sudan black, a characteristic of myeloid cells, and have increased myeloid CFU potential compared with those from heterozygote blood. Interestingly, many cells in the CFU from the knockout blood are immature in morphology as well, similar to what is observed in CFU from leukemic cells. Eosinophils are also not observed, but all of the other lineages, including peripheral blood monocytes and peritoneal macrophages, red blood cells, platelets, and lymphoid cells, appear quantitatively unaffected. Fluorescence-activated cell sorting (FACS) analysis in embryonic and newborn animals confirmed that myeloid markers (Mac-1 and Gr-1) were greatly reduced, with normal B- and T-cell subsets. Expression of the G-CSF receptor mRNA was profoundly and selectively reduced, whereas that of M-CSF receptor and GM-CSF receptor α, βc, and βIL3 were all comparable to wild-type. Interestingly, a fourfold increase in mRNA for Epo receptor was detected, although whether this is due to a negative effect of C/EBPα on Epo receptor expression in precursor cells or alternatively a reflection of an increased number of erythroid cells in C/EBPα −/− livers has not been determined. Compared with wild-type animals, maturation of immature myeloid cells in vivo in response to G-CSF was not observed, as assessed by both morphology and FACS analysis of Gr-1 expression. In addition, no colony-forming units-granulocyte (CFU-G) could be obtained using C/EBPα −/− newborn liver, although other types of colonies (colony-forming unit granulocyte, erythroid, monocyte, megakaryocyte [CFU-GEMM], colony-forming unit-erythroid [CFU-E], colony-forming unit–granulocyte-macrophage [CFU-GM], and colony-forming unit-macrophage [CFU-M]) were quantitatively equal to wild-type animals. These findings suggested that much of the phenotype may be due to decreased or absent G-CSF signaling due to markedly reduced receptor levels. However, G-CSF receptor knockout animals produce mature granulocytes, in contrast to C/EBPα knockouts, strongly suggesting that there must be important C/EBPα target genes in myeloid progenitors in addition to the G-CSF receptor.219 Transplantation of C/EBPα fetal liver into sublethally irradiated animals resulted in detection of T cells but not myeloid cells of donor origin, showing that the defect is indeed in the hematopoietic cells.28
A number of studies have pointed to the role of C/EBPβ in myeloid cells. The avian homolog of C/EBPβ (NF-M) is a myeloid-specific factor that activates the promoter for cMGF, which is related to G-CSF and IL-6.196-199 In mammalian cells, C/EBPβ has been described as having low activity until activated by LPS, IL-1, IL-6, and other inflammatory mediators, which induce its translocation to the nucleus, where it can activate cytokine genes such as IL-6 and G-CSF.185,220 Changes in C/EBPβ phosphorylation (but not in C/EBPα) have also been described during myeloid differentiation of a progenitor cell line.160 Human C/EBPβ is also known as NF-IL6.185 C/EBPβ can also be activated in other cell types by phosphorylation without translocation.221 In one report, targeted disruption of C/EBPβ leads to defects in killing of Listeria and tumor cells by macrophages.222 In a second study, disruption of C/EBPβ led to an increase in IL-6 production (previously it was thought that C/EBPβ, or NF-IL6, was essential for IL-6 production) and hyperproliferation of a number of different hematopoietic lineages, a syndrome resembling Castleman's disease.223 Knockout animals from both of these studies can get this syndrome as the animals get older, but it is not seen in young mice kept in a sterile environment.224 In these younger mice, in the absence of inflammation, myelopoiesis was apparently not adversely affected, consistent with a role of C/EBPβ in cytokine activation rather than lineage development.
As noted above, C/EBPβ (NF-IL6) regulates a number of cytokine genes and may contribute to normal myeloid development as well as activation of mature myeloid cells in response to an inflammatory stimulus. In particular, targeting of C/EBPβ resulted in a reduction of G-CSF production in macrophages but not other cell types.222 C/EBPα has been noted to regulate the promoters for several myeloid granule proteins, including myeloperoxidase and neutrophil elastase.87,88,160 In addition, C/EBP sites, like PU.1 sites, are critical for the activity of a number of myeloid CSF receptor promoters, including the M-CSF receptor,117 the GM-CSF receptor α,54 and the G-CSF receptor promoter.53 In the C/EBPα knockout mice, G-CSF receptor mRNA was selectively and significantly reduced.28 Interestingly, no reduction was noted in the levels of M-CSF receptor and GM-CSF receptor (α and β chain) mRNA, suggesting that perhaps other factors (such as C/EBPβ) can compensate for C/EBPα in some but not all instances.
With respect to the possible role of C/EBP proteins in leukemia, the C/EBP proteins have not been implicated as transforming proteins, with the exception of CHOP10.190,225 Indeed, C/EBPα has been shown to inhibit cell proliferation in fibrosarcoma lines through the cyclin-dependent kinase inhibitor p21183 and can act as a tumor suppressor of hepatoma lines and other cell types.181,182 The human C/EBPα gene maps to chromosome 19q13.1 and C/EBPβ maps to 20q13.1.226 Neither of these is a frequent site of chromosomal translocations found in human myeloid leukemia. Studies of C/EBP expression in primary human leukemias have not been reported. However, given the phenotype of the C/EBPα knockout mouse, in which there appears to be a block in granulocyte maturation, it will be of interest to look for abnormalities of C/EBP expression or mutations in myeloid leukemias, which also represent states with a block in maturation.
In summary, the C/EBP proteins are differentially expressed and can form many interactions with themselves and other transcription factors. These studies show that C/EBPβ (and C/EBPδ) may be involved in activation of cytokines in mature cells, whereas C/EBPα has a major role in granulocytic maturation through regulation of the G-CSF receptor and other as yet not identified target genes.
Other factors binding to CCAAT sites and myeloid development.In addition to the C/EBP proteins, the CCAAT displacement protein (CDP) has also been implicated in myeloid development. This protein was observed to negatively regulate genes by binding to a CCAAT box and displacing the binding of positively acting factors.227 It was subsequently shown to be capable of binding to the gp91phox promoter and repressing this cytochrome component gene expressed specifically in myeloid cells.228,229 At this time no characterization of the positively acting factors that are presumed to be displaced at this site has been made. CDP is expressed at highest levels in undifferentiated myeloid cell lines and is downregulated with differentiation, providing a mechanism for the upregulation of gp91phox observed with myeloid maturation. Importantly, this is one of the few examples explaining the potential mechanism of upregulation of myeloid targets with myeloid differentiation. In most other cases, the mechanism of upregulation is largely unknown. Recently, it has been shown that CDP can also repress the lactoferrin promoter.230 In these studies, overexpression of CDP in myeloid stem cells blocked lactoferrin expression after G-CSF–induced neutrophil maturation without blocking phenotypic maturation. Furthermore, these investigators observed coordinate repression of multiple secondary granule protein genes, including lactoferrin, neutrophil collagenase, and neutrophil gelatinase, suggesting that CDP repression is a common mode of regulation of this class of myeloid promoters (N. Berliner, unpublished observations). Other factors might also act in a similar manner to repress positive activators during differentiation. For example, a factor that is downregulated during adipocyte differentiation and inhibits C/EBPα function, presumably through its similarity to carboxypeptidase, has recently been described,231 but it is not known if it inhibits C/EBPα in hematopoietic cells.
AML1 AND THE CBF FAMILY
An example of a transcription factor that plays an important role in normal monocytic gene expression and in acute myelogenous leukemia is AML1. AML1 is a member of the core binding factor (CBF) or polyoma enhancer binding protein 2 (PEBP2) family of transcription factors. The CBF factors consist of heterodimers between DNA binding α subunits and a β subunit (CBFβ) that does not bind DNA directly but that enhances the binding of the α subunit.232,233 Multiple α subunit genes, including CBFA1, AML1 (CBFA2), and CBFA3, have been detected,233-237 although the role of these other AML1 family members in myeloid development and leukemia is not clear. In addition, alternatively spliced isoforms of the α and β subunits have been detected. In particular, a short form and two long forms of AML1 have been described, and it is the long form that appears to include the transactivation function.235-238 All of the CBFα proteins contain a runt domain, which is similar to the Drosophila pair-rule gene, runt, an early acting segmentation protein that regulates the expression of other segmentation genes.239,240 This runt homology domain (rhd) confers the ability of AML1 to bind to its consensus sequence, TGT/cGGT, and the ability of AML1 to interact with its cofactor CBFβ, which enhances the DNA binding ability.232,233 241
AML1, normally expressed in hematopoietic tissues and during myeloid differentiation,134,237,242 is also expressed in nervous tissue, skeletal muscle, and reproductive tissues as well.243 Knockout studies of AML1 have determined that AML1 is necessary for normal development of all hematopoietic lineages,49,50 and similar effects have been observed with targeted disruption of the CBFβ subunit.244 In these studies, primitive hematopoiesis was apparently normal, but severe impairment of definitive hematopoiesis was observed both in the animals and during in vitro differentiation of −/− ES cells, with defects observed in all lineages examined.
AML1 may act to facilitate the action of other adjacent transcription factors. The AML1 site in the M-CSF receptor promoter is located between the C/EBPα and PU.1 sites, two transcription factors that are important for myeloid gene expression.52,215,245 Significantly, although the C/EBP site is critical for M-CSF receptor activity in transfection experiments,52 C/EBP alone does not transactivate M-CSF receptor promoter activity. The adjacent factor AML1, along with CBFβ, can by itself activate the M-CSF receptor promoter sixfold, but the synergistic activation by C/EBP added to AML1 and CBFβ is much greater (>60-fold). All C/EBP proteins share a highly similar bZIP domain,179,184 and AML1 interacts with the bZIP fragment of C/EBP, explaining why C/EBPα, C/EBPβ, and C/EBPδ can all synergistically activate the M-CSF receptor promoter with AML1. The physical interaction between C/EBP and AML1 could either stabilize binding of both to the M-CSF receptor, or their interaction might provide a surface for another transcription factor to bind and activate the M-CSF receptor promoter. Other examples of interactions of AML1 and other transcription factors include synergism with Ets-1 and c-myb to activate T-cell receptor enhancers.246-249 Because Ets factors, such as PU.1, and c-myb both play an important role in myeloid development, it is likely that these interactions with AML1 will be important in myeloid gene expression as well.
AML1 can be phosphorylated at two serine residues within a proline-, serine-, and threonine-rich region in its carboxyl terminal domain after activation of the extracellular signal regulated kinase (ERK1).250 These serine residues are critical for AML1-dependent transformation of fibroblast cell lines and ERK1-dependent transactivation of a T-cell receptor enhancer target gene. IL-3 treatment of the hematopoietic cell line BaF3 results in phosphorylation of AML1, suggesting that this signalling pathway may play an important role during hematopoietic development.
Role of AML1 in leukemogenesis.AML1 was identified by studying one of the most frequent chromosomal translocation found in AML, t(8; 21)(q22; q22).17,39,241,251,252 AML1 fusion proteins are also involved in other forms of leukemia, including t(3; 21) therapy-related acute myeloid leukemia/myelodysplasia or chronic myelogenous leukemia in blast crisis (AML1/MDS1, AML1/EAP, and AML1/Evi-1) and also the t(12; 21) in childhood B-cell acute lymphoblastic leukemias (TEL/AML1).17,172,251,253-256 The β subunit of CBF, CBFβ, is also involved in a chromosomal inversion, inv(16)(p13; q22), associated with French-American-British (FAB) M4eo AML.257 Therefore, each of the two chains of the CBF heterodimer is directly implicated in the pathogenesis of AML.
As noted above, AML1 is involved in the (8; 21) translocation, associated with the FAB M2 form of AML, which results in the production of the AML1/ETO fusion protein. The mechanism by which AML1/ETO accomplishes leukemic transformation is unknown. In this case, the 5′ part of the AML1 gene, including the amino terminal DNA binding runt domain but lacking the carboxyl terminal activation domain, is fused to almost the entire ETO gene on chromosome 8. Recent studies have indicated that this AML1/ETO fusion protein can act as a dominant negative inhibitor of AML1 transactivation of such AML1 targets as the T-cell receptor β (TCRβ) enhancer.236,238,258 It has recently been reported that the t(12; 21) fusion protein, TEL/AML1, also causes the same interference with AML1 transactivation of the TCRβ enhancer.259 Therefore, the expression of some of the normal AML1 gene targets that play a key role in monocytic differentiation might be adversely affected by AML1/ETO, and these include GM-CSF,236,258 IL-3,260,261 M-CSF receptor,52 and myeloperoxidase and neutrophil elastase.87 262
Recently, it was shown that AML1/ETO and AML1 work synergistically to transactivate the M-CSF receptor promoter, thus exhibiting a different activity than previously described.263 This indicates that AML1/ETO not only can repress, but also can enhance AML1 transactivation of its target genes. Endogenous M-CSF receptor expression was examined in Kasumi-1 cells, derived from a patient with AML-M2 t(8; 21) and the promonocytic cell line, U937. Kasumi-1 cells exhibited a significantly higher level of M-CSF receptor expression than U937 cells. Bone marrow from patients with AML-M2 t(8; 21) also exhibited a higher level of expression of M-CSF receptor compared with normal controls.263 Signaling through the M-CSF receptor promotes cell proliferation by inducing the G1/S transition because of the effect on cyclins D and E.264,265 This effect on proliferation can be tied to the oncogenic nature of the M-CSF receptor, which was first discovered as the cellular counterpart to the transforming oncogene, v-fms.266 Overexpression of the M-CSF receptor causes transformation of NIH3T3 cells and in mice leads to a myeloblastic leukemia.267 268 A premature upregulation of M-CSF receptor expression by AML1/ETO could be one of the factors contributing to leukemogenesis AML.
To study how the fusion protein produced by the t(8:21) translocation leads to the development of myeloid leukemia, knock-in mice have been generated that express the AML1-ETO fusion protein. Mice heterozygous for the AML1-ETO allele die in midgestation with hemorrhaging in the central and peripheral nervous system and exhibit a severe block in fetal liver hematopoiesis.269 This phenotype is very similar to that resulting from homozygous disruption of the cbfα2 (AML1) and cbfβ genes.49,50,244 However, significant differences between AML1-ETO knock-in and cbfα2 −/− or cbfβ −/− mice were observed in hematopoietic CFU assays. Yolk sac from AML1-ETO knock-in mice generated solely homogenous macrophage colonies. The total number of macrophage colonies is similar to the sum of all type of colonies from yolk sacs of wild-type mice. In contrast, no colonies from cbfα2 −/− or cbfβ −/− mice could be detected. These results, like those noted above in which the AML1-ETO fusion protein activates the M-CSF receptor promoter, indicate that the AML1-ETO fusion protein affects hematopoiesis in a manner that is not simply a block of wild-type AML1 function, and these effects could be important in leukemogenesis. Mice heterozygous for a knock-in of the cbfβ-myh11 allele, which mimics the inv(16)(p13; q22) associated with AML-M4Eo form of AML, also die in midgestation from the same pattern of central nervous system hemorrhaging and impairment of fetal liver hematopoiesis. However, instead of preferential macrophage differentiation from early progenitor cells, these embryos exhibit a significant delay in maturation of yolk sac-derived primitive erythrocytes.270 Although t(8:21) and inv(16) disrupt genes encoding different subunits of a single transcription factor complex, the fusion genes have different effects on hematopoiesis, consistent with their association with distinct AML subtypes.
RARα: MYELOID DIFFERENTIATION AND ACUTE PROMYELOCYTIC LEUKEMIA (APL)
Several lines of evidence indicate a key role for the retinoic acid receptor (RAR) in myeloid differentiation. Vitamin A deficiency is associated with defective hematopoiesis,271 and retinoids stimulate granulopoiesis.272,273 Retinoic acid (RA) can induce differentiation of the HL-60 cell line274 and of primary leukemic cells.275 There are three genes encoding RARs; RARα, RARβ, and RARγ (reviewed previously276-278 ). RARα is preferentially expressed in myeloid tissue.279-281 RARα binds to retinoic acid response elements consisting of a direct repeat (A/G)G(G/T)TCA separated by 2 or 5 nucleotides as a heterodimer with the retinoid X receptor RXR282-284 and stimulates transcription in response to all-trans retinoic acid (ATRA).285 RARα was mutated in a retinoid-resistant HL-60 cell line with this deficit corrected by re-expression of wild-type receptor.286-288 Furthermore, introduction of a dominant negative form of RARα into a multipotent hematopoietic cell line changed the fate of these cells from the granulocyte/monocyte lineage to the mast cell line.289 In addition, GM-CSF–mediated myeloid differentiation of these cells was blocked at the promyelocyte stage.45 This basic information was complimented by clinical data indicating that patients with APL could be induced into complete remission with ATRA (reviewed previously19,290 ). It was found that APL associated with t(15:17) disrupted the RARα linking it to the PML gene, yielding the fusion protein PML-RARα.291-296 PML-RARα is an aberrant retinoid receptor with altered DNA binding and transcriptional activities.292,294,296-298 PML-RARα can block the effect of wild-type retinoic acid receptor on many promoters in a dominant negative manner. Two other APL-associated rearrangements of the RARα gene were identified: translocation (11; 17)40,299 fusing the PLZF gene to RARα and t(5; 17)300 linking the nucleophosmin (NPM) gene to RARα. All three situations yield similar chimeric RARα proteins, yet only t(15; 17) patients clearly respond to ATRA therapy.301,302 Expression of the PML-RARα fusion protein in myeloid cells inhibits differentiation in the presence of low levels of ATRA303,304 and actually accelerates differentiation in the presence of pharmacologic doses of RA.304 Expression of PML-RARα in transgenic mice impairs myelopoiesis,113 and work from another group indicated that PML-RARα could induce altered myeloid development, including increased immature and mature myeloid cells, as well as a leukemic syndrome that is ATRA responsive.115 Another group has recently described similar results in PML-RARα transgenic animals using another promoter.109 Together, the experimental and clinical data indicate that RARα function is required to fully proceed through myeloid differentiation. Disruption of RARα function likely selects for the promyelocyte phenotype. However, the nature of the fusion partner and its molecular mechanisms of action apparently play a role in the ability of the leukemic cells to differentiate in response to ATRA.
ATRA treatment drives the expression of multiple myeloid genes, including those for cell surface adhesion molecules, intrinsic host defense systems, extrinsic cytokines, colony-stimulating factor receptors, structural proteins, and enzymes. Some of these are induced with some delay after ATRA treatment, making them unlikely primary targets of RARα. The few identified direct targets of RARα tend to themselves be transcription factors, including the RARs themselves.284 ATRA treatment of fresh APL cells upregulates RARα, correlating with the presence of a RARE in RARα promoter.305,306 Therefore, one way that ATRA may induce myeloid differentiation may be to upregulate the RARα gene, overcoming the dominant negative PML-RARα protein. A recently identified direct target of RAR in myeloid cells is the cyclin-dependent kinase inhibitor p21.307 Another tantalizing set of targets for RARα in myeloid differentiation includes hox family of homeobox-containing transcription factors, which are expressed in myeloid cell lines in a coordinated, dynamic manner.32,33,308-313 In embryonic carcinoma cells homozygous for a targeted disruption in the RARα gene, ATRA treatment fails to induce the hoxb1 and CRABPII expression, indicating that these are true direct targets of RARα.284,314 Identification of further direct target genes of RARα during myeloid differentiation will require use of techniques such as differential screening315 or differential display316 of myeloid cells treated for brief periods with ATRA.
Finally, although the RARα locus and all of the other nuclear receptors have been disrupted by gene targeting in mice (reviewed previously317 ), no major defects in hematopoiesis were described in these animals.318-320 This may reflect the ability of the receptors to compensate for each other in a redundant manner. This is analogous to the way alternative forms of the RAR could rescue mutated HL-60 cells with a RARα mutation.288 Multiple matings of animals deficient in the various RAR, RXR, and other nonsteroid nuclear receptors may yield further insights as to the exact requirements for this class of transcription factor in hematopoiesis.
THE ZINC FINGER FAMILY AND MYELOID CELLS
Transcription factors belonging to the zinc finger family are among the most numerous in vertebrate organisms. These proteins contain cysteine and histidine residues that coordinately bind zinc ions to form a protein loop capable of specifically interacting with DNA.321 The Drosophila Krüppel-related zinc finger proteins are distinct from other Cys/His finger proteins because they contain a relatively invariant amino acid sequence linking the zinc fingers of the form HTGEKP(F/Y)XC. This H/C link is present in a number of zinc finger transcription factor proteins of higher eukaryotes, including Sp1,322 the EGR family of transcriptional activators,323 and the WT-1 tumor suppressor protein.324,325 Myeloid-specific zinc finger proteins were identified by their similarity to the Krüppel sequence (MZF-1),30 by subtraction cloning from myeloid cells (Egr-1),25 by their involvement in chromosomal translocation involved in APL (PLZF),299,302 and by their involvement in apoptosis associated with myeloid differentiation (Requiem).326 In addition, a zinc finger gene not normally expressed in myeloid cells was isolated by its activation in murine models of leukemia by retroviral insertion (Evi-1)327 and has subsequently shown to be involved in human leukemia when fused to the AML1 gene.17,328 329 Interestingly, relatively few of these proteins were identified by traditional promoter analysis, with the notable exception of Sp1. On the contrary, these zinc finger proteins are in general factors in search of molecular targets and mechanisms in myeloid differentiation.
The PLZF gene.The promyelocytic leukemia zinc finger (PLZF) gene, potentially important for myeloid development, was identified by its rearrangement in APL with a variant translocation t(11; 17) (q23; q21).40,299,330 The t(11; 17) APL patients were generally resistant to both chemotherapy and differentiation therapy with ATRA,302 suggesting that the nature of the fusion partner with RARα may determine the clinical course of APL. PLZF on chromosome 11 encodes a zinc finger transcription factor,299 whereas the partner protein with RARα in t(15; 17) APL is PML, a RING finger protein of unknown function (reviewed previously19 ). The t(11; 17) fusion yields two reciprocal transcripts expressed in all patients tested: PLZF-RARα, predicted to encode an aberrant retinoid receptor, and RARα-PLZF, fusing the A-activation domain of RARα to the last 7 zinc finger motifs of PLZF.
In the hematopoietic system, PLZF is expressed in myeloid but not in lymphoid cell lines.299 Upon treatment of NB4 or HL-60 cells with retinoic acid, PLZF levels decline (Chen et al299 and A. Chen, S. Waxman, and J. Licht, unpublished data). The murine homologue of PLZF was found to be expressed at highest levels in multipotent progenitor cell lines such as FDCP-MixA4 and at lower levels in committed cell lines.331 When the multipotent FDCP-MixA4 cell line was induced to differentiate, PLZF levels decreased. Murine PLZF mRNA was also detectable in pro-B–cell lines but not in more mature B- or T-cell lines.331 PLZF was not expressed in undifferentiated embryonic stem cells but was upregulated as ES cells were induced to differentiate into embryoid bodies. In the developing mouse embryo, PLZF is expressed in the aorta, gonadal, and mesonephros region (AGM), a region thought to give rise to hematopoietic stem cells. Finally, CD34+ human progenitor cells could be immunostained with PLZF antisera in a distinct nuclear speckled pattern.331 Together, this information suggests that PLZF may be a marker of and a critical gene for the early hematopoietic stem cell. Downregulation of PLZF may be a necessary step for the committed division and differentiation that accompanies normal bone marrow maturation.
By immunoprecipitation of transfected cells, the PLZF protein was identified as an 81-kD nuclear species331,332 phosphorylated on serine and threonine but not tyrosine residues (R. Shaknovich and J. Licht, manuscript in preparation). A biphenotypic T-cell/macrophage line, presumably representing a primitive transformed cell, expressed high levels of PLZF mRNA and 81-kD PLZF protein when treated with the calcium ionophore A23187, correlating with the induction of a more monocytoid phenotype. Immunofluorescent confocal microscopy showed that PLZF was localized to approximately 50 small nuclear subdomains, whereas the PML protein of t(15; 17) APL was localized in approximately 5 to 10 larger nuclear structures (nuclear bodies or PODs333-335 ) and was present before and after A23187 treatment. It is uncertain if in other cell types these two APL-associated proteins might be localized to the same nuclear domains.
PLZF is a sequence-specific DNA binding protein recognizing a sequence with a core of TAAAT.336 This site was cloned upstream of a reporter gene and was found to mediate transcriptional repression by PLZF. At least one repression domain within PLZF maps to an evolutionarily conserved sequence known as the POZ (poxvirus-zinc finger) or BTB (broad complex-tramtrack-bric-a-brac) domain.337,338 This sequence also mediates self-association by PLZF and is required for the localization of PLZF into a speckled subnuclear pattern.332 339 Whether the domains that mediate homodimerization and transcriptional repression are identical is not known. The last 7 zinc fingers of PLZF that are retained in the RARα-PLZF fusion protein can bind to the same site as the 9 zinc fingers of PLZF, suggesting that the RARα-PLZF protein could act as a dominant negative protein binding and altering transcription of PLZF target genes.
The PLZF-RARα fusion protein has properties quite similar to the PML-RARα fusion protein of t(15; 17)-associated APL. Both fusion proteins can bind as homodimers to a retinoic acid response element (RARE).297,332,339 Homodimerization by PML is mediated by a coiled-coil motif,297 whereas in PLZF the POZ domain mediates self-association.339 In an electrophoretic mobility shift assay (EMSA) both PML-RARα and PLZF-RARα homodimers are chased into novel complexes of higher and lower electrophoretic mobility by addition of RXRα. This suggests that both fusion proteins may work in a similar manner, forming multimeric complexes in the cell that could act to sequester limiting amounts of RXRα, the essential cofactor for RARα function, in the cell. Both PML-RARα and PLZF-RARα can act in a dominant negative manner to inhibit the activity of wild-type RARα294,296,298 and other nuclear receptors such as the vitamin D3 receptor (J. Licht and M. English, unpublished data).297 This effect can be partially relieved by overexpression of RXRα, consistent with the notion that the aberrant receptors generated in APL block myeloid differentiation at least in part through limiting the ability of RARα to bind to its targets in combination with RXR. Dominant negative forms of RARα can block myeloid differentiation at the promyelocyte stage.45,289,340 This correlated with the clinical finding that RARα is rearranged in three translocations, t(15; 17), t(11; 17), and t(5; 17), all yielding APL.300 341 Thus, the block in ATRA-mediated signaling may help select the APL phenotype but may not necessarily determine whether the disease is ATRA-responsive. Further studies of the growth-inhibiting properties of PLZF suggest a reason why t(11; 17) APL may not respond to therapy.
Pools of murine myeloid 32DCL3 cells transduced with a retrovirus to overexpress the PLZF protein were significantly growth inhibited when grown in IL-3 or GM-CSF and retarded in the G1 phase of the cell cycle. In addition, the cells exhibited a higher rate of programmed cell death and a reduced ability to differentiate in response to G-CSF or GM-CSF.342 The high level of expression of PLZF in the hematopoietic precursor cell may play a role in the relative quiescence of these cells. Downregulation of PLZF during myeloid differentiation may be accompanied by cycles of committed cell division. In this way, PLZF does seem similar to PML,343-345 because both proteins can repress cell growth. However, t(11; 17) generates RARα-PLZF, a protein that can potentially interfere with the normal transcriptional functions of PLZF by binding to and dysregulating PLZF target genes. In contrast, the RAR-PML transcript is variably found in t(15; 17) APL19 and may not be able to affect the function of wild-type PML. Consistent with a central role for PLZF disruption in t(11; 17) APL phenotype, RARα-PLZF may confer accelerated growth to 32DCL3 cells. Hence, t(11; 17) may be a resistant disease due to the presence of two oncogenes working through different mechanisms, PLZF-RARα and RARα-PLZF. It remains to be determined what genes are targeted by PLZF and how these play a role in the growth and differentiation of myeloid tissue.
The myeloid zinc finger protein MZF-1.MZF-1 has multiple (13) zinc finger domains, divided in two groups.346 The amino-terminal group has 4 fingers, and the carboxyl-terminal group has 9 fingers. There are acidic regions in the amino-terminal nonzinc finger portion of the protein that may be part of the transcriptional regulatory domain. Interestingly, there is a 24 amino acid glycine-proline–rich link between the first and second zinc finger regions. Glycine-proline–rich domains have also been implicated in transcriptional regulation in a large number of transcription factors, including Oct-3, Rex-1, and CP-1.346
By Northern analysis, MZF-1 was found to be specifically expressed in myeloid cell lines.346 By cRNA in situ hybridization of normal marrow, MZF-1 was found to be expressed in myeloid cells,347 from myeloblasts to metamyelocytes, but in no other marrow cell types, implying a role for MZF-1 in myeloid development. Using in situ chromosomal hybridization, MZF-1 was localized to human chromosome 19q13.4.346 It is the telomeric-most gene on 19q, with its promoter region being less than 10 kb from the telomeric repeats.348 The finding that MZF-1 is located at the telomere of chromosome 19q is significant because there is evidence that aging hematopoietic stem cells lose telomeric size with time.349 In addition, telomeres shorten with transformation of myelodysplasia to leukemia.350 With age, the incidence of stem cell disorders such as myelodysplasia and leukemia increases markedly. It is possible that the regulatory region of MZF-1 or the transcribed sequence is disrupted with age, which could play a role in the increased incidence of hematopoietic stem cell disorders.
The consensus DNA binding sites of MZF-1 were isolated by mobility shift selection of degenerate oligonucleotides and PCR amplification.351 The first finger domain, zinc fingers 1-4 (ZF1-4), bound to a consensus sequence of 5′-AGTGGGGA-3′, whereas zinc fingers 5-13 (ZF5-13) bound to a consensus of 5′-CGGGnGAGGGGGAA-3′. These guanine-rich sites resemble NF-κB binding sites. The full-length recombinant MZF-1 could bind to either sequence. Indeed, the two MZF-1 finger domains, ZF1-4 and ZF5-13, could bind to each other's consensus binding sequence. In addition, it was discovered that ZF5-13 probably needs all 9 fingers to bind to its consensus sequence. Deletion of as few as 3 fingers from either end of ZF5-13 abrogates DNA binding.351
Computer searches of data bases for promoter sequences that contained these consensus MZF-1 binding sites found several possible matches. The promoters of the myeloid genes CD34, myeloperoxidase, c-myb, and lactoferrin all contained consensus MZF-1 binding sites. An intriguing finding about MZF-1 was its bifunctional transcriptional regulatory activity. In the intracellular environment of adherent, nonhematopoietic cells it functions as a transcriptional repressor, and in hematopoietic cells it functions as an activator.352
MZF was found to repress the CD34 promoter and the c-myb promoter in nonhematopoietic cells.352,353 However, in one study using the hematopoietic cell lines K562 and Jurkat, MZF-1 activated transcription from the CD34 promoter.352 In another study, using different techniques, it was found that MZF-1 repressed the CD34 promoter in KG-1 cells.353
The presence of MZF-1 binding sites in several myeloid promoters and the explicit regulation of the CD34 promoter by MZF-1 point out that MZF-1 may have a general role in the regulation of hematopoietic gene expression. Lending evidence to the hypothesis that MZF-1 may be an important regulator of hematopoietic development are studies showing that blocking MZF-1 expression with antisense oligonucleotides inhibits granulopoiesis.347
MZF-1 can also disrupt the normal proliferative behavior of embryonic fibroblasts. When MZF-1 was retrovirally transduced and overexpressed in NIH 3T3 cells, where it is not normally expressed, it rapidly transformed those cells.354 However, two other myeloid transcriptional regulators, PU.1 and PRH, did not transform 3T3 cells when they were similarly overexpressed. In addition, forced overexpression of MZF-1 in IL-3–dependent FDCP1 or HL-60 cells decreased apoptosis induced by IL-3 withdrawal or retinoic acid, respectively.355 356 FDCP1 cells transduced with MZF-1 formed tumors in 70% of congenic hosts, whereas control cells did not. These data imply that the aberrant expression of nodal regulators of development can be neoplastic. It lends evidence for the hypothesis that malignancy represents normal development gone awry.
The MZF-1 promoter was recently isolated.357 There are MZF-1, PU.1, and retinoic acid receptor binding sites within the MZF-1 promoter.
The early response gene Egr-1.Many groups have isolated Egr-1 (NFGI-A) as an early response gene induced by a variety of stimuli, primarily those that induce cell proliferation.358 Egr-1 was also cloned as an early response gene during TPA treatment of human myeloid cell lines and was noted to be upregulated during monocytic and not granulocytic differentiation of these bipotential lines.25 Overexpression and antisense studies implicated Egr-1 as an inducer of monocytic differentiation.25,359 An Egr-1 knockout mouse line has been made, but no abnormalities of monocytic differentiation or function were detected.360 In this mouse model, it is possible that other members of the Egr-1 family can compensate for loss of the Egr-1 gene.
The Wilms' tumor suppressor gene WT-1.An enlarging body of literature suggests a role for the WT1 Wilms' tumor suppressor gene in normal and leukemic myeloid development. WT1 encodes a zinc finger transcription factor that binds to a GC-rich sequence similar to that of the Egr-1 protein (reviewed previously361 ).362 WT1 can both activate and repress transcription and in model systems inhibits cell growth363-365 and can induce programmed cell death.361,366 WT1 is expressed in normal bone marrow, particularly in CD34+ progenitor cells.367,368 CD43+/CD33−/lin− cells, believed to be very early progenitors, express the highest levels of WT1, whereas the CD33+ subset of CD34+ progenitors, reflecting a later point in differentiation, express 100-fold less WT1. WT1 is also expressed in the murine embryonic yolk sac and liver during definitive hematopoiesis.368 WT1 is expressed in a number of myeloid cell lines, and WT1 expression is downregulated in HL-60 and K562 cells at the posttranscriptional level during differentiation of these cell lines.369,370 Given that WT1 can compete for the binding site of Egr-1 and repress transcription and that Egr-1 is an activator that promotes monocytic differentiation,25,359 it is possible that the interplay between these two factors, which have opposing transcriptional effects, may play a role in the tight control of monocytic differentiation. The hypothesized role of WT1 in hematopoiesis must be tempered by the results of Kriedberg et al,371 who performed a targeted disruption of the WT1 gene in the mouse. These animals exhibited complete kidney and genitourinary agenesis, defects in the mesothelium, and cardiac defects, but to date have demonstrated no hematopoietic deficits (J. Kreidberg, personal communication, June 1996).
WT1 mRNA expression has been detected in up to 80% of cases of acute myelogenous leukemia.372,373 Whereas WT1 can be found in the marrow of normal subjects, expression in the peripheral blood is distinctly abnormal and a sign of the presence of leukemic cells.367 In addition, there was found to be a negative correlation between WT1 expression and prognosis.367 Reverse transcription-PCR (RT-PCR) assay for WT1 expression can detect minimal residual leukemic blast cells in leukemia patients and may be more sensitive than the detection of fusion gene products such as the PML-RARα fusion found in APL.367,374 There is an inherent paradox in these findings. WT1 is believed to be a classical tumor-suppressor gene, and yet it is expressed in high levels in actively proliferating leukemia. This expression may reflect the stage of differentiation of the malignant cells at the point of transformation. Alternatively, WT1 may have an oncogenic effect when overexpressed in hematopoietic tissue. Recent data indicate that WT1 expression may be important for leukemic cell growth. When myeloid leukemic cell lines were treated with antisense WT1 oligonucleotides, cellular proliferation was inhibited and programmed cell death was induced.375,376 In addition, fresh human leukemia sample growth was inhibited by these antisense WT1 oligonucleotides.376 Mutation of WT1 may also play a role in leukemogenesis. Initially, a mutation was detected in the remaining WT1 allele of a WAGR patient who had been previously treated for Wilms' tumor.377 This mutation affected a cysteine residue in the zinc finger region and would be predicted to abolish DNA binding by WT1. This suggested that complete loss of WT1 function in the myeloid lineage might be associated with the development of leukemia. More recent data from this same group indicate that 15% of cases of AML have point mutations of WT1.320 These were heterozygous mutations leading to the formation of truncated forms of WT1 devoid of zinc finger DNA-binding motifs. Such truncated proteins may act in a dominant negative manner to inhibit the transcriptional effects of wild-type WT1.378-380 This again suggests that WT1 may play a role in the control of leukemic cell growth.
The mechanism of action of WT1 in affecting myeloid growth is not defined, but one important candidate target gene is c-myb, which contains an Egr-1 site within its promoter. This site acts as a negative regulatory element. WT1 binds to this site in vitro and can repress the c-myb promoter.381 This is intriguing given that forced expression of c-myb blocks myeloid differentiation, and treatment of myeloid cells with antisense c-myb oligonucleotides blocks proliferation.42,382 383 WT1 could therefore affect myeloid growth indirectly through action on c-myb.
HOMEOBOX PROTEINS
The homeobox genes are a group of proteins defined by a specific helix-turn-helix DNA binding motif, the homeo domain.313,384,385 These regulators have been shown by mutational and knockout studies to play a major role in segmental development of many species, from Drosophila to mouse.308,386 The genomic organization of these genes in mammals, including humans, is arranged in four clusters that are expressed in a temporal fashion related to their physical position in the cluster. Recently, a number of exciting studies have shown that this class of proteins may indeed influence myeloid development.308,313 A number of studies have investigated the hematopoietic expression of homeobox genes, including ones expressed in stem cells.32 One of these, HoxA10, was noted to be expressed specifically in myeloid cells.309 Subsequent studies showed that HoxA10 mRNA was expressed at higher levels in human CD34+ cells than other homeobox genes and downregulated during myeloid development.32,33 Expression could not be detected in mature neutrophils and monocytes. These studies suggest that HoxA10 is upregulated and then downregulated as stem cells differentiate into mature myeloid cells.387 Interestingly, HoxA10 was detected in a variety of cases of myeloid leukemia, except for APL (M3), and decreased in lymphoid blast crisis of chronic myelogenous leukemia (CML).33 HoxA10 was localized to chromosome 7p14-21.33 Its expression was detected in several cases of monosomy 7, so complete loss of HoxA10 in this abnormality, frequently seen in AML, is not the mechanism of leukemogenesis. Although the targets of HoxA10 are not known, its function has been investigated by overexpression studies using retroviral infection of murine bone marrow.313 388 Overexpression of HoxA10 led to abnormalities of progenitor cells, with increased numbers of megakaryocytic and blast colonies in vitro and increased myeloid progenitors in vivo, but peripheral blood counts were normal. A substantial number of lethally irradiated congenic transplant recipients developed acute myelogenous leukemia, with high blast counts, 5 to 10 months posttransplantation.
In addition to overexpression studies, the function of HoxA10 has been investigated by knockout experiments. Mice with targeted disruption of HoxA10 are fully viable but have defects in both male and female fertility.389 Investigation of the hematopoietic system of these mice showed an increase in peripheral blood neutrophils and monocytes to a level twofold that of wild-type or heterozygote littermates.387 Interestingly, investigation of CFU in the knockout animals showed no change in numbers of GM, M, or GEMM colonies, but these myeloid CFU were extremely large in size, with a fivefold increase in cell number, and contained a preponderance of immature myeloblastic cells.387 In summary, these studies emphasize the role of HoxA10 in myeloid development and make it likely that HoxA10 plays a critical role in control of proliferation at the myeloblastic or promyelocytic stage. Recently, the effects of targeted disruption of the HoxA9 gene have been described.390 HoxA9 is expressed at high levels in human CD34+ cells and myeloid progenitors, but at lower levels in CD34− cells. It is also expressed in lymphocytes. Disruption of HoxA9 resulted in a 30% to 40% reduction in lymphocytes and granulocytes and a blunted response to G-CSF, suggesting a role for HoxA9 at the level of committed progenitors.390
A number of other homeobox genes have been studied by overexpression and antisense studies, and many of these are of direct relevance to myeloid development. Several of these studies, like those of HoxA10, suggest that downregulation of homeobox genes in progenitors may be important for myeloid maturation. Overexpression of HoxB8, when combined with high levels of IL-3, can induce myeloid leukemia in mice.391 These studies suggested that two events were necessary for leukemogenesis: an increase in progenitor proliferation, mediated by IL-3, and a block in myeloid differentiation, mediated by increased HoxB8 expression.392 Consistent with the notion that downregulation of homeobox genes may be necessary for proper differentiation, antisense studies directed at a diverged homeobox gene expressed in CD34+ cells, HB24, was shown to block IL-3– and GM-CSF–induced proliferation, and overexpression inhibited differentiation. Interestingly, increased expression of HB24 was seen in some cases of AML.393 Another negative regulator of myeloid gene expression that is downregulated with myeloid differentiation, the CDP protein discussed above, is a homeobox protein.229 The HoxB6 gene is expressed in erythroid cells, and blocking its expression with antisense oligonucleotides can induce myeloid differentiation of erythroid lines.312 These studies suggest that HoxB6, like GATA-1, may serve as a switch to direct multipotential progenitors from the myeloid to the erythroid lineage (see discussion below).
In contrast to the studies cited above, in which the homeobox is either downregulated in mature myeloid cells or expressed in other lineages, several studies have looked at homeobox genes expressed in mature myeloid cells. Hlx is expressed in macrophages and neutrophils, and overexpression using a retroviral construct induces myeloid maturation.394 HoxB7 was shown to be induced during vitamin D3 monocytic differentiation of promyelocytic HL-60 cells.395 Overexpression of HoxB7 inhibited neutrophilic differentiation of HL-60 cells, and antisense inhibited GM-CFU.395 PRH (HEX) is an orphan homeobox gene initially identified by three groups using PCR with degenerate primers to conserved homeodomain sequences.359,396,397 PRH is located on human chromosome 10, where HOX 11 is also located.396 PRH is downregulated by TPA-induced monocytic differentiation, but not by retinoic acid-induced granulocytic differentiation.398 PRH is not oncogenic when transduced into 3T3 cells.354
Finally, regulators of homeobox genes may play important roles in myeloid development and leukemia. The MLL (Hrx) gene, a regulator of homeobox genes,399 is implicated in mixed lineage (lymphoid/myeloid) leukemia.399-401 Another modulator of homeobox function is the Pbx1 gene.402 A fusion protein formed between E2A and Pbx contributes to the t(1:19) pre-B–cell ALL, but expression of the E2A-Pbx fusion protein in mice induces myeloid leukemia.403 Pbx1 binds cooperatively to HoxB4, B6, and B7, all of which have been implicated in myeloid development, as described above, but not to HoxA10.402 In summary, there are many studies implicating homeobox genes in normal myeloid development and leukemia, and the next several years should lead to exciting new developments.
OTHER FACTORS INVOLVED WITH MYELOID DEVELOPMENT AND/OR LEUKEMIA
A number of other transcription factors have been implicated in myeloid development or leukemia. Because of space limitations, they shall only be briefly mentioned.
c-myb.Myb was first isolated as a retroviral oncogene and as an Ets fusion protein can transform chicken hematopoietic cells, resulting in a variety of different leukemias.12a,404,405 C-myb is a sequence-specific transcription activator.406 In hematopoietic cells, it is expressed in proliferating progenitors and downregulated with differentiation, with a preferred if not specific expression pattern in myeloid cells.42 This downregulation is important for differentiation of myeloid cell lines.382,383 Knockout of the c-myb gene results in embryonic lethality with a failure of hematopoiesis of fetal liver,29 and c-myb has been shown to be capable of transactivating the CD34 gene in cotransfection studies.407 However, deletion of the c-myb binding sites does not result in significant loss of human CD34 promoter activity,408 and murine CD34 is reportedly expressed at normal levels in c-myb knockout mice,409 so the mechanism by which c-myb regulates CD34 is unclear at this time. Recently, c-myb has been shown to regulate the CD13 gene in myeloid cells410 and downregulate the M-CSF receptor.164 Finally, myb can interact with the AML1 family of transcription factors247 and can synergize with C/EBP to transactivate a myeloid promoter, neutrophil elastase.88 Myb can also cooperate with C/EBP factors to activate target genes in myeloid cells, possibly mediated through the p300/CBP coactivator.197,411,412 All of these results point to an important role of c-myb in myeloid development. In addition, c-myb has been reported to regulate c-myc expression,413 and some of its effects on myeloid development could be mediated through this mechanism (see below).
c-myc.C-myc was isolated as an oncogene whose dysregulation plays a role in avian and human lymphomas. It is a sequence-specific transcription factor and implicated in many processes involving tumorigenesis and apoptosis.12a,373,414,415 Of specific relevance to this review are early studies showing that myc is downregulated in myeloid cell lines as they are induced to differentiate and that treatment of these lines with antisense oligos induces myeloid differentiation.41 Activation of an inducible myc-estrogen receptor construct in a myeloid line can block differentiation.416 Another aspect of c-myc that may be important in myeloid differentiation is that it has been shown to heterodimerize with a number of partners. Dimerization with max leads to transcriptional activation, and dimerization with mad leads to loss of transcriptional activity. It has been shown that mad can be induced with maturation of myeloid cell lines, consistent with the loss of proliferation in these cells.417,418 Finally, a third interesting aspect of myc is that it has been shown to repress the C/EBPα promoter in adipocytes.193 C/EBPα expression increases with adipocyte differentiation as c-myc decreases, and blocking myc function can increase C/EBPα expression and differentiation. Whether c-myc plays a similar role in myeloid expression of C/EBPα is not clear at this time.
NF-κB and rel proteins.In addition to being a major regulator of lymphoid activation, NF-κB has been shown to be activated in myeloid cell lines induced toward monocytic differentiation with TPA. It is thought that this activation may play an important role in human immunodeficiency virus (HIV) replication and production of cytokines, such as macrophage inflammatory protein-1α.419,420 The NF-κB and related rel proteins can interact with C/EBP proteins and as such may modulate the role of C/EBP factors in myeloid cells.210,211,421 Targeted disruption of the relB gene leads to myeloid hyperplasia, suggesting a role for this family in myeloid development.422
Mixed lineage leukemia (MLL) protein.The MLL protein (also known as Hrx or All-1) has been shown to be involved in translocations of 11q23 associated with mixed (myeloid and lymphoid) leukemia.12a,399-401,423,424 MLL is a very large transcription factor with a number of different motifs, including a SET domain found in the trithorax and polycomb genes, which regulate homeobox gene expression in Drosophila. MLL has activation and repression domains, and the translocations of the MLL gene suggest that loss of the activation and retention of the repression domains could alter the expression of as yet unidentified target genes as a mechanism of leukemogenesis.425 The knockout of the MLL gene caused altered Hox expression and segmental identity, with myeloid defects in heterozygote animals.399 In addition, MLL or All-1 −/− ES cell show a block in maturation during CFU formation in vitro, suggesting that normal function of this gene is important for hematopoietic differentiation.426
p53.The p53 protein is a tumor-suppressor gene involved in many types of cancer and thought to play a normal role in elimination of cells that have damaged genomes. p53 has been showed to be mutated in many types of human leukemia, including myeloid leukemias.427,428 Other studies have implicated mutations of p53 in CML, particularly in blast crisis.429 p53 has also been shown to be a sequence specific transcriptional activator that can activate target genes, such as the cyclins.430 Wild-type p53 has been shown to induce differentiation of myeloid cell lines.431 Therefore, it is likely that loss of p53 function plays an important role in leukemogenesis.
c-Jun.The c-jun proto-oncogene forms a heterodimer with c-fos to form the AP-1 transcription factor, which has been shown to be involved in cell proliferation and activation.432 C-jun is an early response gene in differentiating myeloid cells, and its expression is upregulated in myeloid cell lines during differentiation.433,434 One known myeloid target is the macrophage scavenger receptor.166 AP-1 is antagonized by the retinoic acid receptor,435 but the PML/RAR fusion protein found in APL is an activator of c-jun, and this may contribute to the proliferative status of the leukemic cells.436 C-jun can also physically interact with PU.1151; this interaction may be important during myeloid differentiation.
MODELS OF HOW MYELOID TRANSCRIPTION FACTORS WORK ON PROMOTERS TO DIRECT MYELOID DIFFERENTIATION
The study of myeloid promoter elements has suggested several models of how myeloid genes are regulated. In addition, these studies suggest a model of how myeloid differentiation might be directed by transcription factors in normal cells and how abnormal function of transcription factors may play a role in leukemia. There is a strikingly similar promoter geography of three myeloid CSF receptor promoters, all of which use C/EBP and PU.1 sites (Fig 2). There is likely significant redundancy of some CSF pathways, in that different myeloid CSFs can direct myeloid CFU formation in vitro and that knockout studies of some CSFs, such as GM-CSF, did not show significant hematopoietic abnormalities. However, single transcription factors, such as C/EBPα and/or PU.1, could affect the myeloid lineage by regulating expression of multiple CSF receptors. Alternatively, a factor such as AML1, which is expressed at high levels in T cells in addition to myeloid cells,437 could possibly regulate not only myeloid CSF receptors, such as the M-CSF receptor, but also CSFs produced in T cells that bind to those receptors, such as GM-CSF258 and IL-3.260
Structure of the human myeloid CSF receptor promoters. As with many myeloid promoters, relatively small regions direct activity and specificity in transient transfection studies; in addition, there is no well-defined TATA box. The major transcription start site is designated by the horizontal arrow. Shown are the locations of binding sites for PU.1, C/EBP, and AML1. The PU.1 site in the G-CSF receptor promoter is located in the 5′ untranslated region, at bp +3653; a functional PU.1 site is also found in the 5′ UT region of the PU.1 promoter.55 In unstimulated myeloid cell lines, the major C/EBP gene product is C/EBPα.53,54,215 In newborn livers from C/EBPα −/− animals, only expression of the G-CSF receptor is significantly reduced.28 In PU.1 −/− animals and ES cells, expression of the M-CSF receptor is significantly reduced or undetectable.162 163
Structure of the human myeloid CSF receptor promoters. As with many myeloid promoters, relatively small regions direct activity and specificity in transient transfection studies; in addition, there is no well-defined TATA box. The major transcription start site is designated by the horizontal arrow. Shown are the locations of binding sites for PU.1, C/EBP, and AML1. The PU.1 site in the G-CSF receptor promoter is located in the 5′ untranslated region, at bp +3653; a functional PU.1 site is also found in the 5′ UT region of the PU.1 promoter.55 In unstimulated myeloid cell lines, the major C/EBP gene product is C/EBPα.53,54,215 In newborn livers from C/EBPα −/− animals, only expression of the G-CSF receptor is significantly reduced.28 In PU.1 −/− animals and ES cells, expression of the M-CSF receptor is significantly reduced or undetectable.162 163
The M-CSF receptor promoter itself is a good example of how combinations of factors, not exclusively myeloid-specific, can act together to generate a myeloid-specific promoter (Fig 1). In the case of the M-CSF receptor promoter, there are adjacent binding sites for PU.1,117 which is expressed in myeloid and B cells95,124; AML1, expressed in all white blood cells234,437; and C/EBP, expressed in myeloid cells and not lymphocytic cells.52,195,199 The common cell type in which all three of these factors are expressed is myeloid and not lymphocytic cells. In addition to a shared specificity of expression in myeloid cells, AML1 and C/EBP215 can interact physically in a specific manner and can synergize to activate the M-CSF receptor.215 C/EBP and PU.1 can also physically interact in a specific way.88 142 Therefore, the combinatorial effect of these factors is enhanced.
The findings described in this review of how these transcription factors function suggest a model of how they may direct myeloid development. This model suggests that the apparently irreversible pattern of hematopoietic lineage development may proceed in three steps (Fig 3). (1) In stem cells or more likely early multipotential progenitors, processes that are not defined yet result in the activation of a specific transcription factor, such as GATA-1 or PU.1. (2) Increased activation of expression and/or function of this factor leads to increased expression of a specific growth factor receptor and at the same time through an autoregulatory mechanism increases expression of the specific transcription factor. (3) Further differentiation is mediated by the influence of the specific growth factor, augmented by the upregulation or downregulation of other differentiation genes regulated by the transcription factor.
Model of induction of hematopoietic differentiation by specific transcription factors. In this model, transcription factors are expressed at low levels in CD34+ stem cells,134 as are specific growth factor receptors. Under direction of signals that are as yet not defined, such as the influence of stromal interactions or growth factor signalling, specific transcription factors, such as GATA-1 or PU.1,44 are upregulated. Upregulation of specific transcription factors leads to their autoregulation and upregulation of specific growth factor receptors, resulting in increases in proliferation, differentiation, and suppression of apoptosis of specific lineages. Downregulation of specific factors (such as GATA-1 during myeloid development) may also play an important role.
Model of induction of hematopoietic differentiation by specific transcription factors. In this model, transcription factors are expressed at low levels in CD34+ stem cells,134 as are specific growth factor receptors. Under direction of signals that are as yet not defined, such as the influence of stromal interactions or growth factor signalling, specific transcription factors, such as GATA-1 or PU.1,44 are upregulated. Upregulation of specific transcription factors leads to their autoregulation and upregulation of specific growth factor receptors, resulting in increases in proliferation, differentiation, and suppression of apoptosis of specific lineages. Downregulation of specific factors (such as GATA-1 during myeloid development) may also play an important role.
Both GATA-1 and PU.1 activate important growth factor receptors for the erythroid and myeloid lineages, respectively. For example, during erythroid development from multipotential progenitors, GATA-1 is activated (and PU.1 is repressed).44,438 Previous studies have shown that GATA-1, which plays a key role in erythroid development,5 can transactivate its own promoter.439 Among its many gene targets, GATA-1 can stimulate expression of the erythropoietin receptor.440 It is possible that GATA-1 activation initiates an irreversible differentiation process in which GATA-1 stimulates its own expression and that of the erythropoietin receptor, allowing that progenitor to proliferate and survive in the presence of erythropoietin.
Conversely, activation of PU.1 (and repression of GATA-144,438 ) in a progenitor may lead to stimulation of PU.1 expression through an autoregulatory loop,55 as well as the M-CSF receptor,117 allowing that progenitor to proceed along the path of myeloid development. This model would also suggest that downregulation of GATA-1 during myeloid differentiation and of PU.1 during erythroid commitment might also be important. Consistent with this concept are results suggesting that overexpression of GATA-1 in multipotential cell lines result in a block of myeloid differentiation81,441 and the finding that overexpression of PU.1 due to Friend virus insertion near the Spi-1 locus in early erythroblasts leads to a block in erythroid differentiation and murine erythroleukemia.37,38,125,126 A final addition to this model would be findings showing that GATA-1 and PU.1 could downregulate each other. Although there is a GA-rich sequence in the proximal GATA-1 promoter, PU.1 reportedly does not bind to this region.439 There is a site in the proximal PU.1 promoter that binds to GATA-1 and GATA-2, and coexpression of either GATA protein results in a twofold repression in PU.1 expression in myeloid and nonmyeloid lines.55 The role of this site in downregulation of PU.1 in early myeloid cells remains to be determined. Finally, this model is consistent with findings indicating that GATA-1 does not play a major role in myeloid development5 and that GATA-2 is either not expressed in or downregulated in myeloid cells.78 Other myeloid promoters, such as CD11b,76 also have sites in their promoters that bind GATA-1 and GATA-2, but are not functional as assessed either by transient analysis of specific point mutations or by transactivation studies.116
Recent findings suggest that C/EBP factors, notably C/EBPα, may also contribute to myeloid development using mechanisms similar to the model invoking GATA and PU.1. It is not yet known how C/EBP proteins are regulated during multilineage development of CD34+ cells, but it does appear to be selectively expressed at high levels in neutrophilic and not monocytic or erythroid cells (H.S. Radomska and D.G. Tenen, manuscript submitted). If it is upregulated selectively during neutrophilic development, it may also be autoregulated in these cells as it is in adipocytes.192,194 It is known to augment expression of the G-CSF receptor promoter53 and by this mechanism could promote the development of neutrophilic cells in a manner similar to that described above for PU.1 and GATA-1 for the monocytic and erythroid lineages, respectively.
QUESTIONS FOR THE FUTURE
In this review, we have attempted to summarize what is known about myeloid transcription factors, lineage development, and differentiation. Clearly many advances have been made in the last few years, but many very important questions remain unanswered.
How do the combinations of these myeloid transcription factors make a specific lineage? Although we have provided some concepts based on studies of the M-CSF receptor, clearly other genes are regulated by other combinations of factors.
What differences among myeloid subtypes (monocytes, neutrophils, and eosinophils) are determined by transcription factors? An example is the role of C/EBPα in neutrophilic development.
How are the regulators (PU.1, C/EBPα, etc) themselves regulated in stem cells and multipotential progenitors? Are they activated by cytokines or other extrinsic signals, or is the process stochastic and autoregulatory? Are they in turn regulated by other regulators, such as homeobox genes?
Are there truly myeloid locus control regions, scaffold and matrix attachment regions, and other elements that are important for expression of myeloid genes in the proper chromosomal context?
What are the negative regulatory loops? Do repressors like CDP228 or AEBP1231 play a role? What is the role of GATA-1?
Why is it that factors expressed in other lineages (such as PU.1 in B cells) do not induce expression of myeloid genes in those lineages? Is it because combinatorial effects of factors are needed for myeloid expression or because of negative regulation in nonmyeloid cells?
What is the role of myeloid transcription factors in apoptosis? Do they promote myeloid development by inhibition of apoptosis?
How does signalling affect myeloid development? What is the role of Janus kinases (JAKs) and STATs?13,442,443 To date, knockout studies have not shown these factors to be essential for myeloid development. What is the role of hematopoietic cell phosphatase, whose mutation results in increased macrophage production?444,445 What is the role of NF1?446,447
What are the mechanisms of leukemic development? The studies summarized in this review, in which we have characterized targets of myeloid transcription factors and models of how they could induce myeloid development, suggest a number of mechanisms in which abnormalities of myeloid transcription factor function could play a role in myeloid leukemias, which represent a block in early myeloid differentiation. Translating the knowledge of what we understand about myeloid gene regulation to understanding the pathogenesis of leukemia and perhaps developing new therapies will be the major challenge for future studies.
ACKNOWLEDGMENT
The authors apologize in advance to the many investigators whose work we were unable to cite due to space limitations. We also thank David Hume, Hanna Radomska, Laura Smith, Milton Datta, and Gerhard Behre for their careful reading of the manuscript and thoughtful suggestions and Jim Griffin for his patience.
Supported by National Institutes of Health Grants No. CA41456 and DK48660 (to D.G.T.), HL48914 (to R.H.), CA59936 (to. J.D.L.), and CA59589 (to D.-E.Z.) and by American Cancer Society Grants No. DHP-160 (to J.D.L.) and DHP-166 (to D.-E.Z.). R.H. and J.D.L. are Scholars of the Leukemia Society of America.
Address reprint requests to Daniel G. Tenen, MD, Harvard Institutes of Medicine, Room 954, 77 Avenue Louis Pasteur, Boston, MA 02115.