Abstract
The extracellular matrix produced by stromal cells plays a critical role in lympho-hematopoiesis. It was recently discovered that matrix glycoprotein SC1/ECM2 is a component of that matrix and preliminary evidence suggested that it could contribute to the nurturing environment for B-lymphocyte precursors. A fusion protein prepared from the amino terminal portion of SC1/ECM2 and the constant region of human Ig preferentially bound to pre-B cells. Furthermore, the cloning efficiency of interleukin-7–dependent B-cell precursors was increased in a dose-dependent manner by addition of this fusion protein. We now report the complete cDNA sequence for murine SC1/ECM2 and its localization to the central region of chromosome 5. A fusion protein prepared from the full length of SC1/ECM2 and Ig was found to recognize pre-B cells in a divalent cation-dependent manner, and to augment mitogen-dependent proliferation of mature B cells, as well as the cloning of pre-B cells, but to have no influence on myeloid progenitor cells. Although SC1/ECM2 is normally a secreted protein, we show that it is also capable of augmenting lymphopoiesis when expressed as a transmembrane protein on fibroblasts. Although the C-terminal portion of SC1/ECM2 has sequence homology to osteonectin/SPARC, the unique N-terminal one fifth of the protein was sufficient to augment lymphocyte growth.
EXTRACELLULAR matrix molecules within bone marrow play a fundamental role in lympho-hematopoiesis.1-6 They include various collagens, proteoglycans, and glycoproteins. This matrix functions to retain hematopoietic cells within bone marrow (BM), to localize growth factors around cells, and to give signals for proliferation or differentiation to hematopoietic precursors. For example, fibronectin, collagens, and laminin are ligands of integrins that are signal inducers as well as adhesion molecules.3,5 Hyaluronan is a ligand of CD44, and in several systems facilitates cell-cell adhesion and cell migration.4 Such molecules may be important in lympho-hematopoiesis, because antibodies to VLA-4 or CD44 completely blocked the production of lymphoid and myeloid cells in long-term BM cultures.7,8 Proteoglycans also influence the differentiation of hematopoietic cells9 and this may be attributed in part to their ability to immobilize growth factors such as granulocyte-macrophage colony-stimulating factor (GM-CSF ), interleukin-3 (IL-3), and IL-7.10 11
However, the complexity of the matrix provides a major obstacle for detailed functional studies because it is difficult to establish bioassay systems specific to each molecule. We recently developed a new cloning strategy that is capable of identifying cell-surface or soluble factors that recognize hematopoietic cells.12 With this approach we identified several stromal cell-derived matrix components whose N-terminal portions could bind to pre-B cells. One of them was matrix glycoprotein SC1/ECM2. SC1/ECM2 cDNA was originally isolated from a rat brain expression library using polyclonal antibodies directed against synaptic junction glycoproteins.13 The deduced amino acid sequence suggested that SC1/ECM2 is a secreted and calcium-binding protein, and it is considered to be a matrix component in brain. A human homolog of SC1/ECM2 was isolated by means of a monoclonal antibody (MoAb) directed to high endothelial venule cells.14 It is associated with basal, lateral, and apical surfaces of endothelial venule cells and was shown to be anti-adhesive for endothelial cells on fibronectin-coated surfaces.15 Our studies showed that SC1/ECM2 is also expressed by stromal cells and that a fusion protein containing the amino terminal portion of SC1/ECM2 is recognized by pre-B cells. Furthermore, the preliminary analysis suggested that SC1/ECM2 might represent a matrix protein in BM that is important for lymphopoiesis. This possibility has now been explored with a more detailed investigation of SC1/ECM2 structure and function.
MATERIALS AND METHODS
Cells. 293T, a human renal cell carcinoma cell line, was kindly provided by Dr T. Hirano (Osaka University, Osaka, Japan) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; GIBCO, Grand Island, NY). DW34, a mouse stromal cell–dependent pre-B cell line, was kindly provided by Dr S. Nishikawa (Kyoto University, Kyoto, Japan) and maintained on a mouse stromal cell line, P5, in RPMI1640 medium supplemented with 5% FCS and 5 × 10−5 mol/L 2-mercaptoethanol.16 Mouse stromal cell clones (OP42, BMS2, and NIH3T3), a mouse fibroblast cell line (L), mouse pre-B cell clones (1A9, BCB8, BCB9, and BCB10), mouse lymphoma cell lines (7OZ/3, WEHI231, WEHI279, AKR1, and EL4), a mouse erythroid cell line (GM86), and a mouse myelomonocytic leukemia cell line (WEHI3) were maintained as previously described.17 18
Cloning of full-length SC1/ECM2 cDNA. The isolation and sequencing of a 5′-portion cDNA of murine SC1/ECM2 was described in our previous report.12 Sense and antisense primers, 5′-ATCCAGCCACCTCTCCGCAGATCT-3′ and 5′-CTTCAGGTCCACCTCGAAGCTGTA-3′, were designed from this sequence to perform polymerase chain reactions (PCR). Because BMS2 stromal cells expressed this gene, a cDNA library was made from mRNA of BMS2 cells using oligo dT and a TimeSaver cDNA synthesis kit (Pharmacia, Uppsala, Sweden) and ligated into pEFBOS vector. Suspensions of pools containing several thousand colonies were subjected to PCR reactions with the above two primers. After 35 cycles of PCR (94°C for 1 minute, 55°C for 2 minutes, 72°C or 3 minutes), PCR samples were separated in 1.5% agarose gels and screened with a specifically amplified band (307 bp). A positive pool was divided into progressively smaller pools and rescreened until a single clone was isolated. The nucleotide sequence of the insert was then determined using an automated DNA sequencer (Aplied Biosystems, Foster City, CA).
Northern blot analysis. Total RNAs from various cell lines were isolated using TRIzol Reagent (GIBCO), electrophoresed through a formaldehyde agarose gel, and transferred onto a nylon membrane (Amersham, Arlington Heights, IL). A blot containing poly(A)+ RNAs from adult mouse tissues was acquired from Clontech (Palo Alto, CA). The SC1/ECM2– or β-actin–containing cDNA fragments were labeled with [32P]dCTP using a random primed DNA labeling kit (Boehringer Mannheim, Indianapolis, IN) and hybridized to the membrane overnight. Blots were then washed and autoradiographed.
Amino acid sequence of murine homolog of SC1/ECM2. The box marks a potential signal peptide. Potential N-glycosylation sites are indicated with circles. The osteonectin-like sequence is underlined with dashes, and the EF-hand Ca-binding site is underlined with a bold line.
Amino acid sequence of murine homolog of SC1/ECM2. The box marks a potential signal peptide. Potential N-glycosylation sites are indicated with circles. The osteonectin-like sequence is underlined with dashes, and the EF-hand Ca-binding site is underlined with a bold line.
Interspecific backcross mapping. Interspecific backcross progeny were generated by matching (C57BL/6J × Mus spretus ) F1 females and C57BL/6J males as described.19 A total of 205 N2 mice were used to map the SC1/ECM2 locus (see text for details). DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as described.20 All blots were prepared with Hybond-N+ nylon membrane (Amersham). The probe, an ∼2.7-kb Xba I fragment of mouse cDNA, was labeled with [32P] dCTP using a nick-translation labeling kit (Boehringer Mannheim); washing was done to a final stringency of 1.0× SSCP (phosphate-buffered sodium chloride sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 65°C. A major fragment of 7.4 kb was detected in Sca I–digested C57BL/6J DNA and a major fragment of 9.0 kb was detected in Sca I–digested M spretus DNA. The presence or absence of the 9.0-kb Sca I M spretus–specific fragment was followed in backcross mice. A description of the probes and RFLPs for the loci linked to SCI/ECM2 including Fgf5, Gfil, and Adrbk2 has been reported previously.21,22 Recombination distances were calculated as described23 using the computer program SPRETUS MADNESS. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Fusion proteins. An Ig/pEFBOS vector was used to produce Ig fusion proteins.12 This Ig cassette was mutated to diminish Fc-mediated bindings.24 An HT/pEFBOS vector was used to produce fusion proteins composed of an HPC4 epitope tag and a transmembrane domain of tissue factor.12 The full-length cDNA of SC1/ECM2 was amplified by PCR with 5′-CCCGCGGCCGCCACCTCTCCGCAGATCTAGCCAGC-3′ and 5′-CCCCTCGAGAAGAGGTTTTCATCTATATCCTC-3′. PCR samples were digested with Not I and Xho I, and ligated into the Not I and Xho I sites of Ig/pEFBOS and HT/pEFBOS vectors (F-SC1-Ig/pEFBOS and F-SC1-HT/pEFBOS). In some experiments, an N-SC1-HT/pEFBOS plasmid was used to produce fusion proteins containing N-terminal portion of SC1/ECM2 (93 amino acids). All constructs were confirmed by sequencing.
Immunoprecipitation and immunoblotting. Supernatants of cultures of 293T cells transfected with CD44- or F-SC1-Ig/pEFBOS plasmids were immunoprecipitated with proteinG-sepharose (Pharmacia). Western blot was performed according to our previously reported methods using horseradish peroxidase–conjugated goat antihuman Ig (Southern Biotechnology Associates, Birmingham, AL) and the ECL chemiluminescence detection system (Amersham).7
Flow cytometry analysis. Antibody incubation and washing steps were performed at room temperature in Hanks' balanced salt solutions (GIBCO) containing 1% bovine serum albumin (Sigma, St Louis, MO) and 0.02% sodium azide. Cells were stained with supernatants of cultures of 293T cells transfected with the F-SC1-Ig/pEFBOS plasmid. Fluorescein isothionate (FITC)-conjugated goat antihuman IgG (Southern Biotechnology Associates) was used as a second antibody. Supernatants containing soluble CD44-Ig were used for negative controls. In some experiments, fresh or cultured BM cells were stained with FITC-conjugated antimouse Mac1 (Boehringer Mannheim) and phycoerythrin (PE)-conjugated anti-mouse CD45RA (Pharmingen, San Diego, CA). Surface expression of fusion proteins composed of SC1/ECM2, an HPC4 epitope tag, and a transmembrane domain of tissue factor was evaluated by the stainings with an HPC4 antibody.25 The surface immunofluorescence was evaluated by a FACScan flow cytometer (Becton Dickinson, Mountain View, CA).
Amino acid sequence homology among SC1/ECM2, osteonectin/SPARC, and QR1. Periods indicate gaps introduced to align sequences. Boxes with dashed lines represent amino acid identities. Boxes with solid lines represent positions of cysteine residues.
Amino acid sequence homology among SC1/ECM2, osteonectin/SPARC, and QR1. Periods indicate gaps introduced to align sequences. Boxes with dashed lines represent amino acid identities. Boxes with solid lines represent positions of cysteine residues.
Colony-forming cell assays. Mouse BM cells were prepared and suspended in 1 mL of assay medium as previously described.26 The semisolid agar colony-forming unit assay for B-lymphocyte precursors (CFU-IL7) was performed with 1 ng recombinant murine IL-7 (R&D Systems, Minneapolis, MN). Clonable B cells (CFU-B) were enumerated in semisolid agar containing 25 μg lipopolysaccharide (LPS). The granulocyte-macrophage progenitor assay (CFU-c) was performed with 25 μL of 10-fold concentrated WEHI3-conditioned medium. CD44- and F-SC1-Ig fusion proteins were purified with protein A columns (Pierce, Rockford, IL) and added to cultures at the indicated concentrations. All colony assays were performed in 35-mm dishes and incubated at 37°C, 5% CO2 . Colonies were scored on day 6.
Cell adhesion assay. NIH3T3 cells that overexpressed N-terminal or full length of SC1/ECM2 were obtained by stable transfection with N-SC1-HT/pEFBOS or F-SC1-HT/pEFBOS plasmids, respectively. Each NIH3T3 clone was plated at 105 per well in 24-well plates and allowed to grow for 2 days before adhesion assays. BCB10 cells were suspended in Hanks' balanced salt solution supplemented with 3% FCS and adjusted to a concentration of 106/mL before being applied to NIH3T3 monolayers. After a 30-minute coculture, unbounded cells were removed by three time washes. Before each aspiration, plates were agitated on a vortex mixture. Bound cells were then recovered and counted. The percentage of bound cells was determined as follows: Percent Bound = 100 × No. of Bound Cells/No. of Applied to Each Well.
Coculture of BM cells and fibroblasts. A BM cell coculture assay was performed as previously described with minor modifications.27 Each stably transfected NIH3T3 clone was plated on a 24-well plate at subconfluent density. BM cells (2 × 104/well) were then added to cultures in the presence of 1 ng/mL IL-7. NIH3T3 cells as well as nonadherent populations were obtained from each well on day 5. Viable BM-derived cells were easily distinguished from NIH3T3 cells by size and were counted by trypan blue dye exclusion.
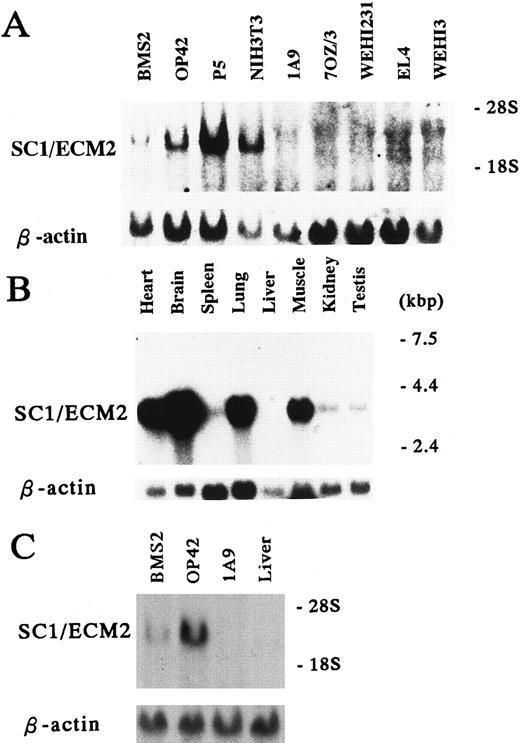
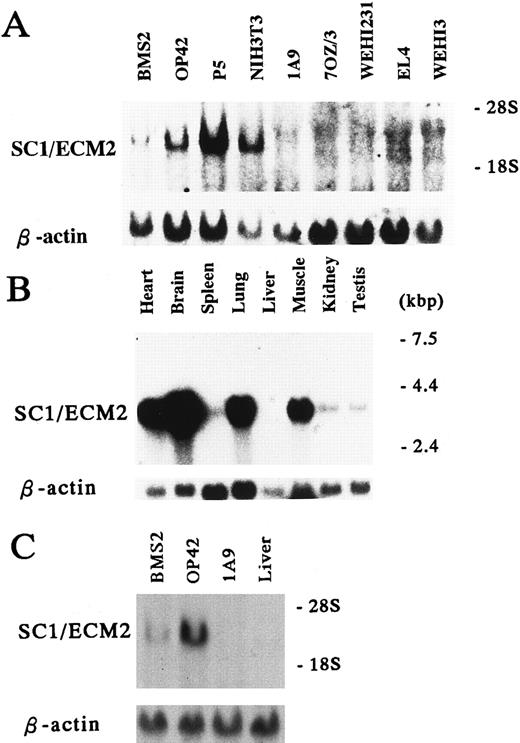
Expression of SC1/ECM2 in various cells and tissues. (A and C) Total RNA (15 μg/lane) and (B) Poly(A)+ RNA (2 μg/lane) were isolated from the indicated cell lines and tissues, and subjected to Northern blot analysis. The lower panel is a control for equal loading where the same blot was probed with β-actin.
Expression of SC1/ECM2 in various cells and tissues. (A and C) Total RNA (15 μg/lane) and (B) Poly(A)+ RNA (2 μg/lane) were isolated from the indicated cell lines and tissues, and subjected to Northern blot analysis. The lower panel is a control for equal loading where the same blot was probed with β-actin.
RESULTS
Isolation of murine homolog SC1/ECM2 cDNA and primary structure of the predicted protein. Although our initial studies suggested that SC1/ECM2 might be important for support of B lymphopoiesis, functional experiments were only conducted with the amino terminal portion of the molecule expressed as an Ig fusion protein. Furthermore, a complete sequence of the murine homologue of SC1/ECM2 had not been previously described. Therefore, we determined the full-length cDNA sequence by screening a BMS2 library by PCR with sense and antisense primers designed from the original 5′-portion cDNA sequence of murine SC1/ECM212 (Fig 1, Genbank/EMBL accession number AB000127). The insert cDNA sequence was 2,730 bp and contained an open reading frame that encoded 650 amino acids. The protein possesses a highly hydrophobic stretch of 17 amino acids at the extreme amino-terminal end that is appropriate for a signal peptide,28 but lacks an internal hydrophobic membrane spanning domain (Fig 1). There are four potential N-glycosylation sites at positions 29, 148, 168, and 462. In addition, an EF hand Ca-binding domain is present near the carboxy terminal end (DPNKDKHITLKEW), suggesting that SC1/ECM2 may bind calcium.29 The deduced amino acid sequence of murine SC1/ECM2 showed high homology with rat SC1/ECM2 (86.2% identity), and 232 carboxy-terminal amino acids (amino acid 419-650) showed especially high homology with osteonectin (62.2%) and QR1 (72.5%). All 14 of the cysteines in this area were completely conserved among these three molecules30 31 (Fig 2).
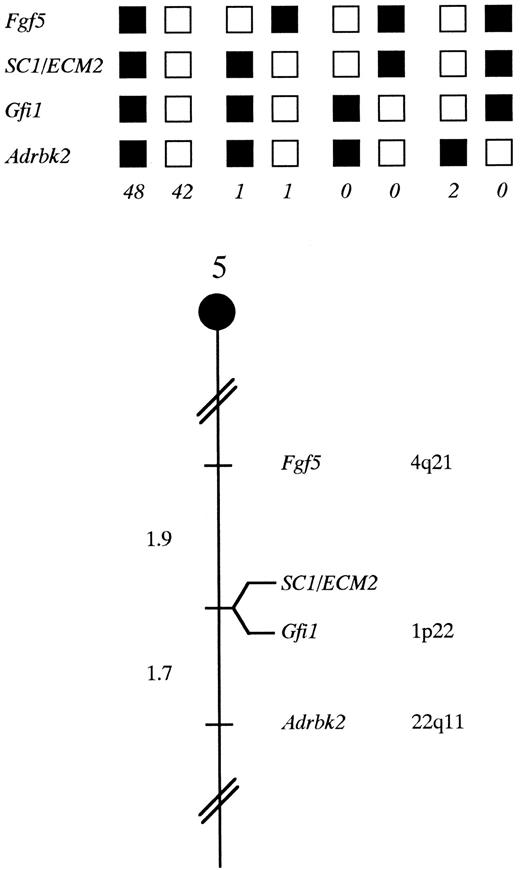
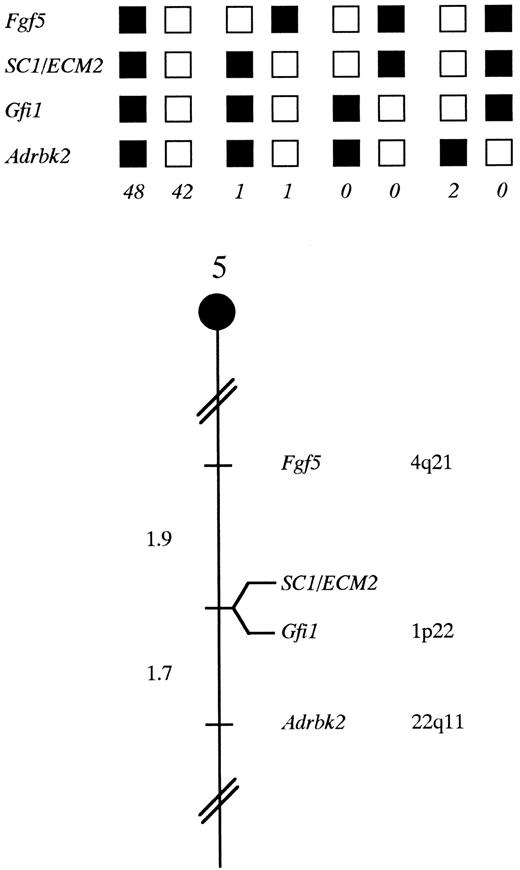
SC1/ECM2 maps in the central region of mouse chromosome 5. SC1/ECM2 was placed on mouse chromosome 5 by interspecific backcross analysis. The segregation patterns of SC1/ECM2 and flanking genes in 94 backcross animals that were typed for all loci are shown at the top of the figure. For individual pairs of loci, more than 94 animals were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M spretus ) F1 parent. The shaded boxes represent the presence of a C57BL/6J allele and white boxes represent the presence of a M spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 5 linkage map showing the location of SC1/ECM2 in relation to linked genes in shown at the bottom of the figure. Recombination distances between loci in centiMorgans are shown to the left of the chromosome, and the position of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from GDB (Genome Data Base), a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD).
SC1/ECM2 maps in the central region of mouse chromosome 5. SC1/ECM2 was placed on mouse chromosome 5 by interspecific backcross analysis. The segregation patterns of SC1/ECM2 and flanking genes in 94 backcross animals that were typed for all loci are shown at the top of the figure. For individual pairs of loci, more than 94 animals were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M spretus ) F1 parent. The shaded boxes represent the presence of a C57BL/6J allele and white boxes represent the presence of a M spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 5 linkage map showing the location of SC1/ECM2 in relation to linked genes in shown at the bottom of the figure. Recombination distances between loci in centiMorgans are shown to the left of the chromosome, and the position of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from GDB (Genome Data Base), a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD).
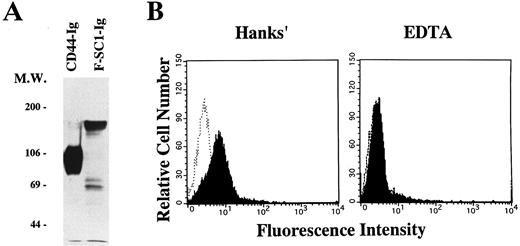
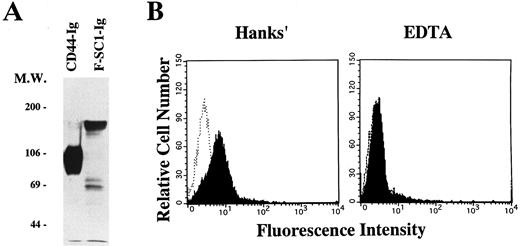
A full length of F-SC1-Ig fusion protein recognizes the BCB10 pre-B cell line. Either the F-SC1- or CD44-Ig/pEFBOS plasmid was transfected into 293T cells. (A) Supernatants from each transfected sample were immunoprecipitated with proteinG-Sepharose. Western blot was perfomed by using horseradish peroxidase–conjugated goat antihuman Ig and the ECL chemiluminescence detection system. Samples were evaluated under reducing conditions. (B) BCB10 cells were stained with supernatants from each transfected sample, followed by FITC-goat antihuman IgG (shaded histograms). Hanks' solution with (right) or without (left) 5 mmol/L EDTA was used as washing buffer. Negative control stainings obtained with CD44-Ig are also shown (open histograms).
A full length of F-SC1-Ig fusion protein recognizes the BCB10 pre-B cell line. Either the F-SC1- or CD44-Ig/pEFBOS plasmid was transfected into 293T cells. (A) Supernatants from each transfected sample were immunoprecipitated with proteinG-Sepharose. Western blot was perfomed by using horseradish peroxidase–conjugated goat antihuman Ig and the ECL chemiluminescence detection system. Samples were evaluated under reducing conditions. (B) BCB10 cells were stained with supernatants from each transfected sample, followed by FITC-goat antihuman IgG (shaded histograms). Hanks' solution with (right) or without (left) 5 mmol/L EDTA was used as washing buffer. Negative control stainings obtained with CD44-Ig are also shown (open histograms).
Cell and tissue distribution of SC1/ECM2. Because of very limited information available concerning SC1/ECM2 expression in rodents,13 we conducted Northern blot analyses of mRNA obtained from panels of cell lines and tissues (Fig 3). This showed that SC1/ECM2 was expressed by two BM-derived stromal cell lines (BMS2 and P5), as well as a splenic stromal cell clone (OP42) and a fibroblast cell line (NIH3T3). In contrast, SC1/ECM2 transcripts were not detectable in any lympho-hematopoietic cell lines (1A9,7OZ/3, WEHI231, EL4, and WEHI3). SC1/ECM2 was widely expressed in various tissues including heart, brain, spleen, lung, muscle, kidney, and testis. Although SC1/ECM2 transcripts were particularly abundant in heart, brain, and lung, expression was not detected in liver. This pattern of expression is similar in most respects to that described for a human SC1/ECM2 homolog.14 The kidney represents the most notable exception and it contained SC1/ECM2 transcripts only in the mouse.
Chromosomal mapping of the murine SC1/ECM2 gene. The mouse chromosomal location of SC1/ECM2 was determined by interspecific backcross analysis using progeny derived from matings of [(C57BL/6J × M spretus )F1 × C57BL/6J] mice. This interspecific backcross mapping panel has been typed for over 2,300 loci that are well distributed among all the autosomes as well as the X chromosome.19 C57BL/6J and M spretus DNAs were digested with several enzymes and analyzed by Southern blot hybridization for informative restriction fragment length polymorphisms (RFLPs) using a mouse cDNA SC1/ECM2 probe. The 9.0-kb Sca I M spretus RFLP (see Materials and Methods) was used to follow the segregation of the SC1/ECM2 locus in backcross mice. The mapping results indicated that SC1/ECM2 is located in the central region of mouse chromosome 5 linked to Fgf5, Gfi1, and Adrbk2. Although 94 mice were analyzed for every marker and are shown in the segregation analysis (Fig 4), up to 156 mice were typed for some pairs of markers. Each locus was analyzed in pairwise combinations for recombination frequencies using the additional data. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are: centromere- Fgf5 - 3/156 - SC1/ECM2 - 0/135 - Gfi1 - 2/121 - Adrbk2. The recombination frequencies (expressed as genetic distances in centiMorgans [cM]) ± the standard error) are - Fgf5 - 1.9 +/− 1.1 - [SC1/ECM2,Gfi1] - 1.7 +/− 1.2 - Adrbk2. No recombinants were detected between SC1/ECM2 and Gfi1 in 135 animals typed in common, suggesting that the two loci are within 2.2 cM of each other (upper 95% confidence limit). Therefore, SC1/ECM2 maps to the central region of mouse chromosome 5. As summarized in Fig 4, this shares homology with human chromosomes 4q, 1p, and 22q. In particular, Gfi1 has been placed on human 1p22. The tight linkage between SC1/ECM2 and Gfi1 in mouse suggests that SC1/ECM2 will reside on 1p as well.
Recognition of BCB10 cells by SC1/ECM2. We cloned the N-terminus of SC1/ECM2 on the basis of its ability to bind to the BCB10 pre-B cell line.12 To evaluate whether the native form of SC1/ECM2 also recognizes pre-B cells, we made a fusion protein composed of the full length of SC1/ECM2 and a human IgG1 cassette (F-SC1-Ig). The protein was confirmed by Western blot, and the molecular weight of this fusion protein was approximately 150 kD (Fig 5A). As shown in Fig 5B, F-SC1-Ig could bind to BCB10 cells, when the cells were stained in Hanks' solutions that contained Ca2+ and Mg2+. Because this staining was completely blocked by addition of EDTA, divalent cations facilitated recognition of BCB10 cells by F-SC1-Ig. Because integrins require divalent cations for binding to their ligands, some integrins could be the receptors for SC1/ECM2 on pre-B cells. However, monoclonal antibodies to α4 (PS/2), α5 (MFR5), αL (FD441.8), β1 (9EG7), or β2 (M18/2) did not block the bindings of F-SC1-Ig to BCB10 cells (data not shown). Therefore, the receptor for SC1/ECM2 was not attributed to any of these integrins.
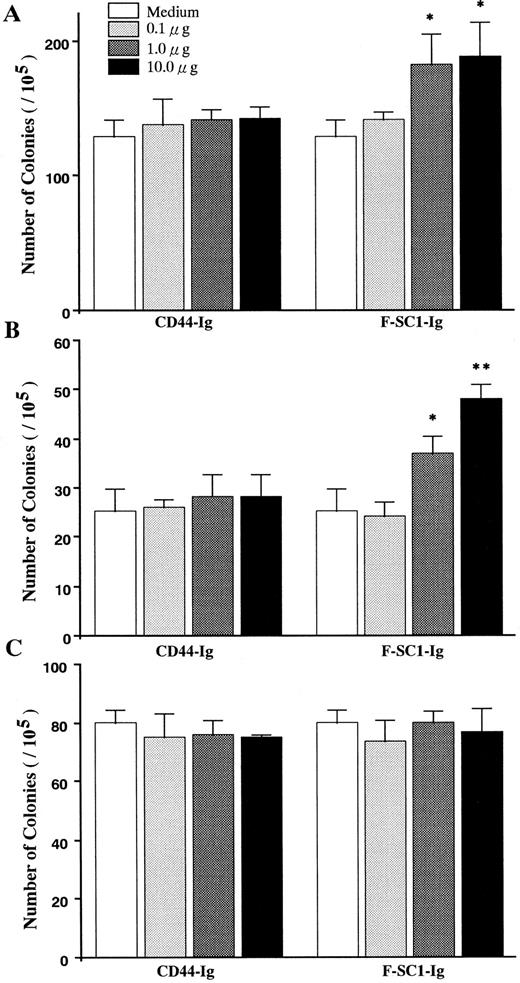
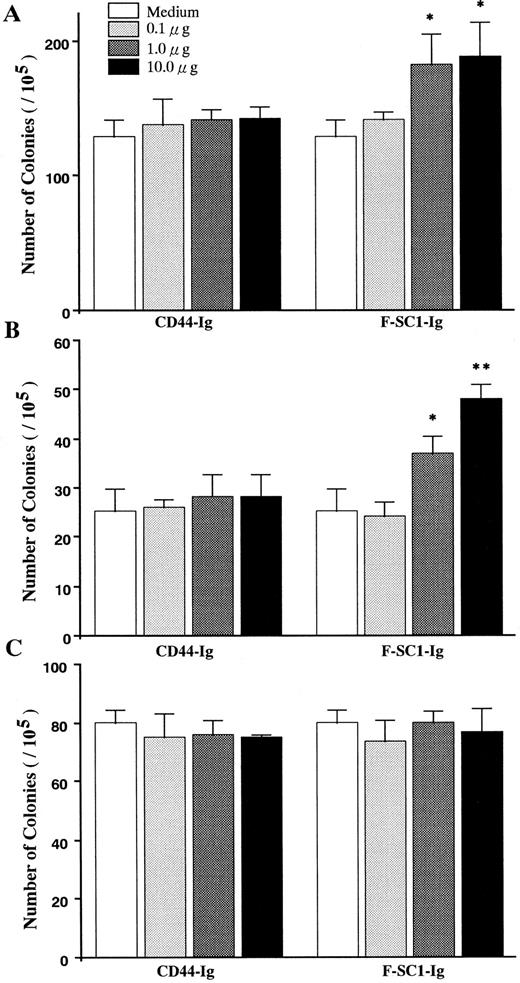
Influence of SC1/ECM2 on production of B-lineage lymphocytes. Semisolid agar cloning assays were used to evaluate the direct influence of SC1/ECM2 on lympho-hematopoiesis. As shown in Fig 6, the cloning efficiency of IL-7 responding pre-B cells (CFU–IL-7) was increased in a dose-dependent manner by addition of full length of F-SC1-Ig. Mature mitogen responsive B cells (CFU-B) were even more dramatically influenced and clonal proliferation was increased approximately 100%. In contrast, F-SC1-Ig had no influence on the responsiveness of myeloid progenitor cells (CFU-c) to colony-stimulating factors and CD44-Ig fusion protein had no significant effect on any of these cultures and served as a negative control. F-SC1-Ig did not elicit lymphocyte colony formation when cultures were established in the absence of IL-7 or mitogen (data not shown). Therefore, SC1/ECM2 augments survival and/or proliferation of B-lineage lymphocytes and their precursors, but has no obvious influence on myeloid lineage cells.
The effects of F-SC1-Ig on colony assays. A F-SC1-Ig/pEFBOS plasmid or a CD44-Ig/pEFBOS plasmid was transfected into 293T cells. The F-SC1-Ig and the CD44-Ig were purified with a protein A column and added to (A) CFU-IL7, (B) CFU-B, and (C) CFU-c colony assays of mouse BM cells at the indicated concentrations. Results represent mean numbers of colonies per 105 BM cells ± SD (n = 3). Statistically significant differences from control (CD44-Ig) values are indicated by one (P < .05) or two (P < .01) asterisks. Similar results were observed in three independent experiments.
The effects of F-SC1-Ig on colony assays. A F-SC1-Ig/pEFBOS plasmid or a CD44-Ig/pEFBOS plasmid was transfected into 293T cells. The F-SC1-Ig and the CD44-Ig were purified with a protein A column and added to (A) CFU-IL7, (B) CFU-B, and (C) CFU-c colony assays of mouse BM cells at the indicated concentrations. Results represent mean numbers of colonies per 105 BM cells ± SD (n = 3). Statistically significant differences from control (CD44-Ig) values are indicated by one (P < .05) or two (P < .01) asterisks. Similar results were observed in three independent experiments.
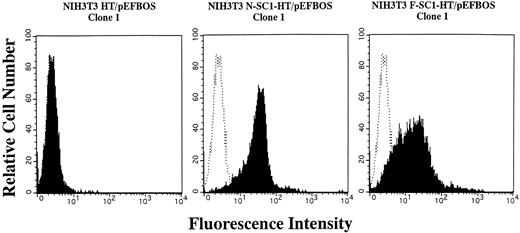
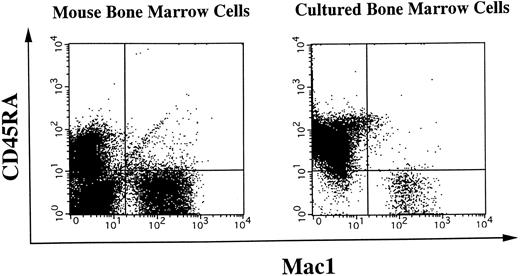
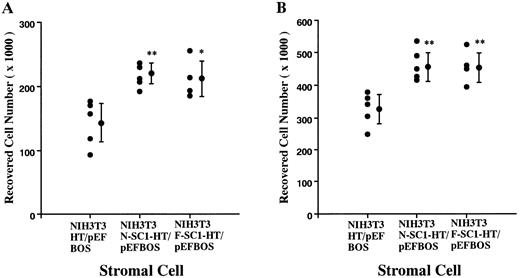
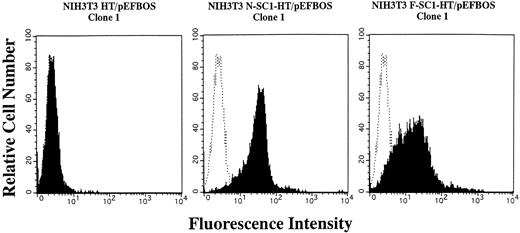
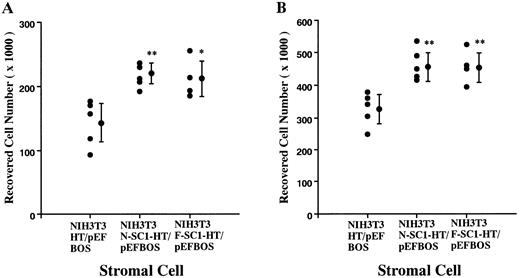
A membrane-anchored form of SC1/ECM2 augments lymphopoiesis. SC1/ECM2 is normally a secreted protein but might be immobilized in tissues through interaction with transmembrane or extracellular matrix molecules. Therefore, we investigated whether an immobilized form of the molecule would have functional activity. NIH3T3 fibroblasts that normally express the soluble form were transfected with N-SC1- or F-SC1-HT/pEFBOS plasmids. This resulted in large quantities of the protein localized to the cell surface, where it could be detected by means of the HPC4 MoAb (Fig 7). There was no obvious influence of this treatment on growth or morphology of the fibroblasts. In addition, overexpression of membrane-anchored N-terminal portion or full length of SC1/ECM2 on NIH3T3 cells did not change adhesion to BCB10 cells (Table 1). However, the transfected fibroblasts had increased ability to support growth of pre-B cells. Proliferating foci appeared within 5 days when murine BM was cultured on control fibroblasts in the presence of IL-7. The recovered cells were determined to be B-lymphocyte lineage by flow cytometry (presence of CD45RA and absence of Mac1, Fig 8). Significantly more lymphocytes were recovered from cocultures initiated with NIH3T3 cells that expressed immobilized SC1/ECM2 (Fig 9A). This response was similar when the full length or just the amino terminal portions of SC1/ECM2 were present. It would have been possible in this model for immobilized SC1/ECM2 to influence lymphocyte precursors indirectly via some other cell type present in the initial marrow inoculum. Therefore, cocultures were also performed with the stromal cell dependent DW34 pre-B cell line.16 Survival and expansion of DW34 cells was again much more efficient on transfected fibroblasts (Fig 9B). Therefore, an immobilized form of this matrix glycoprotein can deliver signals in addition to those provided by the constitutively produced soluble SC1/ECM2 made by this fibroblast cell line.
Expession of membrane-anchored SC1/ECM2 on stably transfected NIH3T3 cells. The N- or F-SC1-HT/pEFBOS plasmid or HT/pEFBOS vector was stably transfected into NIH3T3 cells. Each clone was stained with an HPC4 antibody. Shaded histograms depict stainings obtained with the HPC4 antibody, and open histograms represent results obtained with the second antibody alone.
Expession of membrane-anchored SC1/ECM2 on stably transfected NIH3T3 cells. The N- or F-SC1-HT/pEFBOS plasmid or HT/pEFBOS vector was stably transfected into NIH3T3 cells. Each clone was stained with an HPC4 antibody. Shaded histograms depict stainings obtained with the HPC4 antibody, and open histograms represent results obtained with the second antibody alone.
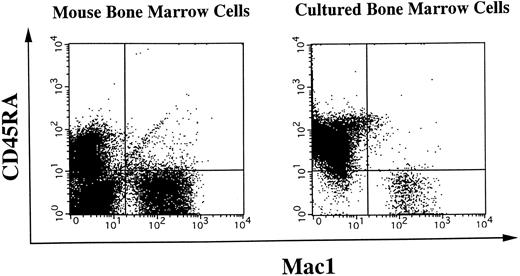
Surface phenotypes of freshly isolated or cultured BM cells. Freshly isolated mouse BM cells or BM cells cocultured on NIH3T3 cells were subjected to two-color flow cytometry analysis. Myeloid cells were simultaneously detected by anti-Mac1 antibody, and B-lineage cells were stained for a CD45RA marker. Quadrants are indicated to show levels of background stainings observed with appropriate irrelevant control antibodies.
Surface phenotypes of freshly isolated or cultured BM cells. Freshly isolated mouse BM cells or BM cells cocultured on NIH3T3 cells were subjected to two-color flow cytometry analysis. Myeloid cells were simultaneously detected by anti-Mac1 antibody, and B-lineage cells were stained for a CD45RA marker. Quadrants are indicated to show levels of background stainings observed with appropriate irrelevant control antibodies.
Augmentation of pre-B cell growth by SC1/ECM2 overexpressed on the surface of stromal cells. An N-SC1-HPC4-TF/pEFBOS plasmid or an F-SC1-HPC4-TF/pEFBOS plasmid was stably transfected into NIH3T3 cells. (A) BM cells (2 × 104) or (B) a pre-B cell line, DW34 (1 × 103), were cocultured in the presence of 1 ng/mL IL-7. Each dot represents means of recovered nonadherent cells from cocultures with each transfected clone. Statistically significant differences from control (cocultures with NIH3T3 transfected with HPC4-TF/pEFBOS plasmid) values are indicated by one (P < .05) or two (P < .01) asterisks. Similar results were observed in four independent experiments.
Augmentation of pre-B cell growth by SC1/ECM2 overexpressed on the surface of stromal cells. An N-SC1-HPC4-TF/pEFBOS plasmid or an F-SC1-HPC4-TF/pEFBOS plasmid was stably transfected into NIH3T3 cells. (A) BM cells (2 × 104) or (B) a pre-B cell line, DW34 (1 × 103), were cocultured in the presence of 1 ng/mL IL-7. Each dot represents means of recovered nonadherent cells from cocultures with each transfected clone. Statistically significant differences from control (cocultures with NIH3T3 transfected with HPC4-TF/pEFBOS plasmid) values are indicated by one (P < .05) or two (P < .01) asterisks. Similar results were observed in four independent experiments.
DISCUSSION
A thorough understanding of the marrow microenvironment should eventually make it possible to propagate stem cells and their progeny ex vivo and substantial progress has been made in identifying transmembrane, secreted, and extracellular matrix molecules that could be important. Of particular interest are those which promote survival of blood cell precursors, and a preliminary study suggested that SC1/ECM2 might represent such a protein.12 We now describe the complete coding sequence of the murine SC1/ECM2 gene and localize it to chromosome 5. The amino terminal portion of the molecule was sufficient for recognition of lymphocytes and it promoted clonal proliferation of mature, as well as immature cells of the B lineage. Furthermore, SC1/ECM2 augmented B lymphopoiesis when present in either soluble and immobilized forms. Detailed information is not available about mechanisms through which SC1/ECM2 has this influence, but structural similarity to the well-studied osteonectin/SPARC protein provides some basis for speculation.
The cDNA sequence of SC1/ECM2 (Fig 2) is well conserved between mice and rats (86.2% identity at the amino acid level), and especially in the C-terminal portion (98.3% identity in a stretch of 232 amino acids). This C-terminal portion of SC1/ECM2 also has high homology with osteonectin, QR1, testican and transforming growth factor-β–induced protein TSC36/glioma-secreted follistatin-related protein, and all of 14 cysteine residues were completely conserved among these molecules.30-34 This part of osteonectin consists of three domains, a follistatin-like domain, a high α-helix content, and an EF-domain.35 The EF region is an autonomously folding and crystalliable domain that binds calcium and collagen IV.36
The high degree of sequence homology in the C-terminal portion of SC1/ECM2 suggests that the protein may have many functions in common with osteonectin/SPARC. For example, it might be expected to be anti-adhesive in some circumstances, to regulate cell proliferation, and to bind cytokines.37 However, the amino terminal portion of SC1/ECM2 had similarity to neither osteonectin nor QR1 and might be expected to mediate unique functions. Indeed, approximately one fifth of the molecule (93 amino acids at N-terminus) was sufficient for recognition by lymphoid cells and augmentation of their expansion. Interactions between SC1/ECM2 and lymphocytes are divalent cation dependent,12 but the calcium-binding EF domain is localized near the C-terminus and need not to be involved. Divalent cations may therefore be important for function of the counter-receptor for SC1/ECM2 on lymphocytes.
Osteonectin/SPARC is known to bind cytokines such as platelet-derived growth factor (PDGF ).38 Therefore, it will be important to learn if this is true for SC1/ECM2 and which domains are required. It is interesting that growth of the DW34 pre-B cell clone was augmented by both full-length and truncated forms of SC1/ECM2. This clone has previously been shown to be responsive to a chemokine known as PBSF/ SDF-1, which is the natural ligand of fusin.39-42 Lymphocyte development is disrupted in mice that have been targeted for the PBSF gene.43 One possibility to be explored is that SC1/ECM2 binds and presents this essential chemokine to developing and activated B lymphocytes.
The DW34 lymphocyte clone was also used as a biological indicator to identify the stromal cell-surface ectoenzyme known as BST1.44 Although the function of BST1 has not been fully characterized, BM stromal cell lines derived from patients with multiple myeloma and rheumatoid arthritis had greatly elevated expression of BST1. It will be interesting to investigate SC1/ECM2 production in such conditions because abnormalities of BM stromal cells have been reported in some diseases with monoclonal or polyclonal B-cell proliferation, and the BM microenvironment plays important roles in the pathogenesis of those diseases.45
Although we originally isolated SC1/ECM2 on the basis of its recognition of pre-B cells, this matrix molecule may also influence activities of mature B lymphocytes. Northern blot analysis of spleen did not show high SC1/ECM2 expression, but the signal could have been substantially diluted by lymphocyte-derived mRNA, that does not contain SC1/ECM2 transcripts. A monoclonal antibody to murine SC1/ECM2 will be useful in identifying positive cells in peripheral lymphoid tissues inasmuch as the human homolog is relatively restricted to endothelial cells within the tonsil.14 Functions have yet to be demonstrated for SC1/ECM2 in that tissue and possible roles should be investigated in heart, brain, and lung, because these tissues have particularly high levels of expression.
Collagens, laminin, fibronectin, hyaluronan, proteoglycans, and a variety of glycoproteins are present within the extracellular matrix of BM and there is reason to believe that this is much more than an inert framework.3,5,6,46 In addition to the ability to immobilize growth factors, the matrix may deliver signals for survival and expansion, as appears to be the case with SC1/ECM2. The possibility will need to be investigated whether SC1/ECM2 delivers signals related to differentiation and if it influences lymphocyte migration. Changes in transfected fibroblast morphology were not obvious. Moreover, the amounts of F-SC1-Ig fusion proteins that bind to B-lineage lymphocytes are not substantial, and overexpression of SC1/ECM2 on a fibroblast did not change adhesion to a pre-B cell line (Fig 5 and Table 1). Therefore, SC1/ECM2 may not function as an important adhesion molecule and it will be interesting to learn if it can be antiadhesive with respect to lymphocytes, as it has been shown to be for endothelial cells.15 Our findings showed that, in contrast to B-lineage precursors, growth of myeloid progenitors from BM was not influenced by SC1/ECM2. This does not exclude possible roles in myeloid cell maturation, and it is still possible that SC1/ECM2 might interact with megakaryocytic and erythroid progenitors, as well as stem cells in that organ.
We compared our interspecific map of chromosome 5 with a composite mouse linkage map that reports the map location of many uncloned mouse mutations (provided by Mouse Genome Database, a computerized database maintained at the Jackson Laboratory, Bar Harbor, ME). SC1/ECM2 mapped in a region of the composite map that lacks mouse mutations with a phenotype that might be expected for an alteration in this locus (data not shown). Mapping of human SC1/ECM2 has not been reported, but tight linkage to the murine Gfi1 gene suggests that it might be located on 1p. An examination of human mutations in this location (OMIM) did not reveal an obvious candidate for a condition that can be attributed to SC1/ECM2-related defects.
NIH3T3 cells produce SC1/ECM2 transcripts and presumably secrete the mature protein. Nonetheless, transfection of these fibroblasts with membrane-bound forms of SC1/ECM2 markedly increased their ability to support lymphocyte expansion. As additional information accrues about the molecular requirements for lympho-hematopoiesis, it should become possible to engineer ever more efficient supporting microenvironments.
ACKNOWLEDGMENT
We thank Mary Barnstead for excellent technical assistance.
Supported in part by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories, Inc. Supported in part by grants from the Ministry of Education, Science and Culture, the Inamori Foundation, Senri Life Science Foundation, and Mochida Memorial Foundation as well as Grant No. AI-33085 from the National Institutes of Health.
Address reprint requests to Kenji Oritani, MD, The Second Department of Internal Medicine, Osaka University Medical School, 2-2 Yamadaoka, Suita, Osaka 565, Japan.