Abstract
We have developed a large-scale, expression-based gene trap strategy to perform genome-wide functional analysis of the murine hematopoietic and vascular systems. Using two different gene trap vectors, we have isolated embryonic stem (ES) cell clones containing lacZreporter gene insertions in genes expressed in blood island and vascular cells, muscle, stromal cells, and unknown cell types. Of 79 clones demonstrating specific expression patterns, 49% and 16% were preferentially expressed in blood islands and/or the vasculature, respectively. The majority of ES clones that expressedlacZ in blood islands also expressed lacZ upon differentiation into hematopoietic cells on OP9 stromal layers. Importantly, the in vivo expression of the lacZ fusion products accurately recapitulated the observed in vitro expression patterns. Expression and sequence analysis of representative clones suggest that this approach will be useful for identifying and mutating novel genes expressed in the developing hematopoietic and vascular systems.
THE PHYSICAL AND genetic analysis of mammalian genes is yielding a vast amount of information concerning the 80,000 or so proteins encoded by our genomes. Clearly, a major challenge for the immediate future will be to understand the individual functions of each of these proteins and to place them within the correct molecular circuitry in the appropriate cell types. These insights will provide the basis for understanding both normal physiological processes and human disease. Although many mammalian proteins are related in sequence and biochemical functions to proteins found in other more experimentally tractable organisms, knowledge of the biological function of these proteins will require direct analysis within the context of an intact mammal, because the different members of large mammalian gene families function in diverse cell types during development and in the adult. In addition, sequence similarity or homology is neither a necessary nor a sufficient predictor of similarity of biological function.
The large amount of genetic information in mammalian organisms and the cellular complexity of the developing embryo require new experimental approaches that can rapidly and efficiently identify and analyze genes. In the mouse, the existing repertoire of naturally occurring and induced mutations has provided important insights into the molecular mechanisms that regulate embryonic development and cellular differentiation within the hematopoietic system.1-3However, positional cloning of each of these mutations is still a significant and labor-intensive undertaking.
The contemporaneous development of totipotent embryonic stem (ES) cell lines and homologous recombination in mammalian cells has provided an entirely new approach to generate new mutations in vitro before introduction of these mutations into the mouse germline.4 5However, targeted mutagenesis is also labor intensive and time-consuming. In addition, gene targeting requires prior detailed knowledge of the sequence and genomic organization of each gene and hence is not easily applicable to genome-wide approaches.
A third strategy involves the random insertion of exogenous DNA into single sites in the mammalian genome.6 When applied to ES cells, this insertional mutagenesis approach, or gene trapping, provides a genome-wide strategy for functional genomics in a mammal. In this report, we have combined gene trapping with the developmental potential of ES cells to differentiate in vitro into a wide variety of distinct cell lineages7-9 to trap genes that are expressed in hematopoietic and vascular endothelial cells. This experimental approach, which we have termed expression trapping, offers a novel strategy to identify, mutate, and characterize large numbers of genes on the basis of their cell lineage-specific expression.
MATERIALS AND METHODS
Vectors.
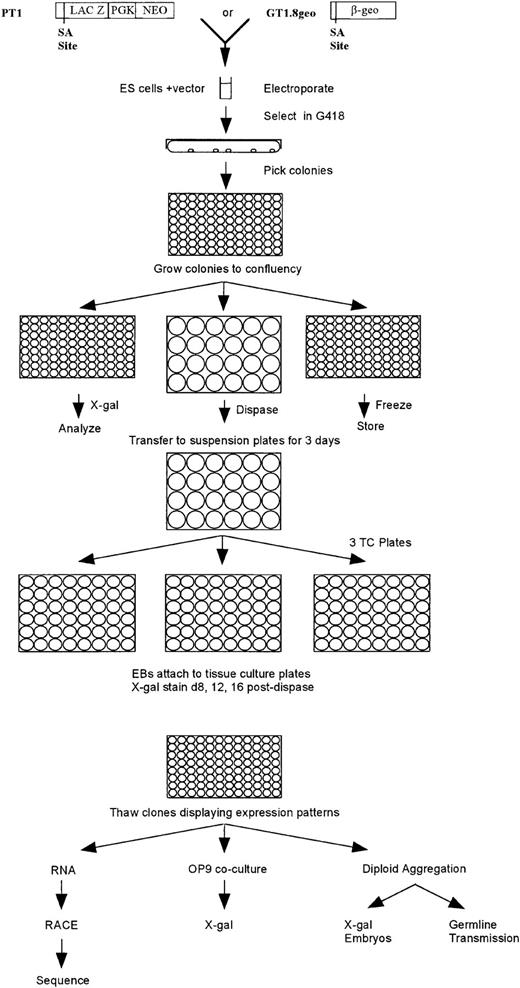
Two gene trap vectors were used for this study and are shown in Fig 1. The vector PT1-ATG (PT1 henceforth) contains the En-2 splice acceptor site positioned immediately upstream of the lacZ reporter gene with an ATG translational start site.10 Immediately downstream of the lacZgene, the phosphoglycerate kinase-1 (PGK-1) promoter drives the bacterial neomycin-resistance (neo) gene. The vector GT1.8geo contains the En-2 splice acceptor site immediately upstream of a lacZ-neo fusion gene.11The point mutation in the neo fragment of SAβgeo12 is not contained in GT1.8geo vector, thereby allowing neomycin resistance at a lower level of endogenous gene expression than the SAβgeo vector.
Schematic diagram depicting gene trap screening strategy. Two gene trap vectors were used. The PT1 vector contains a promoterlesslacZ gene immediately downstream of a splice acceptor (SA) site and the neoR gene driven by the PGK-1promoter. Although not all neoR colonies represented trapped genes, all genes could be trapped regardless of their expression in undifferentiated ES cells using the PT1 vector. The GT1.8geo vector contains a promoterless lacZ-neoR(β-geo) fusion gene immediately downstream of an SA site. Although all neoR colonies represented trapped genes, only genes expressed in undifferentiated ES cells could be trapped. ES cells were electroporated with either vector and G418R clones were picked into 96-well plates and grown to confluency. The clones were passaged 1:3 into two 96-well plates and one set of 24-well plates. The cells from one 96-well plate were frozen, and the cells from the second 96-well plate were assayed forlacZ expression. The colonies in the 24-well plates were treated with dispase and then transferred to suspension 24-well plates and grown in suspension for 3 days; EBs were then transferred to 48-well tissue culture (TC) plates. Cultures were fed every other day and analyzed for lacZ expression 8, 12, and 16 days after dispase treatment. Clones exhibiting expression patterns were thawed and grown for RNA isolation, RACE polymerase chain reaction (PCR) and sequencing, and/or OP9 coculture and subsequent lacZexpression, and/or diploid aggregation for in vivo lacZexpression analysis and germline transmission.
Schematic diagram depicting gene trap screening strategy. Two gene trap vectors were used. The PT1 vector contains a promoterlesslacZ gene immediately downstream of a splice acceptor (SA) site and the neoR gene driven by the PGK-1promoter. Although not all neoR colonies represented trapped genes, all genes could be trapped regardless of their expression in undifferentiated ES cells using the PT1 vector. The GT1.8geo vector contains a promoterless lacZ-neoR(β-geo) fusion gene immediately downstream of an SA site. Although all neoR colonies represented trapped genes, only genes expressed in undifferentiated ES cells could be trapped. ES cells were electroporated with either vector and G418R clones were picked into 96-well plates and grown to confluency. The clones were passaged 1:3 into two 96-well plates and one set of 24-well plates. The cells from one 96-well plate were frozen, and the cells from the second 96-well plate were assayed forlacZ expression. The colonies in the 24-well plates were treated with dispase and then transferred to suspension 24-well plates and grown in suspension for 3 days; EBs were then transferred to 48-well tissue culture (TC) plates. Cultures were fed every other day and analyzed for lacZ expression 8, 12, and 16 days after dispase treatment. Clones exhibiting expression patterns were thawed and grown for RNA isolation, RACE polymerase chain reaction (PCR) and sequencing, and/or OP9 coculture and subsequent lacZexpression, and/or diploid aggregation for in vivo lacZexpression analysis and germline transmission.
Generation of trapped ES cell lines.
R1 ES cells were maintained on primary embryonic fibroblasts as previously described.13 After electroporation and selection in G418, drug-resistant colonies were transferred to 96-well plates and expanded to confluency. Clones were passaged to two 96-well plates and one set of 24-well plates. Once clones reached confluency, one 96-well plate was frozen, the second 96-well plate was assayed for β-galactosidase (β-gal) expression, and the 24-well plates were used for attached EB differentiation cultures. Expression of thelacZ reporter gene was carefully determined both in undifferentiated and differentiated ES cells. Clones with observable expression patterns were refrozen and, in some cases, reanalyzed. In addition, the expression patterns were photographed and cataloged.
Reporter gene expression.
β-gal activity of cultures was detected as follows. Cells were rinsed in 100 mmol/L Na2HPO4 (pH 7.5) and then fixed in 0.2% glutaraldehyde, 5 mmol/L EGTA, 2 mmol/L MgCl2, and 100 mmol/L Na2HPO4 for 5 minutes. The cells were washed 3 times for 5 minutes each in 2 mmol/L MgCl2, 0.02% NP-40, and 100 mmol/L Na2HPO4. The cells were stained with X-gal overnight at 37°C. β-gal activity was detected in embryos as described above, except that the fixative included 1.5% formaldehyde and embryos were fixed for 30 minutes to 1 hour and washed 3 times for 15 minutes each wash.
Attached EB screen.
ES cells were allowed to differentiate into attached EBs as previously described,14 with several modifications. Clones were grown to confluency in 24-well plates, treated with dispase (1:1 dilution in phosphate-buffered saline [PBS]; Collaborative Research VWR, Mississauga, Ontario, Canada), washed 3 times in PBS, and grown in suspension in Ultra Low Cluster 24-well plates (COSTAR, Cambridge, MA) in ES media without Leukemic Inhibitory Factor (LIF). On day 3 after dispase treatment, 5 to 10 embryoid bodies were transferred to 48-well tissue culture plates (Falcon, Mississauga, Ontario, Canada). Cultures were fed every other day with fresh media. β-gal activity was determined on days 8, 12, and 16 after dispase.
OP9 induction assay.
ES cells were allowed to differentiate on the OP9 stromal cell line as previously described,9 with several modifications. ES clones were differentiated on OP9 stroma in replica wells of 6-well plates (104 ES cells/well) for 5 days to generate mesodermal colonies. A single-cell suspension was prepared using trypsin from one well for each clone, and 105 mesodermal cells were replated onto OP9 stroma in two wells of a 6-well plate and grown for 3 days. On day 8, nonadherent hematopoietic cells were transferred from both wells to one new well for an additional 3 days. β-gal activity was determined on mesodermal cells on the duplicate day 5 OP9 plate and on adherent hematopoietic cells on days 8 and 11.
5′ RACE.
RNA was prepared from either undifferentiated or differentiated cells using Trizol (GIBCO/BRL, Grand Island, NY) according to the manufacturer’s instructions. 5′ RACE was performed using the 5′ RACE kit (GIBCO/BRL), according to manufacturer’s instructions with modifications previously described.155′ RACE products were subcloned into the CloneAmp plasmid (GIBCO/BRL) and sequenced using the Sequenase kit (Pharmacia, Uppsala, Sweden). Sequences were analyzed by comparison to the nonredundant GenBank and EST of NCBI using the BLASTN program (www.ncbi.nlm.nih.gov/BLAST/).
Generation of chimeras.
ES cells were aggregated with diploid embryos as described,16 transferred into pseudo-pregnant ICR females, harvested at embryonic day (e) 9.5 to 14.5, and stained for β-gal activity. About half of the diploid embryos were allowed to mature to term for germline transmission. Chimeric males were bred to ICR females, and tail DNA of F1 and F2 offspring was analyzed by Southern blotting and hybridization to En-2 or RACE fragment probes.
RESULTS
Identification of trapped gene expression patterns.
In the absence of leukemic inhibitory factor, ES colonies spontaneously differentiate into embryoid bodies (EBs) in suspension culture. The complex structure of the EB contains all three germ layers and resembles the extra-embryonic yolk sac both morphologically and transcriptionally.7,17-19 As in the yolk sac, the mesoderm of the EB gives rise to angioblastic cords that form morphologically distinquishable blood islands containing primitive hematopoietic cells surrounded by vascular endothelium.8 Because of the developmental potential of EBs, the differentiation of ES cells into EBs has provided an excellent model to study the effects of targeted mutations on hematopoietic, vascular, and myoblast lineages.20-22 Thus, the EB should provide an excellent in vitro expression screen of gene trap clones for insertions in genes expressed in hematopoietic and vascular lineages. However, EBs grown in suspension are difficult to manipulate in clonal cultures and the outer layer of visceral endoderm precludes the identification of small numbers of lacZ positive cells. Therefore, we modified the EB culture system so that EBs grow attached to tissue culture plastic.14 This attached or flat culture method places the visceral endoderm layer beneath the blood islands and renders the EB more accessible to observation and experimental manipulation.
The gene trap screening strategy that we used is shown in Fig 1. The PT1 gene trap vector, which contains a splice acceptor site immediately upstream of a promoterless lacZ reporter gene and aneoR gene driven by PGK-1 promoter, was introduced into ES cells (clone R1) by electroporation. After G418 selection, drug-resistant colonies were transferred to 96-well plates and expanded to confluency. Clones were replica plated to two 96-well plates and one set of 24-well plates. Once clones reached confluency, one 96-well plate was frozen, the second 96-well plate was assayed for β-gal expression, and the 24-well plates were used for attached EB differentiation cultures. Each neoR colony represented a vector integration event. If the vector integrated within an intron, a spliced fusion transcript between lacZ and the endogenous gene would be generated upon transcriptional activation of the trapped gene. Because all ES cells that had an integrated PT1 vector were G418 resistant regardless of whether the integration occurred within a gene, genes that were not expressed in undifferentiated ES cells could be trapped using this vector. Five percent (37/779) of the neoR clones tested expressed lacZ in undifferentiated ES cells, of which 30 clones continued to be expressed in at least some cells during EB differentiation (Table 1). By comparison, 61 clones (8%) that did not express lacZ as undifferentiated ES cells demonstrated lacZ expression during EB differentiation (Table 1). Of the neoR clones that expressedlacZ as undifferentiated or differentiated ES cells, one-third (32 clones) exhibited a restricted pattern of expression (Table 1). The expression patterns of these clones can be grouped into seven categories (Table 2). More than one third of these clones were expressed in blood islands and/or the vasculature; in contrast, stromal and muscle cells each represented only 3% of the clones displaying restricted expression patterns. In addition, 9% of these clones expressed lacZ constitutively in virtually all undifferentiated and differentiated cells. The remaining clones exhibited restricted patterns of expression in a cell type(s) that has not yet been identified.
In a second series of experiments, we used the GT1.8geo vector (Fig 1) that contains a splice-acceptor site immediately upstream of a promoterless β-gal-neo fusion gene (or geo). Thus, unlike the PT1 vector, all neoR clones selected after introduction of the GT1.8geo vector represented integrations into genes that were transcriptionally active in undifferentiated ES cells. Accordingly, a much higher proportion of the GT1.8geo clones (34%v 5% for PT1) expressed detectable levels of β-gal activity in undifferentiated ES cells (ie, Blue; Table 1). Of those, 159 clones continued to express lacZ in at least some cells during EB differentiation. Of the 337 neoR clones that expressed geo at levels too low to detect lacZ expression (ie, White; Table 1) as undifferentiated ES cells, more than half upregulated expression of lacZ in a portion of differentiated cells in EB cultures. Of the 353 GT1.8geo clones that expressed lacZ, 47 clones displayed an obvious pattern of expression (Tables 1 and 2). The majority of the pattern-expressing clones expressed lacZ in the blood islands and/or the endothelium (Table 2).
In contrast to EB body differentiation in which ES cells differentiate into all three germ layers that eventually give rise to many lineages, including hematopoietic and vascular cells, ES cells grown in coculture with OP9 stromal cells differentiate into mesodermal colonies that, when replated, differentiate into hematopoietic cells.9,23All gene trap cell lines that demonstrated lacZ expression in blood islands were reanalyzed by differentiating ES cells in replicate OP9 stromal cell cultures. ES-derived mesodermal colonies expressing brachury (M. Hidaka, unpublished observations) were apparent by day 3 of culture. On day 5, a single-cell suspension of a replicate culture was prepared and replated onto OP9 cells. Primitive erythrocytes and multipotential precursors differentiated from the mesodermal precursors within the next 2 to 3 days and single lineage precursors predominated the cultures by day 119 23 (M. Hidaka, unpublished observations). Cultures were assayed for lacZ expression at days 5, 8, and 11. The majority of blood island positive clones (70%) expressedlacZ in hematopoietic cells when cultured on an OP9 feeder layer (Table 2).
Identification of trapped genes.
To determine the DNA sequence of the trapped genes, RNA was prepared from either differentiated or undifferentiated ES clones and used to perform 5′ RACE.24 The RACE products of 11lacZ fusion transcripts were cloned and sequenced. Table 3 summarizes the lacZexpression pattern, the gene trap vector, and sequence information for each clone. Eight of the RACE product sequences corresponded to novel genes, of which four shared similarity with EST sequences. The sequences of three of the trapped genes corresponded to genes that encode known protein products: Mena, Karyopherin β3, and 5′GMP synthetase. Clone K18E2 encodes Mena, the mammalian homologue of Drosophilia Enabled(ena), which was originally cloned by a genetic screen for suppressors of Abl-dependent phenotypes.25,26 In clone K18E2, the PT1 vector has integrated into the first intron of Mena, which is downstream of the initiation codon and, therefore, should result in a null mutation. Clone B2C3 encodes the murine homologue of karyopherin/importin β3 and yeast Pse1p,27 proteins that are involved in the transport of proteins and mRNA across the nuclear membrane.28,29 The RACE product suggests that a fusion protein was generated from the N-terminal 312 amino acids andlacZ. Mutational analysis of Xenopus karyopherin-β suggests that this fusion protein should bind weakly to the nuclear pore complex and to RanGTP but not to karyopherin-α28 and may act as a weak dominate negative mutation. In ES clone GC10G7, the GT1.8geo vector has integrated within the 3′ coding region of the gene for guanosine 5′-monophosphate (GMP) synthetase. GMP-synthetase catalyzes the amination of xanthosine 5′-monophosphate to form GMP in the presence of glutamine and ATP. Although GMP-synthetase is expressed in many cell types in the adult, we observed high levels of β-gal activity only in endothelial cells and a population of hematopoietic cells (Table 3).
In vitro and in vivo expression of selected clones.
Previous studies using these vectors have demonstrated that the lacZ fusion protein expression accurately represents wild-type expression of the trapped gene.15,30 31 To determine if the patterns of expression in vitro were a good predictor of in vivo expression, selected ES clones were aggregated with diploid embryos to generate chimeric mice. Analysis of lacZ reporter gene expression was performed first on chimeric embryos to assess quickly expression patterns and subsequently was confirmed in F1 embryos (summarized along with sequence analysis in Table 3). In this report, we present three clones that correspond to a sequence homologous to several ESTs (K17G2), a completely novel gene (GC11E10), andMena (K18E2). K17G2 was isolated using the PT1 vector and displayed significant sequence similarity to several ESTs from human, rodent, Drosophilia, and yeast cDNA libraries. K17G2-lacZ was expressed at low to medium levels in undifferentiated ES cells (Fig 2A), whereas its expression was restricted to blood islands (labeled bi) and some associated endothelial cells (arrows) in attached EBs (Fig 2B). Differentiation on OP9 stromal cells showed that K17G2-lacZ was expressed in some mesodermal (labeled M) and hematopoietic cells (arrow; Fig 2C and D, respectively). To analyze the expression pattern of K17G2-lacZin vivo, K17G2 ES cells were used to generate chimeric mice. As predicted by in vitro expression, K17G2-lacZ was expressed by embryonic vasculature (arrow) including the pericardium (arrow) as well as circulating blood cells (Fig 2F and G) in e12.5 embryos. In the adult, K17G2-lacZ expression was observed in splenocytes, thymocytes, and bone marrow cells and in the vasculature, including the endocardium as well as the pericardium. In addition, analysis of K17G2 F1 embryos showed additional tissues that expressed theK17G2-lacZ fusion product (Fig 2E). For example, thelacZ fusion product was expressed in the myocardium and the dorsal root ganglia (Fig 2H and I, respectively). Brother-sister matings of K17G2 heterozygous littermates failed to produce viable homozygous mice, indicating that the trapped K17G2 gene is essential for embryogenesis (data not shown).
K17G2-lacZ expression in vitro and in vivo. Overnight X-gal staining showed fusion transcript expression at medium intensity in most undifferentiated K17G2 ES cells (A). The fusion transcript was expressed in the blood island (bi) and some of the associated vascular endothelium (arrows) in attached EB culture (B). Differentiation of clone K17G2 on OP9 stromal cells demonstrated lacZ expression in mesodermal (M) colonies (C) and hematopoietic clusters (arrow; D). X-gal staining of an e10.5 F1 embryo demonstrated limitedlacZ expression in the embryo (whole mount; E). An X-gal stained e12.5 F1 embryo demonstrated lacZexpression in the pericardium (F) and vascular endothelium and circulating hematopoietic cells (G). In addition, the myocardium (H) and the dorsal root ganglia (I) also express the lacZ fusion protein.
K17G2-lacZ expression in vitro and in vivo. Overnight X-gal staining showed fusion transcript expression at medium intensity in most undifferentiated K17G2 ES cells (A). The fusion transcript was expressed in the blood island (bi) and some of the associated vascular endothelium (arrows) in attached EB culture (B). Differentiation of clone K17G2 on OP9 stromal cells demonstrated lacZ expression in mesodermal (M) colonies (C) and hematopoietic clusters (arrow; D). X-gal staining of an e10.5 F1 embryo demonstrated limitedlacZ expression in the embryo (whole mount; E). An X-gal stained e12.5 F1 embryo demonstrated lacZexpression in the pericardium (F) and vascular endothelium and circulating hematopoietic cells (G). In addition, the myocardium (H) and the dorsal root ganglia (I) also express the lacZ fusion protein.
Clone GC11E10 was isolated using the GT1.8geo vector and represents a novel ORF. The GC11E10-geo fusion protein was expressed at medium to high levels in undifferentiated ES cells (Fig 3A). In attached EBs, expression appeared within blood islands (bi) and the vasculature (arrow) associated with these structures (Fig 3B). Differentiation of GC11E10 ES cells on OP9 stromal cells demonstrated lacZ expression within mesodermal (M) colonies (Fig 3C) and high levels of expression within hematopoietic cell clusters (long arrow) and large hematopoietic cells that may be megakaryocytes (short arrows; Fig 3D). In vivo,lacZ was expressed in the yolk sac, dorsal aorta, heart, the developing liver, and vasculature (Fig 3E and F). Further analysis demonstrated lacZ expression within blood cells circulating throughout the embryo and blood islands in the yolk sac (Fig 3G and H). The GC11E10-geo fusion protein was also expressed in endothelial cells throughout the embryo, including the intersomitic vessels (arrow) shown in Fig 3I.
GC11E10-lacZ expression. Overnight X-gal staining showed fusion transcript expression at medium to high levels in most undifferentiated ES cells (A). In attached EB cultures, lacZwas expressed within blood islands (bi) and the associated vascular endothelium (arrows; B). Differentiation of clone GC11E10 on OP9 stromal cells demonstrated lacZ expression in mesodermal (M) colonies (C) and a proportion of hematopoietic clusters long arrow as well as all large hematopoietic cells (short arrows; D). Overnight whole mount X-gal staining of an e9.5 chimeric embryo and yolk sac demonstrated lacZ expression in the dorsal aorta, heart, liver, and vasculature (E). LacZ expression in the yolk sac was confined to endothelial and hematopoietic cells (F and G). LacZwas expressed by the endocardium and circulating blood cells in the heart (H) and by the intersomitic endothelial cells (arrow; I).
GC11E10-lacZ expression. Overnight X-gal staining showed fusion transcript expression at medium to high levels in most undifferentiated ES cells (A). In attached EB cultures, lacZwas expressed within blood islands (bi) and the associated vascular endothelium (arrows; B). Differentiation of clone GC11E10 on OP9 stromal cells demonstrated lacZ expression in mesodermal (M) colonies (C) and a proportion of hematopoietic clusters long arrow as well as all large hematopoietic cells (short arrows; D). Overnight whole mount X-gal staining of an e9.5 chimeric embryo and yolk sac demonstrated lacZ expression in the dorsal aorta, heart, liver, and vasculature (E). LacZ expression in the yolk sac was confined to endothelial and hematopoietic cells (F and G). LacZwas expressed by the endocardium and circulating blood cells in the heart (H) and by the intersomitic endothelial cells (arrow; I).
As discussed above, clone K18E2 (a PT1 clone) represents an integration into the first intron of Mena. Mena has been implicated in actin assembly and cell motility; thus, although its embryonic expression has not been previously described, its ubiquitous expression in rapidly dividing cells was expected. Mena-lacZ was expressed at very high levels in nearly all undifferentiated ES cells (Fig 4A) and virtually all cells in attached EBs including the blood island (bi) shown in Fig 4B. Differentiation of K18E2 on OP9 stromal cells demonstrated high levels of Mena-lacZ expression in mesodermal cells (Fig 4C), but only low level expression in a minority of hematopoietic cells (long arrow; Fig 4D). This pattern and level of lacZ expression was reproduced in F1 embryos. Mena-lacZ was expressed by almost all cells in the developing embryo with the exception of hepatocytes and some hematopoietic cells (Fig 4E and F and data not shown).
Mena-lacZ (K18E2) expression. Overnight X-gal staining demonstrated high-level lacZ expression in undifferentiated ES cells (A) and in virtually all cells in the attached EB culture, including blood islands (bi) and their associated vasculature (arrow; B). Differentiation of clone K18E2 on OP9 stromal cells followed by overnight X-gal staining demonstrated high-level lacZexpression in mesodermal (M) colonies (C), whereas most hematopoietic cells did not express lacZ (short arrows), although low-level expression was observed in some isolated hematopoietic cells (long arrows; D). Mena-lacZ was expressed at high levels in vivo, as demonstrated by strong X-gal staining in less than 90 minutes in an e10.5 F1 embryo (E). Overnight X-gal staining of an e13.5 F1 embryo showed strong lacZ expression in all tissues except the liver (F).
Mena-lacZ (K18E2) expression. Overnight X-gal staining demonstrated high-level lacZ expression in undifferentiated ES cells (A) and in virtually all cells in the attached EB culture, including blood islands (bi) and their associated vasculature (arrow; B). Differentiation of clone K18E2 on OP9 stromal cells followed by overnight X-gal staining demonstrated high-level lacZexpression in mesodermal (M) colonies (C), whereas most hematopoietic cells did not express lacZ (short arrows), although low-level expression was observed in some isolated hematopoietic cells (long arrows; D). Mena-lacZ was expressed at high levels in vivo, as demonstrated by strong X-gal staining in less than 90 minutes in an e10.5 F1 embryo (E). Overnight X-gal staining of an e13.5 F1 embryo showed strong lacZ expression in all tissues except the liver (F).
DISCUSSION
We have developed an expression-based strategy to identify and mutate genes that are preferentially expressed in cells of the hematopoietic and vascular lineages. Gene trap vectors were introduced into ES cells by electroporation and sibling clones were allowed to differentiate into attached EBs to identify expression patterns. Clones exhibiting reporter gene expression in blood islands were then differentiated on OP9 stromal cells to determine if hematopoietic cells expressed the reporter gene. From almost 1,300 clones, we isolated 79 clones with identifiable expression patterns, of which 33 were preferentially expressed in hematopoietic and/or endothelial cells. These in vitro patterns of expression, which can be analyzed relatively quickly and in large numbers, were reliable predictors of in vivo patterns of expression as subsequently determined in chimeric and F1embryos. ES clones with expression patterns of interest were then used to clone and sequence the upstream coding region of the trapped gene by 5′ RACE. Three of the clones corresponded to known genes and eight were novel. The three known genes, Mena, Karyopherin β3, and 5′GMP synthetase, have diverse biological and biochemical functions, yet little is known about their developmental expression or the consequences of mutations in these genes on mammalian development or function in the adult. We are currently introducing these mutations into the mouse germline to determine their biological function in vivo. Detailed in vivo analysis of mutant phenotypes, combined with sequence analysis and expression pattern, should provide valuable insight into the biological functions of these genes.
In vivo analysis of several of the novel hematopoietic and vascular genes has demonstrated tissue-specific expression of these genes.GC11E10 expression is confined to hematopoietic and endothelial lineages (Fig 3 and data not shown). The GC11E10 mutation has been transmitted through the germline. Brother-sister matings of heterozygous littermates are currently being performed to analyze gene function. K17G2 is expressed exclusively in the hematopoietic, vascular, heart, and sensory nervous systems in the embryo. In the adult, 17G2 expression is dramatically upregulated in the vascular system and is expressed at low levels by most hematopoietic cells and is expressed at high levels by approximately 1% of bone marrow cells (data not shown). Preliminary analysis of K17G2 homozygous embryos has determined that a significant percentage of the homozygous embryos are edematous and develop hemorrhages. More thorough investigation of expression and homozygous phenotype is required to assess the function of K17G2.
Gene trapping in ES cells is a powerful technique because it simultaneously integrates gene identification and structure, expression, and functional analysis into one process. Previously published gene trap screens have used one of these three types of analysis as the primary determinant to select clones for further study. The first group of screens used no preselection to study mutant phenotypes. Collectively, these studies have determined that nearly 40% of gene trap mutants result in recessive embryonic lethality.12,32-34 Several sequence-based screening strategies have been developed to either rapidly isolate 5′RACE sequences,35-37 isolate 3′RACE sequences,38,39 or clone proviral integration sites by plasmid rescue.40 In addition, Skarnes et al11modified the GT1.8geo vector to trap specifically genes that encode secreted or transmembrane proteins. Several groups, including ourselves, have performed screens based on regulated expression. Each of these screens analyzed clones that contained integrations into genes that were transcriptionally active in ES cells. The expression of the fusion transcripts was either analyzed by in vivo expression30 or regulation by exogenous factors15,41,42 or by in vitro differentiation.43-45 The previous in vitro prescreening strategies identified clones with regulated expression in cardiomyocyte, neuronal, and chondrocyte lineages.
Thus, the expression trapping approach described here complements and extends the previous expression-based gene trap screens by specifically identifying integrations into genes preferentially expressed in hematopoietic and endothelial lineages. In addition, we have designed the screen to perform large-scale, genome-wide scans for genes of interest. All integrations with identifiable expression patterns in vitro were catalogued to generate a biological resource of gene-trap insertions based on expression pattern, cDNA sequence, and mutant phenotypes.
The attached EB differentiation assay used here as the primary screen enabled us to identify a large number of genes with a spatially or cell-type restricted expression in several cell lineages, including hematopoietic, endothelial, stromal, and myocyte. Many cell types develop in these cultures; thus, a screen based on in vitro expression patterns would be feasible provided the patterns accurately reflect the pattern of gene expression within the context of a developing embryo or adult. Therefore, an important result from the studies reported here was the observation that the patterns of expression observed in vitro accurately paralleled the expression of the gene in vivo. Clones that exhibited lacZ expression patterns in unknown cell types are currently being tested in vivo to determine the cell lineages in which they are expressed. This information should widen the range of cell types and corresponding lineage-restricted genes that can be trapped by this strategy.
We were particularly interested in comparing the expression trapping parameters of the PT1 vector, in which the neo gene is under the control of an autonomous promoter and GT1.8geo, which will only give rise to a G418-resistant colony if the vector has integrated into an actively expressed gene. A priori, each vector has its own advantages and disadvantages. PT1 can trap genes that are not expressed in undifferentiated ES cells but that may only be activated in differentiated derivatives. On the other hand, the PT1 vector will also give rise to G418-resistant colonies after integration within intergenic regions. These integration events would be expected to lower the numbers of trapped genes per ES clones. In contrast, GT1.8geo will only give rise to G418-resistant colonies after integration within a gene, providing that the gene is already expressed in undifferentiated ES cells. Interestingly, one third of the blue PT1 clones but only one eighth of the blue GT1.8geo clones exhibited identifiable expression patterns (Table 1). Thus, although integration of the PT1 vector in both intragenic and intergenic regions can result in G418-resistant ES clones, a higher percentage of β-gal–positive PT1 clones have identifiable expression patterns. Therefore, provided that the screening method is efficient, the PT1 vector, or similar vectors, can be used to trap genes not expressed in ES cells. In addition, because the neoR gene is driven by the PGK-1promoter, it should be possible to use high G418 selection46 to produce homozygous ES cells for each gene trap line at a high throughput. Because of differing levels ofneoR activity in GT1.8geo cell lines, high G418 selection would be more difficult and labor-intensive. The homozygous ES cell lines could be used to perform chimeric analysis as well as a functional in vitro gene trap screen to assay mutations affecting development of hematopoietic, endothelial, or myoblast lineages. We are currently pursuing these strategies.
Finally, it should be possible to increase the efficiency and versatility of the screen by incorporating multiplex assays to identify various gene families. Related strategies based on the ability of ES cells to differentiate and respond to exogenous factors in vitro will also make it possible to identify genes that are differentially regulated in many distinct cell lineages in vivo. When combined with the potential of ES cells to give rise to germ cells, this approach simultaneously should provide expression, sequence, and phenotypic information on a very wide spectrum of genes. Thus, expression trapping in ES cells provides a useful complement to other strategies designed to analyze at a functional level the very large number of proteins encoded by the mammalian genome.
ACKNOWLEDGMENT
The authors thank Caryn Ito and Janet Rossant for critical reading of the manuscript; Bill Skarnes for the GT1.8geo vector; Ken Harpel for the preparation of histological sections; Sandra Tondat, Marina Gertsenstein, Lois Schwartz, and Sarang Kulkarni for transgenic work; and Michelle Tam for technical assistance.
Supported by grants from Bristol-Myers Squibb (Princeton, NJ), the Leukemia Society of America (New York, NY), the National Institutes of Health (Bethesda, MD), and the Terry Fox Foundation (Vancouver, British Columbia, Canada).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Alan Bernstein, PhD, Samuel Lunenfeld Research Institute, Mount Sinai Hospital, 600 University Ave, Toronto, Ontario, M5G 1X5, Canada; e-mail: bernstein@mshri.on.ca.