Abstract
This study evaluated whether relapse of acute promyelocytic leukemia (APL) patients from clinical remissions achieved and/or maintained with all-trans retinoic acid (RA) in combination with intensive chemotherapy is associated with leukemic cellular resistance to RA and with alterations in the PML-RAR fusion gene. We studied matched pretreatment and relapse specimens from 12 patients who received variable amounts of RA, primarily in nonconcurrent combination with daunorubicin and cytarabine (DA) on Eastern Cooperative Oncology Group (ECOG) protocol E2491, and from 8 patients who received DA only on protocol E2491. Of 10 RA-treated patients evaluable for a change in APL cell sensitivity to RA-induced differentiation in vitro, 8 showed diminished sensitivity at relapse, whereas, of 6 evaluable patients treated with DA alone, only 1 had marginally reduced sensitivity. From analysis of sequences encoding the principal functional domains of the PML and RAR portions of PML-RAR, we found missense mutations in relapse specimens from 3 of 12 RA-treated patients and 0 of 8 DA-treated patients. All 3 mutations were located in the ligand binding domain (LBD) of the RAR region of PML-RAR. Relative to normal RAR1, the mutations were Leu290Val, Arg394Trp, and Met413Thr. All pretreatment analyses were normal except for a C to T base change in the 3′-untranslated (UT) region of 1 patient that was also present after relapse from DA therapy. No mutations were detected in the corresponding sequences of the normal RAR or PML (partial) alleles. Minor additional PML-RAR isoforms encoding truncated PML proteins were detected in 2 cases. We conclude that APL cellular resistance occurs with high incidence after relapse from RA + DA therapy administered in a nonconcurrent manner and that mutations in the RAR region of the PML-RAR gene are present in and likely mechanistically involved in RA resistance in a subset of these cases.
© 1998 by The American Society of Hematology.
ALL-TRANS RETINOIC ACID (RA), which functions by inducing terminal granulocytic differentiation of acute promyelocytic leukemia (APL) cells,1-4 is a highly effective agent for inducing complete hematologic and clinical remission (CR) in APL patients.5-13 Variations in the reported incidence of RA-induced CR in APL patients (72% to 100%) have been related to early disease complications, to RA toxicity, or to other extenuating circumstances (such as inadequate diagnostic criteria).5-12 We are not aware of any well-documented case of primary clinical resistance to RA in a previously untreated APL case determined either to have the classical 15;17 chromosome translocation and/or to express the pathognomonic PML-RARα fusion gene product. On the other hand, acquired clinical resistance universally develops within a few months in APL cases treated with RA as a single agent.5-7,9 This universality logically suggests some regulatory systemic mechanism of acquired resistance.14This hypothesis has been supported by the finding that there is a rapid decrease in achievable plasma RA levels within a few days of beginning RA therapy.15-17 This, in turn, has been related to the induction of cytochrome P-450 enzymes and lipid peroxidases that enhance the metabolism of RA15,16,18 and to the induction of a cytoplasmic RA binding activity, cellular retinoic acid binding protein-II (CRABP-II), in various types of cells, including skin17 and bone marrow cells,19,20 that may then act as a systemic sump to diminish the amount of RA able to reach crucial molecular response sites in the nucleus of APL cells.4,14 However, some observations suggest that these systemic considerations may not be a sufficient explanation for the development of clinical ATRA resistance. Of particular note are observations that APL cells from many patients who relapse after RA therapy have reduced sensitivity to RA-induced differentiation in vitro,6,20,21 suggesting the presence of significant intrinsic APL cellular RA resistance. Most of these reported cases were in second relapse, having previously relapsed from chemotherapy-induced remissions, and many were treated with various types of low-dose consolidation or maintenance chemotherapy. Thus, it is of interest to determine the incidence of leukemic cellular resistance in previously untreated APL cases who relapse from remissions achieved on RA in combination with intensive consolidation chemotherapy, which has been demonstrated to be necessary for high long-term disease-free survival rates.4,8 12
Two molecular alterations have previously been associated with the development of intrinsic APL cellular RA resistance. First, CRABP-II protein RA binding activity was reported to be markedly increased in fresh bone marrow specimens from 4 RA-treated APL patients after clinical relapse compared with absent levels in pretreatment specimens.20 From these observations, it was suggested that cytoplasmic sequestration and catabolism of RA might produce intrinsic APL cellular RA resistance by impeding RA nuclear access.4,20 However, we have consistently found constitutive expression of CRABP-II mRNA and of cytoplasmic RA protein binding activity in the leukemic cells of previously untreated APL cases (Hallam et al22 and Zhou et al, manuscript submitted), raising uncertainty about the general importance of this proposed mechanism. Second, in the established APL cell culture line NB4, abnormalities of RA binding and of nuclear interprotein complex formation by the APL-specific PML-RARα fusion gene product have been reported in several sublines selected for RA resistance in vitro.23-27 In two sublines, these were related to missense mutations in the ligand binding domain (LBD) of the RARα region of PML-RARα.25,28 Notably, mutations in the LBD of normal RARα have also been related to the development of RA resistance in independently selected sublines of the RA-sensitive myeloid leukemia cell line HL-60.29-31 However, in the only reported analysis of fresh APL specimens, no mutations were found in the LBD of RARα in either the chimeric or normal allele in 20 APL cases, including 5 cases with acquired RA resistance.32
In the current study, we tested matched pretreatment and relapse APL cells from 19 patients treated on Eastern Cooperative Oncology Group (ECOG) protocol E2491 with RA + chemotherapy (daunorubicin and cytarabine [DA]) or, as controls, with DA alone12 for in vitro sensitivity to RA-induced differentiation and for gross or fine abnormalities of PML-RARα mRNA. In addition, we made this assessment in 1 protocol patient and 1 non-protocol patient who were further treated with intravenous liposomal RA after relapse from oral RA-containing therapy. The scope of the analysis was affected not only by the possibility of finding LBD mutations in RARα sequences in a manner analogous to those in resistant sublines of HL-60 or NB4, but also in consideration of reports that the PML portion of PML-RARα is importantly involved in the APL cell response to RA33-37and of a report that the RING finger and second B-box domains of the PML-region of PML-RARα were consistently mutated in a PML-RARα–induced avian leukemia model.38 The results of our analysis indicated that APL cellular RA resistance occurs with high incidence in patients relapsing from nonconcurrently administered RA + DA and that in a subgroup of these patients this is likely due to missense mutations in the RARα region of PML-RARα. These are the first reported mutations of PML-RARα under clinical conditions and, more generally, are the first reported mutations of RARα sequences in naturally occurring tumor cells in vivo. Identification of these mutations in only a subgroup of patients with APL cellular RA resistance also indicates that the molecular mechanisms of resistance are heterogeneous.
MATERIALS AND METHODS
Patients and pretreatment clinical laboratory studies.
Nineteen of the 20 patients involved in this study were participants in ECOG clinical trial E2491, which is a subset of previously untreated adult APL patients studied on intergroup protocol INT 0129.12 One patient was a non-protocol patient from the Montefiore Medical Center Oncology Service. Patient materials were obtained under an ECOG protocol-approved consent form and/or individual institutional review board approval. Baseline patient data, eg, white blood cell (WBC) count, for ECOG cases were obtained from the central data registry.
Cell preparation and in vitro tests for RA sensitivity.
Heparinized bone marrow (BM) and peripheral blood (PB) specimens were received by overnight express mail from ECOG institutions at room temperature. A low-density WBC fraction (density ≤1.077 g/mL) was isolated and evaluated as previously described.39 Isolates with ≥75% myeloblasts/promyelocytes were used for the assesment of in vitro APL cell sensitivity to RA, as previously described.30 39
Reverse transcription-polymerase chain reaction (RT-PCR) and allele-specific nested PCR analysis.
Total cellular RNA was prepared by a modified protocol of the guanidine isothiocyanate extraction-cesium chloride gradient ultracentrifugation procedure, as previously described,30 or using the TRIzol reagent (Life Technologies, Inc, Gaithersburg, MD), according to the supplier’s instructions. Reverse transcription of RNA from random hexamer primers (Pharmacia, Piscataway, NJ) and PCRs were performed essentially as previously described,39 except that amplification conditions were modified to be optimal for each primer pair and 2% formamide was included to reduce nonspecific signals.40 Primer pairs were designed using Oligo software (National Biosciences, Inc, Plymouth, MN) and synthesized by Perkin Elmer (Norwalk, CT) via the Albert Einstein College of Medicine.
For allele-specific nested PCR, an aliquot of first-round PCR product was diluted (10- to 100-fold), and 1 to 2 μL was used as template for the second-round PCR reaction following the same protocol as above. All PCR products were analyzed through 3% agarose gels run in TAE (0.04 mol/L Tris-acetate, pH 7.2, 1 mmol/L disodium EDTA) buffer.
DNA sequence analysis.
PCR products were purified either with a QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany) or recovered from a low-melting temperature agarose gel with the Wizard PCR Preps DNA Purification System (Promega, Madison, WI). These DNA fragments were directly cycle-sequenced with the corresponding PCR primers on an automated DNA sequencer using dye terminators and AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA). Sequencing results were edited using the Gene Inspector software (Textco, West Lebanon, NH).
PML-RARα RT-PCR analysis.
To classify the PML-RARα mRNA type, primer pair I (Fig 1) was used: IS (sense), 5′-ACCGATGGCTTCGACGAGTTC-3′; IAS (antisense), 5′-AGCCCTTGCAGCCCTCACAG-3′. For V-form PML-RARα cases, nested PCR with primer pair II was applied and further verified by sequence analysis, as described39: IIS, 5′-CAATACAACGACAGCCCAGAA-3′; IIAS(1), 5′-TCACAGGCGCTGACCCCATA-3′; IIAS(2), 5′-GACCCCATAGTGGTAGCCTGAGGA-3′.
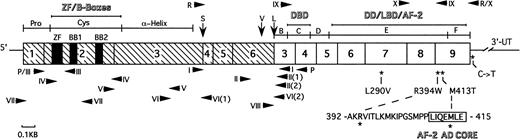
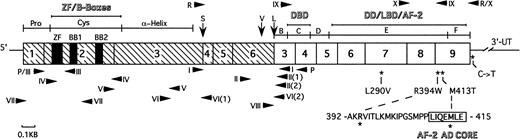
Schematic structure of PML-RAR mRNA showing the major functional domains. The PML portion is hatched and the RAR portion is clear. Numbers in the boxes indicate exons. Vertical arrows indicate the positions of break/fusion sites in the translocated PML gene that result in the formation of three different forms of PML-RAR mRNA: S-, V-, and L-forms. Arrowheads show the sites of PCR primers used in this study. Primers used for screening of the PML region are shown beneath the schematic bar, whereas those for the RAR region are shown above it. The asterisks indicate the positions of base changes found in the PML-RAR allele, and the coding region amino acid changes are shown in expanded form below the schematic bar. The abbreviations are as follows: Pro, proline-rich domain; Cys, cysteine/histidine clusters; -Helix, -helical coiled-coil dimerization domain; B-F, B- to F-region of RAR; ZF, zinc-finger RING motif; BB1 and BB2, two B-boxes; DBD, DNA binding domain; DD, dimerization domain; LBD, ligand binding domain; AF-2, activator function-2 domain; AF-2 AD CORE, 7 amino acid long AF-2 activation domain; 3′-UT, 3′ untranslated region.
Schematic structure of PML-RAR mRNA showing the major functional domains. The PML portion is hatched and the RAR portion is clear. Numbers in the boxes indicate exons. Vertical arrows indicate the positions of break/fusion sites in the translocated PML gene that result in the formation of three different forms of PML-RAR mRNA: S-, V-, and L-forms. Arrowheads show the sites of PCR primers used in this study. Primers used for screening of the PML region are shown beneath the schematic bar, whereas those for the RAR region are shown above it. The asterisks indicate the positions of base changes found in the PML-RAR allele, and the coding region amino acid changes are shown in expanded form below the schematic bar. The abbreviations are as follows: Pro, proline-rich domain; Cys, cysteine/histidine clusters; -Helix, -helical coiled-coil dimerization domain; B-F, B- to F-region of RAR; ZF, zinc-finger RING motif; BB1 and BB2, two B-boxes; DBD, DNA binding domain; DD, dimerization domain; LBD, ligand binding domain; AF-2, activator function-2 domain; AF-2 AD CORE, 7 amino acid long AF-2 activation domain; 3′-UT, 3′ untranslated region.
Strategy for PML sequencing.
PML-RARα allele-specific RT-PCR was performed first by primer pair P (Fig 1): PS, 5′-CCCAGCCCCAGCCCCAGCCCTACA-3′; PAS, 5′-AGCGGTTCCGGGTCACCTTGTTGA-3′. For nested PCR, four sets of inner primers (III through VI) were used for exon 2 and 3 segmental amplifications of the PML-RARα allele. These primers were as follows: IIIS, same as PS; IIIAS, 5′-GCGCCGCTGCAGACTCTCGAAAAAGA-3′; IVS, 5′-CCCGCTTCGGAGGAGGAGTTCCA-3′; IVAS, 5′-ACGAGCAGCACAGCGGCTTGGAACA-3′; VS, 5′-ACCCGCAAGACCAACAACAT-3′; VAS, 5′-CGACTGGCCATCTCCTCGTAG-3′; VIS, 5′-GTAGCTCACGTGCGGGCTCAGGAG-3′; VIAS(1), 5′-CTCTGGGCTGGCTTTCTTGGATACAG-3′; VIAS(2), 5′-GCGAGGGAGGGCTGGGCACTATCT-3′.
The PCR segments generated from these primer sets were sequenced as described above. PML exon 1 and 2 (partial) sequences of both alleles were also screened by direct PCR followed by sequencing with primers VII: VIIS, 5′-CCCCTTCAGCTTCTCTTCAC-3′; VIIAS, 5′-TCGCACTCAAAGCACCAGAA-3′. For the boundary region of PML-RARα, primer pairs I and II, as well as VIIIS versus IAS, were used separately in the tests of S-, V-, and L-form cases. In addition, 11 of 20 cases were also tested with primers VIIIS and VIIIAS (not shown in Fig 1) for PML exon 7a region sequence: VIIIS, 5′-CTTCCTGCCCAACAGCAACC-3′; VIIIAS, 5′-GCCCCAGGAGAACCCACTTT-3′.
Strategy for RARα sequencing.
Bi-allelic screening of the RARα C- to F-regions was first performed with the following two primer sets, which incorporate overlapping gene sequences (Fig 1): IXS, 5′-CCCCCTCTACCCCGCATCTACAAG-3′; IXAS, 5′-CGGGATCTCCATCTTCAGCGTGAT-3′; XS, 5′-TGATGCGGAGACGGGGCTGCTCAG-3′; XAS, 5′-GGGGCGGAGGGCGAGGGCTGTGTC-3′. Four heterozygous base changes detected by bi-allelic screening were further tested by nested PCR with the above-mentioned primers, after first-round PML-RARα allele-specific PCR with primer pair R: RS, 5′-GCGCACCGATGGCTTCGACGAGTTC-3′ and RAS (same as XAS).
RESULTS
Patient clinical characteristics.
Selected clinical features of each relapse case analyzed in this study are summarized in Tables 1 and2, in which the cases have been ordered on the basis of the treatment sequence with RA and/or DA chemotherapy. This systemization follows the two randomization points in protocol E2491, which involved remission induction therapy with either RA or DA and, after 2 courses of DA consolidation therapy, re-randomization to either RA maintenance therapy for 1 year or to simple observation (Obs).12 Cases no. 1 through 19 were previously untreated APL patients who were enrolled on protocol E2491, whereas case no. 20 was a non-protocol case with secondary APL that developed 2 years after treatment of breast cancer with doxorubicin and cyclophosphamide. The latter case as well as protocol case no. 19 (due to the development of a toxic complication) received only RA therapy before relapse. All other RA-treated cases received combined, nonconcurrent DA therapy on a variety of schedules as detailed in Table1 and below. Three of the E2491 patients failed to achieve CR on initial protocol therapy, either because of primary drug resistance (cases no. 12 and 15) or an intercurrent complication (case no. 16) and, thus, are considered to be salvage cases related to the analysis of relapse samples from CRs that were achieved on alternative, cross-over therapy.
Eighteen of the E2491 patients received treatment following the four possible protocol randomization sequences, ie, DA-Obs, DA-RA, RA-Obs, or RA-RA. Perhaps related to a significantly higher relapse rate of patients randomized to the DA-Obs protocol arm,12 a disproportionate number of cases in our subset never received RA therapy (8/19 [42%]). This group included 5 cases (no. 1 through 5) who achieved CR on DA and were randomized to Obs after consolidation therapy; 1 case (no. 6) who was originally randomized to receive RA but who was crossed-over to the DA arm before receiving RA; and 2 cases (no. 7 and 8) who achieved only transient CRs and who relapsed before completing or before beginning consolidation therapy, respectively (Table 1). Four of the 10 combined therapy patients followed the DA-RA treatment sequence. In cases no. 9 through 11, this was accomplished according to protocol specification. In case no. 12, initial DA therapy resulted in prolonged BM hypoplasia with failure to achieve CR status. This patient was then treated off-protocol with RA and achieved a CR after prolonged induction therapy (82 days). RA was then continuously administered until relapse 99 days later. The RA-Obs sequence (with intervening consolidation DA therapy) was followed in 4 cases; however, in only 2 of these cases (no. 13 and 14) was this accomplished per protocol. Two salvage cases (no. 15 and 16), as noted above, were also included in this category, because they were treated with RA before receiving any chemotherapy. These 2 cases each received an extra course of DA for cross-over CR induction in addition to the standard two rounds of consolidation DA therapy (Table 1). Finally, 2 cases (no. 17 and 18) followed the RA-RA sequence per protocol.
In all, 12 of 20 relapse cases received oral RA therapy (45 mg/m2/d) for quite variable periods of time (19 to 415 days) before the acquisition of relapse specimens for biological and molecular testing. In addition, 2 of these cases (no. 12 and 20) were treated with intravenous liposomal RA (90 mg/m2 every other day) for 72 and 15 days, respectively, after relapse from oral RA; in these cases, biological and molecular testing were performed on samples collected after relapse from oral RA and, additionally, after failure of liposomal RA.
For the 19 E2491 patients detailed in Table 2, the median age was 30 years (range, 16 to 68 years); 10 cases (53%) were male and 9 cases (47%) were female. The median pretreatment peripheral WBC count was 6,800/μL (range, 400 to 77,400/μL), and the median percentage of the circulating blasts plus promyelocytes was 29.5% (range, 0% to 95%). In 2 salvage cases tested, the WBC count was lower than before presentation, which may have been related to hydroxyurea administration (case no. 15) or to partially effective ATRA therapy (case no. 16). The percentage of cases assigned to the French-American-British (FAB) class M3v41 was 32% (6/19 cases). Although it is not possible to make statistical comparisons with the parent E2491/INT 0129 cohort of 221 adult PML-RARα–positive APL patients in view of the small and incomplete number of relapse cases, it seems noteworthy that the median values for the WBC count, the percentage of circulating blasts plus promyelocytes, and the percentage of M3v cases were higher for the relapse group than the corresponding median values for the overall group.42
In vitro tests for sensitivity to RA-induced differentiation.
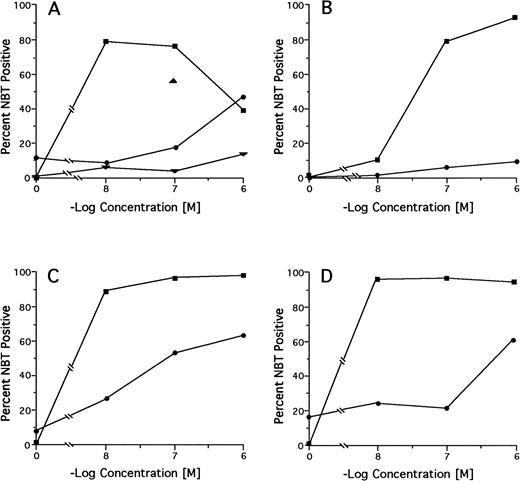
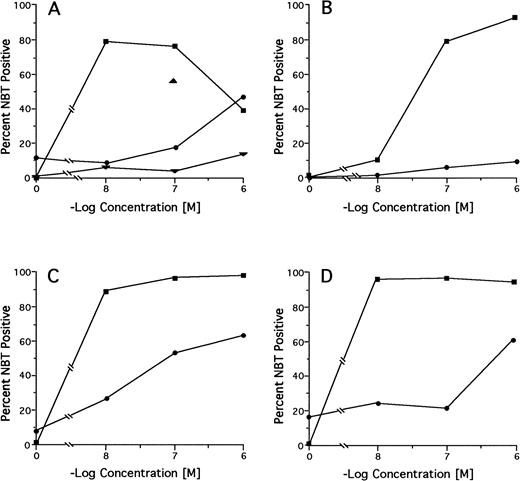
Among 18 pretreatment specimens tested by the NBT dye reduction test, we found 2 cases (no. 15 and 20) with markedly reduced and 2 cases (no. 16 and 19) with moderately reduced in vitro RA sensitivity (NBT positivity <20% at 100 nmol/L RA and <50% at ≤100 nmol/L RA, respectively; Table 3). However, in the latter 2 cases, there was no reduction in differentiation response as alternatively measured by the differentiation marker CD11b. Of 6 cases tested after relapse from treatment with DA alone, 5 (no. 1, 2, 5, 6, and 7) had preserved and 1 (no. 8) had marginally reduced in vitro RA sensitivity. Of 11 cases tested after relapse from regimens containing RA in some treatment phase, 2 had preserved or slightly reduced (no. 11 and 13), 2 had moderately reduced (no. 17 and 18), and 6 (no. 9, 10, 12, 14, 16, and 20) had markedly reduced in vitro RA sensitivity compared with pretreatment sensitivity. Neither the pretreatment nor the relapse leukemic cells from case no. 15 were responsive to RA. Serial testing of specimens from case no. 12 showed a progressive decrease in RA sensitivity from pretreatment to primary DA refractoriness to relapse from a CR achieved on oral RA to refractoriness to intravenous liposomal RA (Fig 2A). Complete RA dose-response curves for the pretreatment and relapse specimens from the 3 RA-treated cases in which mutations were found in the PML-RARα gene from the relapse specimens (no. 9, 14, and 17; see below) are shown in Fig 2B through D.
In vitro RA-induced differentiation of pretreatment and relapse APL cells from selected patients determined by the NBT test. (A) Case no. 12; (B) case no. 9; (C) case no. 17; (D) case no. 14. (▪) pretreatment sample; (•) relapse sample. Case no. 12 only: (▴) primary DA refractoriness; (▾) postintravenous liposomal RA.
In vitro RA-induced differentiation of pretreatment and relapse APL cells from selected patients determined by the NBT test. (A) Case no. 12; (B) case no. 9; (C) case no. 17; (D) case no. 14. (▪) pretreatment sample; (•) relapse sample. Case no. 12 only: (▴) primary DA refractoriness; (▾) postintravenous liposomal RA.
PML-RARα RT-PCR analysis.
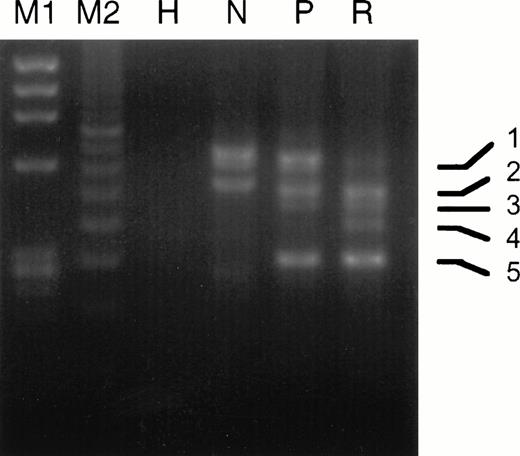
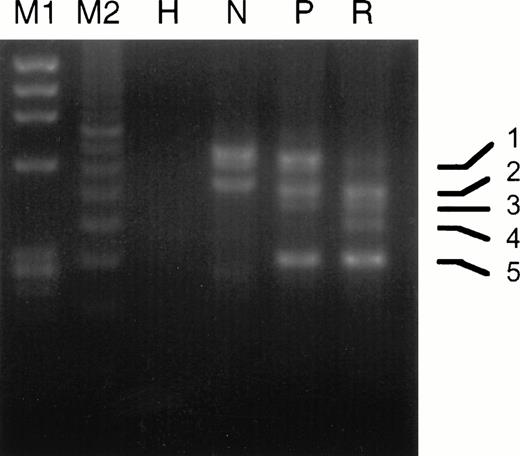
The experimental strategy used for analyzing PML-RARα by RT-PCR is shown in Fig 1 and described in Materials and Methods. As listed in Table 4, we found the following breakdown of PML-RARα mRNA types in our 20 relapse study cases: 6 L-form (30%), 9 S-form (45%), and 5 V-form (25%). This is a considerable underrepresentation of L-form cases and overrepresentation of V-form cases, which constituted 55% and 8%, respectively, of 221 overall INT 0129 protocol cases classified by PML-RARα type.42 Ten of 14 cases (71%) analyzed expressed the reciprocal RARα1-PML fusion gene mRNA, which approximates the percentage of protocol cases previously reported to express RARα1-PML.43 Four of the 5 V-form PML exon 6 break/fusion site cases noted in Table 4 (no. 6, 9, 15, and 16) were previously reported.39 In case no. 9, two additional atypical bands were noted by gel electrophoresis of RT-PCR products from the diagnostic specimen, which were increased relative to the major V-form gel bands at relapse (Fig3). Sequence analysis of these atypical bands showed alternative breaksites in exon 6 at nucleotides 1576 and 1581, which coded for truncated PML proteins, in contrast to the full-length isoform with the break/fusion site at nucleotide 1685, which, like all other major V-form isoforms, encoded a modified PML-RARα protein.39In case no. 15, in which the PML exon 6 break/fusion site was also at nucleotide 1685, a minor additional RT-PCR band was observed in 3 serial samples collected during the unsuccessful induction phase with RA, which was shown by sequence analysis to lack all of PML exon 6 and which encoded a truncated PML peptide (data not shown). Notably, in the relapse specimen from this patient after a DA-induced CR, we were unable to detect PML-RARα mRNA. No other consistent changes in the expression pattern of PML-RARα isoforms due to alternative exon splicing, as judged by gel electrophoretic analysis of RT-PCR product DNAs, were observed in any other relapse specimen.
Comparison of the gel electrophoretic pattern of RT-PCR products from the pretreatment and relapse specimens of case no. 9. PCR amplification used primer pair I as described in the Materials and Methods. M1, Hae III-digested ΦX 174 DNA; M2, 100-bp size standard; H, HL-60 cell RNA; N, NB4 cell RNA; P, pretreatment RNA; R, relapse RNA. PCR band 1, full-length V-form with PML breaksite at nt 1685; band 2, full-length product of comigrating atypical isoforms with breaksites at nts 1576 and 1581; band 3, same as band 1 but lacking exon 5 due to alternative splicing; band 4, same as band 2 but lacking exon 5; band 5, isoform lacking exons 5 and 6 due to alternative splicing.
Comparison of the gel electrophoretic pattern of RT-PCR products from the pretreatment and relapse specimens of case no. 9. PCR amplification used primer pair I as described in the Materials and Methods. M1, Hae III-digested ΦX 174 DNA; M2, 100-bp size standard; H, HL-60 cell RNA; N, NB4 cell RNA; P, pretreatment RNA; R, relapse RNA. PCR band 1, full-length V-form with PML breaksite at nt 1685; band 2, full-length product of comigrating atypical isoforms with breaksites at nts 1576 and 1581; band 3, same as band 1 but lacking exon 5 due to alternative splicing; band 4, same as band 2 but lacking exon 5; band 5, isoform lacking exons 5 and 6 due to alternative splicing.
Sequence analysis of the PML-region of PML-RARα.
The primary objective of this analysis was to determine if PML-RARα mRNA from any of the matched pretreatment-relapse APL specimens contained base changes in PML exons 1 through 3, which are conserved in all normal PML mRNA isoforms44 and which encode the proline-rich, RING/B-box elements, and coiled-coil (α-helical) dimerization domain of PML (see Fig 1 and Borden et al46and de The et al47). From unambiguous, bi-directional automated sequencing results in all 20 cases, no base changes were observed in PML exons 1 through 3 of PML-RARα or in normal PML in exon 1 or exon 2 sequences coding for the RING finger and first B-box (see Fig 1). Additionally, in 11 relapse cases tested, no base changes were observed in PML exon 7a, which is conserved in all PML isoforms and contains a casein kinase-II phosphorylation site.44
Sequence analysis of the RARα-region of PML-RARα.
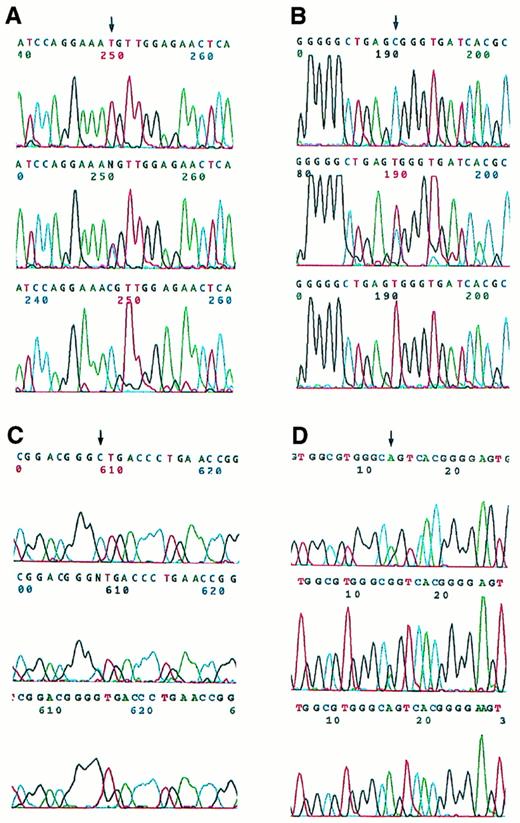
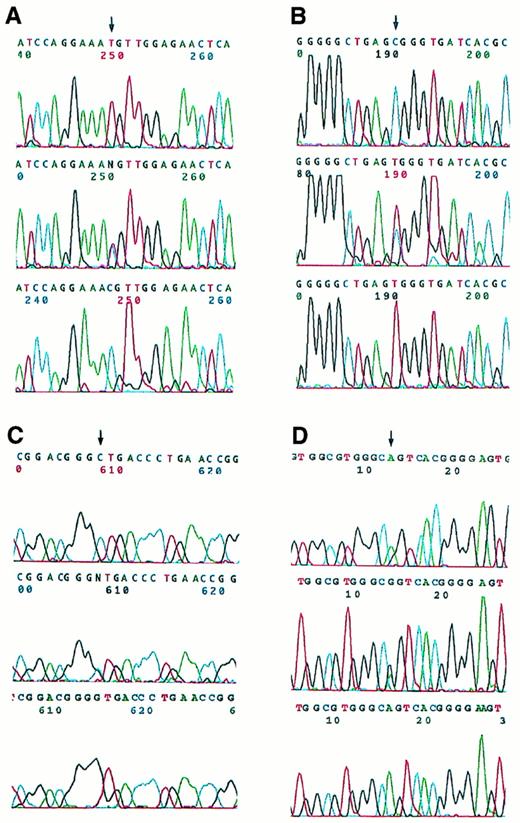
Bi-allelic sequence analysis was used to investigate RARα exons 4 through 9 (C- to F-regions) as well as 16 nucleotides of the 3′-UT region (Fig 1). Heterozygous base changes were detected in 4 relapse specimens, which were then further studied by allele-specific amplification and found to reside in the PML-RARα allele and not the normal RARα allele (Fig 4). In 3 cases (no. 9, 14, and 17), these base changes were detected in the coding region of PML-RARα mRNA from the relapse specimen but not the pretreatment specimen. In 1 case (no. 2), a C to T substitution was found in the second nucleotide beyond the PML-RARα coding region stop codon (S-form nucleotide 2478),47 ie, at the beginning of the PML-RARα mRNA 3′-UT region, in the pretreatment as well as the relapse specimen. No base changes were observed in the RARα B-region or PML-region adjacent to the RAR-region boundary in the PML-RARα allele of these cases.
Automated DNA sequence analysis of PCR products from four APL cases with single nucleotide changes. The upper panels show bi-allelic sequencing results for the pretreatment samples, which contain normal signals as indicated by the arrows in (A), (B), and (C), whereas (D) demonstrates a heterozygous pattern. The middle panels show heterozygous bi-allelic signals for the corresponding relapse samples. The lower panels show the results of nested, PML-RAR allele-specific amplification of PCR products from the relapse cases. (A) Case no. 9, T→C (Met→Thr). (B) Case no. 17, C→T (Arg→Trp). (C) Case no. 14, C→G (Leu→Val). (D) Case no. 2, C→T in 3′-UT region (illustrated from antisense strand sequence analysis, ie, shown as G→A; in the upper panel the heterozygotic mutant A appears on the shoulder of the normal G).
Automated DNA sequence analysis of PCR products from four APL cases with single nucleotide changes. The upper panels show bi-allelic sequencing results for the pretreatment samples, which contain normal signals as indicated by the arrows in (A), (B), and (C), whereas (D) demonstrates a heterozygous pattern. The middle panels show heterozygous bi-allelic signals for the corresponding relapse samples. The lower panels show the results of nested, PML-RAR allele-specific amplification of PCR products from the relapse cases. (A) Case no. 9, T→C (Met→Thr). (B) Case no. 17, C→T (Arg→Trp). (C) Case no. 14, C→G (Leu→Val). (D) Case no. 2, C→T in 3′-UT region (illustrated from antisense strand sequence analysis, ie, shown as G→A; in the upper panel the heterozygotic mutant A appears on the shoulder of the normal G).
Three of the PML-RARα RARα region base changes were present in the LBD and represented point mutations resulting in amino acid codon changes. These are described with reference to normal RARα1, because their position is variable in PML-RARα depending on the type and isoform of PML-RARα mRNA. In case no. 9, a T to C change was noted in nucleotide 2745 of this shortened V-form PML-RARα mRNA (using the sequence published by de The et al47 for L-form PML-RARα as a reference). Relative to normal RARα1, this corresponds to nucleotide 1340 (reference sequence, Giguere et al48) and produces a change from methionine to threonine in codon 413 (Met413Thr). This codon is centrally located in the highly conserved core of the activator function-2 (AF-2) activation domain (Fig1).49 In case no. 14, a C to T change was detected in S-form nucleotide 1955,47 which changed the codon specificity from leucine to valine. This corresponds to RARα 1 codon 290 (Leu290Val)48 and is located more proximally in the LBD (Fig 1). In case no. 17, a C to G transversion was detected in the PML-RARα S-form nucleotide 2267,47 which changed the amino acid triplet code from arginine to tryptophan. This mutational site corresponds to RARα 1 codon 394 (Arg394Trp)48 and is located just upstream from the conserved AF-2 activation domain (Fig1).
DISCUSSION
A lead finding of this study was that 8 of 10 evaluable RA-treated relapse cases, including 6 of 8 cases that relapsed after DA consolidation therapy, had reduced APL cell sensitivity to RA-induced differentiation in vitro (Table 3). This indicates that diminished APL cellular RA sensitivity occurs with high frequency after relapse of previously untreated APL cases treated with RA in combination with intensive consolidation chemotherapy. This is similar to previously reported findings in patients studied in second relapse after failing both chemotherapy and RA and/or in previously untreated cases that received less intensive postremission chemotherapy with or without continuous maintenance RA.6,20,21 In the current study, relapse cases with reduced RA sensitivity were seen after a variety of RA treatment schedules, including patients who received ≥120 days of RA (5/6), who received ≤52 days of RA (2/3), who received RA only before DA (2/3), who received RA only after DA (3/4), who received RA both before and after DA (2/2), who relapsed while taking RA (4/4), who relapsed within 90 days after stopping RA (2/2), and who relapsed after ≥174 days after stopping RA (2/4). Clearly, with such small case numbers it is not possible to make any conclusions about whether any of these conditions favored the development and/or maintenance of APL cellular RA resistance. However, they do suggest that there was no tight association between the amount of RA and the development of RA resistance and that even relatively short periods of RA treatment can lead to durable APL cellular resistance in some cases. These results seem remarkably different from our observation that only 1 of 6 patients treated with DA alone had reduced APL cellular RA sensitivity (relatively minor) at relapse. These data, then, imply that RA was a positive selective force for RA resistant relapse APL cells and, conversely, that DA alone, as administered in protocol E2491, was not a potent selective factor for cellular RA resistance. However, the DA could have had a facilitative role in conjunction with RA. Our results fundamentally disagree with early reports that relapse APL cells from RA-treated patients retain in vitro ATRA sensitivity,16,50which has been partly attributed to testing with a single high concentration of RA (1 μmol/L) that could have overlooked reduced sensitivity.14,20 Even so, in our study, 2 of 10 relapse patients who received substantial amounts of RA had either retained or recovered APL cell in vitro sensitivity to RA at relapse (cases no. 11 and 13), indicating that the stable loss of APL cell RA sensitivity is not universal in RA treated patients, a finding also in accord with previous reports.6,20 21
This study also provided some information about primary APL cell RA resistance. When RA-naive patients have been documented to be t(15;17)-positive and/or PML-RARα–positive, ie, to have genetically defined APL, the APL cells are almost invariably found to be highly sensitive to RA-induced differentiation in vitro.4,14,39 Notably, 2 of 18 of the current PML-RARα–positive cases tested had markedly reduced sensitivity to RA-induced differentiation in vitro at presentation (cases no. 15 and 20; Table 3). In case no. 20, a non-protocol case with secondary APL, this seems likely related to chemotherapy with doxorubicin and cyclophosphamide 2 years previously for breast cancer, because drug resistance in secondary leukemias is well-recognized51 and because diminished APL RA sensitivity has been documented after APL relapse from extensive chemotherapy.2,21 Perhaps most remarkably, this patient relatively promptly achieved CR (in 39 days) that lasted for almost 1 year on RA maintenance therapy alone. This suggests an alternative mechanism of RA-selected resistance, because patients with APL cells resistant in vitro after relapse from RA therapy are not frequently durably responsive to retreatment with RA in vivo.5,6,9,20,21 In case no. 15, there also may be an unusual explanation for the primary RA resistance that included failure to respond clinically to RA induction therapy. At relapse after achieving CR on cross-over DA therapy, the APL cells of this patient no longer had detectable PML-RARα, as has been reported for at least 1 other documented PML-RARα–positive APL case.52 Thus, we speculate that the lack of pretreatment APL cell RA sensitivity in this case could have been due to an initial mixed cell or chimeric leukemia at presentation. Consistent with this possibility, 72% of the presentation leukemic cells were positive for CD11b antigen expression,39 which is typically expressed at very low levels on APL cells.53 By this reckoning, the RA induction therapy could have selected against the PML-RARα–positive subpopulation. Against this possibility, PML-RARα mRNA remained detectable until the time RA therapy was discontinued on day 44; however, this is not conclusive, because PML-RARα mRNA signals frequently remain positive by sensitive RT-PCR tests in RA-treated APL cases through RA induction treatment and at CR.11,54 In addition to these 2 unusual cases, 4 other cases (cases no. 9, 11, 16, and 19; Table 3) showed some apparently reduced responsiveness to 10 or 100 nmol/L RA, as measured by the NBT test, in comparison to the high degree of responsiveness of the other cases in the current patient cohort and in a larger cohort of 85 pretreatment APL patients.39 Although it is unclear if these reductions are significant, it seems notable that they all occurred in the RA-treated relapse group and that 2 of these cases (cases no. 9 and 16) were prospectively identified as possibly high-risk cases with a short V-form of PML-RARα.39 In sum, these results suggest that primary RA resistance or a predisposition to develop RA resistance exists in a small subset of APL patients and that the reasons for this are heterogeneous.
The central molecular finding of this study was point mutations in the RARα coding region of the PML-RARα fusion gene from 3 of 12 patients after relapse from treatment regimens containing RA. Conversely, no coding region mutations were found in APL cells from 8 patients treated with DA alone, the same chemotherapy used in a sequential manner for induction, and/or consolidation therapy in 10 of 12 of the RA-treated patients. Although the number of cases is too small to test statistically, the finding of mutations in 3 of 12 RA-treated versus 0 of 8 of non–RA-treated patients is consistent with the hypothesis that the PML-RARα mutations were related to RA therapy and to RA resistance. This hypothesis is also supported by descriptive features of the 3 mutant cases, including the lack of PML-RARα mutations in the pretreatment APL cells, the diminished RA sensitivity of the relapse APL cells, and, most importantly, the nature of the mutations in the relapse cells.
All three PML-RARα mutations were missense mutations resulting in amino acid changes in the LBD of the RARα region. Both the Met413Thr mutation in case no. 9 and the Arg394Trp mutation in case no. 17 constitute 2 of the 24 amino acids that have been identified by crystallographic analysis of the RAR LBD to contribute directly to stabilization of the RA binding pocket through the formation of hydrogen bonds or van der Waals contacts with the bound RA molecule.55 However, other data indicate that the Met413Thr mutation likely has only a minor effect on the binding affinity of RA for RARα.56 Rather, the major dysfunction of this mutation almost certainly arises because of its central location in the 7-amino acid core of the AF-2 activation domain (AD), which resides in the 12th and final α-helix of the LBD near the carboxy-terminus of RARα.49,55 After RA binding, the 12th α-helix has been demonstrated to undergo a crucial configurational change that brings the AF-2 AD core sequence into apposition with specific amino acid motifs in coactivator nuclear proteins that are essential for RAR-mediated transcriptional activation.49,55,57,58Site-directed mutagenesis studies in which Met 413 and the adjacent hydrophobic residue Leu 414 were deleted or replaced with the small, neutral amino acid alanine extinguished both binding to coactivator sequences and positive transcriptional activation function.56-59 Although studies have not been performed with mutations of Met 413 alone, it seems highly probable that replacement of the hydrophobic, sulfhydryl-containing R-group of methionine with the small polar group of threonine would be inactivating, as was demonstrated by site-directed mutagenesis for the nearby Met 406 residue.56 The Met413Thr mutation probably has only a relatively minor effect on RA binding affinity because it only comes into contact with the RA molecule after the RA-induced configurational shift of the 12th α-helix over the binding pocket, where it serves as a lid for the presituated ligand.55 On the other hand, the Arg394Trp mutation in case no. 17, which is situated in the 11th α-helix that also undergoes important configurational changes on ligand binding,49,55 seems quite likely to produce significantly altered RA binding, as was recently reported for a nearby Leu398Pro mutation in PML-RARα in an RA-resistant subline of NB4 cells.25 Furthermore, the amino acid change involves a radical shift from the most hydrophilic (Arg) to the most hydrophobic (Trp) of amino acids.60 The Leu290Val mutation in case no. 14 is located in the β-turn between α-helices 5 and 6, and nearby residues in this β-turn also are involved in forming the binding pocket for RA.55 Although the amino acid change from leucine to valine is much more conservative than the other two mutations, marked changes in protein interactions have, nevertheless, been associated with this relatively minor shift in R-group length on hydrophobic amino acids (eg, see Heery et al58). Studies are in progress to evaluate the protein interactions and functional properties of the three mutant PML-RARα proteins.
In addition to the coding region mutations, a C to T change was found in the noncoding region of PML-RARα in both the pretreatment and relapse specimens from 1 chemotherapy treated patient whose cells remained RA sensitive at relapse (case no. 2). This likely functionally neutral base change raises the possibility that the development of mutations selectively in the PML-RARα allele rather than the normal RARα allele, both in our in vivo study and in NB4 cell line mutants,25 28 could be related to modification of the PML-RARα allele (or of the RARα allele involved in the translocation) in a way that increases susceptibility to mutational events.
To our knowledge, the PML-RARα mutations described here are the first reported in this fusion gene in APL cells under naturally occurring conditions. More generally, they appear to be the first report of clinical mutations in an RAR sequence, aside from the integration of a hepatitis B virus in the B-C region of RARβ in a human hepatocellular carcinoma.61 In a previous report using single-strand conformational polymorphism (SSCP) analysis after PCR amplification from DNA of the last 3 coding exons from the E/F region of RARα, no RAR mutations were observed in 118 specimens from a variety of human cell lines and fresh cells from patients with acute myeloid leukemia or myelodysplastic syndromes.32 This sampling included 20 APL patients, 5 of whom had acquired and 1 of whom reportedly had primary APL cellular RA resistance, and in 7 newly diagnosed cases, the PCR-SSCP analysis included all RARα exons encoding the B- to F-regions. These results in no way conflict with our results that were obtained in a highly select group of APL patients using direct sequencing of RT-PCR products, a technique that has a lower rate of false-negatives than PCR-SSCP analysis. Our sequencing results were also in the great majority negative, ie, among PML-RARα–positive relapse cases no mutations were found in the RARα region of PML-RARα in 16 of 19 overall cases, in 8 of 11 RA-treated cases, or in 5 of 8 RA-treated cases with diminished APL cell RA sensitivity. Furthermore, from the bi-allelelic sequence analysis, we were able to conclude that no mutations were present in any case in the C- through F-regions of the normal RARα allele, which encode the DNA binding domain, hinge/corepressor region, and the ligand binding/coactivator domain of RARα. In addition, the PML region of PML-RARα that specifies the proline-rich amino-terminus, the RING zinc-finger, both B-boxes and the coiled-coil sequence, which appear to be the principal functional domains of PML and which contained two consistent mutations in an avian PML-RARα leukemia model,38 were shown to be free of mutations. Finally, from analysis of the gel electrophoretic pattern of the RT-PCR DNA products, we found no evidence of abnormal PML-RARα splice variants that might code for competitive inhibitory proteins in any of the relapse cases that lacked PML-RARα mutations. However, in case no. 9, which harbored the Met413Thr mutation, we did observe minor isoforms of PML-RARα due to alternative break/fusion sites in PML exon 6 that increased in the relapse APL cells (Fig 3), and we cannot exclude the possibility that these novel isoforms, which encode truncated PML proteins, contributed to the RA resistance in this case. From the overall analysis, we conclude that it is improbable that primary structural alterations of PML-RARα–encoded or normal RARα-encoded proteins account for RA resistance in the majority of APL relapse cases. Given the reported findings that independent subclones of RA-resistant NB4 cells consistently form variable abnormal protein complexes involving RA-bound PML-RARα,25-27 it still must be considered that abnormal posttranslational modification of PML-RARα or aberrations in the other nuclear proteins that form part of the interactive protein complex with PML-RARα could contribute to selectable APL cell RA resistance in such cases.
The apparent rarity of RAR sequence mutations in tumor cell populations enhances the possibility that the RAR region mutations in PML-RARα were acquired during the treatment course. However, our study did not discriminate between this possibility and the alternative that the mutations were present in a small fraction of APL cells before treatment that were selected during RA therapy. A strong consideration in the 2 cases that relapsed during or shortly after RA maintenance therapy (cases no. 9 and 17) is that the chemotherapeutic agents administered during the induction and/or consolidation phase of protocol treatment could have generated the PML-RARα mutations, which were then selected by continuous RA maintenance treatment. This consideration does not apply in mutant case no. 14 in which RA was administered for a 52-day course leading to CR before the administration of consolidation DA. Because RA has mutational suppressive rather than inductive activity,62 it seems more plausible in this case that RA induction therapy might have selected a pre-existing PML-RARα mutant subpopulation, which was also associated with other drug-resistance properties leading to the early relapse shortly after finishing consolidation chemotherapy. Resolution of the timing of the development of PML-RARα mutations selective for RA resistance is an important issue, because it has practical implications for the scheduling of the administration of these two classes of agents that have been demonstrated to have clinical synergy.8,12In future studies, it will also be of interest to determine the effect of two therapeutic modifications associated with improved clinical outcome on the incidence of RA resistance and PML-RARα gene mutations in relapse APL cells: (1) after the use of higher doses of anthracycline (daunorubicin), reported to improve disease-free survival in chemotherapy-alone regimens,63 in combination with RA; and (2) after the administration of concurrent RA + chemotherapy, reported to improve disease-free survival compared with sequential, nonconcurrent RA and chemotherapy.13
ACKNOWLEDGMENT
The authors thank Dr Janet Andersen (Dana Farber Cancer Institute, Boston, MA) for careful reading of the manuscript and Susan Allen (ECOG, Brookline, MA) for assistance with patient data analysis.
Supported by Public Health Service Grants No. CA56771, CA17145, CA21115, and P30CA13330 and by the National Leukemia Research Association, Inc.
This study was in the majority based on protocol E2491, “Phase III Randomized Study of All-trans Retinoic Acid versus Cytosine Arabinoside and Daunomycin for Patients with Previously Untreated Acute Promyelocytic Leukemia,” conducted by the Eastern Cooperative Oncology Group (Robert L. Comis, MD, Chair). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
Address reprint requests to Robert E. Gallagher, MD, Department of Oncology, Montefiore Medical Center, 111 E 210th St, Bronx, NY 10467.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.