Abstract
The mi locus encodes a member of the basic-helix-loop-helix-leucine zipper protein family of transcription factors (hereafter called MITF). We have reported that expression of several genes was impaired in cultured mast cells (CMCs) ofmi/mi mice due to a defective transactivation ability of mutant MITF (mi-MITF). Because attachment of mi/mi CMCs to fibroblasts is impaired, we examined the expression of integrin genes in mi/mi CMCs in the present study. Among the integrin genes examined, the expression of integrin 4 subunit was barely detectable in mi/mi CMCs, and the 4 protein was not detected by flow cytometry either. The specific adhesion to vascular cell adhesion molecule-1 (VCAM-1), the ligand for 4 subunit, was observed in +/+ CMCs but not in mi/mi CMCs, indicating that the expression of integrin 4 subunit at a functional level did not occur in mi/mi CMCs. In the promoter region of the 4 subunit gene, there was a CACTTG motif to which normal MITF (+- MITF) bound. The coexpression of +-MITF but not of mi-MITF transactivated the promoter of the 4 subunit gene. The deletion or mutation of the CACTTG motif abolished the transactivation by +-MITF, suggesting that +-MITF directly transactivated the gene encoding 4 subunit of integrin.
© 1998 by The American Society of Hematology.
THE mi LOCUS OF MICE encodes a member of the basic-helix-loop-helix-leucine zipper (bHLH-Zip) protein family of transcription factors (hereafter calledmi-transcription factor [MITF]).1,2 The MITF encoded by the mutant mi allele (hereafter mi-MITF) deletes 1 of 4 consecutive arginines in the basic domain.1,3,4 The mi-MITF is defective in the DNA binding activity and the nuclear localization potential,5,6and it does not transactivate target genes.6-11 Themi/mi mice show microphthalmia, depletion of pigment in both hair and eyes, osteopetrosis, and a decrease in the number of mast cells.12-16 In addition to the decrease in number, the phenotype of mast cells is abnormal in mi/mimice.17-20 The expression of the mouse mast cell protease 6 (MMCP-6) and c-kit receptor tyrosine kinase genes was remarkably reduced in skin mast cells of mi/mimice.18 We have also demonstrated the involvement of +-MITF in the transactivation of the MMCP-6, c-kit, MMCP-5, and p75 nerve growth factor receptor genes in cultured mast cells (CMCs).7-10
We previously reported that attachment of CMCs derived frommi/mi mice (mi/mi CMCs) to Swiss albino/3T3 fibroblasts was impaired when compared with that of normal (+/+) CMCs.17 Since we demonstrated that the interaction of c-kit receptor with its ligand (stem cell factor [SCF]) plays an important role in the attachment of CMCs to fibroblasts,21 the impaired attachment of mi/miCMCs appears partly attributable to the low expression of c-kit. On the other hand, several members of the integrin family are known to be involved in the attachment of mast cells.22-28 Integrins are heterodimeric (αβ) adhesion molecules, and mast cells express α3, α4, α5, αv, β1, and β7 integrin subunit genes.25 In the present study, we examined whether the expression of integrin genes was impaired inmi/mi CMCs. We found that the expression of integrin α4 subunit was deficient in mi/mi CMCs. There was a CACTTG motif in the promoter region of the α4 subunit gene, and the binding of +- MITF to the CACTTG motif transactivated the α4 gene.
MATERIALS AND METHODS
Mice.
The original stock of C57BL/6-mi/+ (mi/+) mice was purchased from the Jackson Laboratory (Bar Harbor, ME). The mice were maintained in our laboratory by consecutive backcrosses to our own inbred C57BL/6 colony (more than 15 generations at the time of the present experiment). Female and male mi/+ mice were crossed together, and the resulting mi/mi mice were selected by their white coat color.12,13 Mast cell-deficient (WB × C57BL/6) F1-W/Wv(W/Wv) mice were purchased from the Japan SLC (Hamamatsu, Japan).29
Cells.
Pokeweed mitogen-stimulated spleen cell conditioned medium (PWM-SCM) was prepared according to the method described by Nakahata et al.30 Mice of mi/mi,W/Wv, and control C57BL/6-+/+ (+/+) genotypes were used at 2 to 3 weeks of age to obtain CMCs. The mice were killed by decapitation after ether anesthesia and the spleens were removed. Spleen cells were cultured in α-minimal essential medium (α-MEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% PWM-SCM and 10% fetal calf serum (FCS; Nippon Bio-supp Center, Tokyo, Japan). Half the medium was replaced every 3 days. Cells were harvested at various times after the initiation of the culture. Cytospin preparations of cells were fixed with Carnoy’s solution and stained with alcian blue and nuclear fast red. The proportions of alcian blue-positive cells were determined under the microscope. IC-2 cells were provided by Dr I. Yahara31 (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) and maintained in α-MEM supplemented with 10% PWM-SCM and 10% FCS. CHO cells (American Type Culture Collection, Manassas, VA) were maintained in α-MEM supplemented with 10% FCS. In one experiment, recombinant mouse SCF (rmSCF; a generous gift of Kirin Brewery Co Ltd, Tokyo, Japan) was added to the α-MEM containing 10% PWM- SCM and 10% FCS.
Semiquantitative reverse transcriptase modification of polymerase chain reaction (RT-PCR).
Total RNA (5.0, 0.5, and 0.05 μg) obtained from +/+ or mi/miCMCs was reverse transcribed in 20 μL of the reaction mixture containing 20 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim GmbH Biochemica, Mannheim, Germany) and random hexamer. One microliter of each reaction product was amplified in 25 μL of PCR mixture containing 0.125 U of Taq DNA polymerase (Takara Shuzou, Kyoto, Japan) and 12.5 pmol each of sense and antisense primers by 30 cycles of 1 minute of denaturation at 94°C, 2 minutes of annealing at 55°C, and 2 minutes of synthesis at 72°C. Ten microliters of the PCR products was electrophoresed in 2.0% agarose gel containing ethidium bromide. The following oligonucleotide primers were used: α3: sense primer, 5′- ATTGACTCAGAGCTGGTGGAGGAG (3223 through 3246), antisense primer, 5′-TACTTGGGCATAATCCGGTAGTAG (3849 through 3872); α4: sense primer, 5′-CTTCTGTCTGCGCTGTGGAC (1081 through 1100), antisense primer, 5′-CCATTTCAACCATCACAGCT (1202 through 1221); α5: sense primer, 5′-CATTTCCGAGTCTGGGCCAA (2836 through 2855), antisense primer, 5′-TGGAGGCTTGAGCTGAGCTT (3136 through 3155); αv: sense primer, 5′-GTTGGGAGATTAGACAGAGGA (2787 through 2810), antisense primer, 5′-CAAAACAGCCAGTAGCAACAA (3031 through 3054); β1: sense primer, 5′-TGTTCAGTGCATATCCGGCA (2045 through 2064), antisense primer, 5′-CCTCATACTTCGGATTGACC (2454 through 2473); β7: sense primer, 5′-ATGGTGGATTCATCAACTGTT (33 through 53), antisense primer, 5′-TGGCTGGCAGGATCCCTGCA (153 through 173); β-actin: sense primer, 5′-TAAAGACCTCTAT GCCAACAC (950 through 970), antisense primer, 5′-CTCCTGCTTGCTGATCCACAT (1143 through 1163). The numbers represent the nucleotide numbers on the complementary strands of each cDNA sequence.23 32-34
Flow cytometry.
CMCs were harvested and washed once with cold phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin (BSA) and 0.1% sodium azide. The cells were incubated with PS/2 anti-integrin α4 rat monoclonal antibody35 (MoAb; Chemicon, Temecula, CA) at 4°C for 30 minutes, rinsed, developed with fluorescein isothiocyanate-conjugated mouse antirat IgG, and then analyzed on a FACScan (Becton Dickinson, Los Angeles, CA).
Construction of expression plasmids.
Bluescript KS(−) plasmid (pBS; Stratagene, La Jolla, CA) containing the whole coding region of +-MITF or mi-MITF (hereafter called pBS-+-MITF and pBS-mi-MITF, respectively) had been constructed in our laboratory.5 The vascular cell adhesion molecule-1 (VCAM-1) cDNA was obtained by PCR according to the sequence reported by Araki et al36 and was verified by sequencing. The VCAM-1 cDNA was subcloned into pBS (pBS-VCAM-1). The pEF-BOS expression vector was kindly provided by Dr S. Nagata37 (Osaka University Medical School, Osaka, Japan). The Sma I-HincII fragment of pBS-+-MITF, pBS-mi-MITF, or pBS-VCAM-1 was introduced into the bluntedXba I site of pEF-BOS (hereafter called BOS-+-MITF, BOS-mi-MITF, and BOS-VCAM-1, respectively). We also produced pEF-BOS containing the antisense VCAM-1 cDNA (BOS-control).
Adhesion assay.
Using electroporation apparatus (BioRad, Hercules, CA), the BOS-VCAM-1 or BOS-control was cotransfected into CHO cells with pSTneo, the expression plasmid containing the neomycin-resistance gene.38 The neomycin-resistant CHO cell clones were selected by culturing α-MEM supplemented with 10% FCS and G418 (1.0 mg/mL; GIBCO BRL, Grand Island, NY) for 4 weeks. The CHO cells transfected with either BOS-VCAM-1 or BOS-control were grown overnight in 96-well polystyrene plate to form a confluent monolayer. CMCs were collected and resuspended in α-MEM containing 10% PWM-SCM and 10% FCS at a final concentration of 1.0 × 106 cells/mL. CMCs were labeled by [3H] thymidine (10 μCi/mL; Amersham, Arlington Heights, IL). After incubating for 24 hours at 37°C, labeled CMCs were washed twice with α-MEM without PWM-SCM and FCS and resuspended in α-MEM at the concentration of 4.0 × 105 cells/mL before using the adhesion assay. The radioactivity of labeled CMCs (4.0 × 104) was measured by a liquid scintillation counter (Amersham). The CMCs were added to each well containing the confluent monolayer of CHO cells transfected with either BOS-VCAM-1 or BOS-control. The plates were then incubated at room temperature for 30 minutes. CMCs that did not bind to CHO cells were removed by washing with 0.2 mL of α-MEM three times, and the radioactivity that remained with CHO cells was determined. The necessity of the α4 subunit for the binding was confirmed by incubating CMCs with PS/2 anti-α4 MoAb for 30 minutes before the coculture with CHO-VCAM-1 cells.
Promoter of integrin α4 subunit gene and reporter plasmids.
The promoter region of integrin α4 subunit gene was obtained with PCR.39 The isolated promoter region (−949 to +221, +1 shows transcription initiation site) was cloned into pBluescript and sequenced. The luciferase gene subcloned into pSP72 (pSPLuc) was generously provided by Dr K. Nakajima40 (Osaka University Medical School). To construct reporter plasmids, a DNA fragment containing the promoter region and the first exon (noncoding region) of the integrin α4 subunit gene (−949 to +221) was cloned into the upstream of the luciferase gene in pSPLuc. The deletion of the integrin α4 promoter was produced by PCR using the appropriate primers. The mutation was introduced by PCR with mismatch primers. Deleted or mutated products were verified by sequencing.
Transfection of reporter plasmids and luciferase assay.
Reporter plasmids (40 μg) were transfected into IC-2 cells by electroporation (380 V, 975 μF). The cells were harvested 18 hours after the transfection and lysed with 0.1 mol/L potassium phosphate buffer (pH 7.4) containing 1% Triton X-100. Soluble extracts were then assayed for luciferase activity with a luminometer LB96P (Berthold GmbH, Wildbad, Germany). The cotransfection of the reporter (40 μg) and expression (5 μg) plasmids was also performed.
EGMSA.
The production and purification of glutathione-S-transferase (GST)-+-MITF or GST-mi-MITF fusion protein was described previously.5 Oligonucleotides were labeled with α-[32P]-dCTP by filling 5′-overhangs and used as probes of EGMSA. DNA-binding assays were performed in a 20 μL reaction mixture containing 10 mmol/L Tris-HCl (pH 8.0), 1 mmol/L ethylenediaminetetraacetic acid (EDTA), 75 mmol/L KCl, 1 mmol/L dithiothreitol, 4% Ficoll type 400, 50 ng of poly (dI-dC), 25 ng of labeled DNA probe, and 1.0 μg of GST-+-MITF fusion protein. In some experiments, 1.0, 2.0, or 4.0 μg of GST-mi-MITF was added to the reaction mixture containing 1.0 μg of GST-+-MITF. After incubation at room temperature for 15 minutes, the reaction mixture was subjected to electrophoresis at 14 V/cm at 4°C on a 5% polyacrylamide gel in 0.25× TBE buffer (1× TBE is 90 mmol/L Tris-HCl, 64.6 mmol/L boric acid, and 2.5 mmol/L EDTA, pH 8.3). The polyacrylamide gels were dried on Whatman 3MM chromatography paper (Whatman, Maidstone, UK) and subjected to autoradiography.
RESULTS
Impaired expression of integrin α4 subunit in mi/mi CMCs.
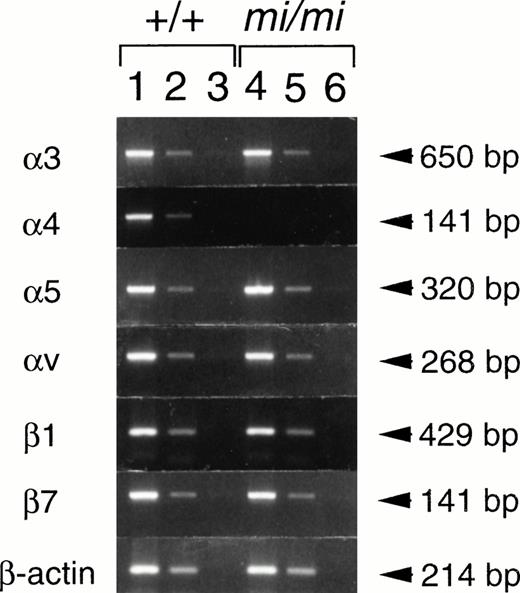
We compared the expression of integrin genes between +/+ andmi/mi CMCs. CMCs were used 4 weeks after the initiation of the culture, because the mRNA expression of integrins reached maximum levels at this time.23,24 RNAs were extracted from +/+ andmi/mi CMCs, and semiquantitative RT-PCR was performed to estimate the expression levels of integrin subunits. We first examined the expression of integrins of α subunit family (α3, α4, α5, and αv) that were expressed by CMCs.25 Expression levels of α3, α5, and αv genes were comparable between +/+ andmi/mi CMCs. In contrast, the expression of the α4 gene was significantly lower in mi/mi CMCs than in +/+ CMCs (Fig 1). We next examined the expression of integrin β1 and β7 subunits, because only these can associate with the α4 subunit.27 Comparing +/+ and mi/mi CMCs, no significant difference in the expression of β1 and β7 genes was detectable (Fig 1).
Semiquantitative RT-PCR for the expression of various integrins in CMCs 4 weeks after the initiation of culture. RNAs obtained from +/+ CMCs (lanes 1 through 3) or from mi/miCMCs (lanes 4 through 6) were reverse-transcribed and PCR-amplified with 3, 4, 5, v, β1, β7, or β-actin primer. PCR products were electrophoresed in 2.0% agarose gel containing ethidium bromide. Amounts of RNA used for the reverse transcription were 5.0 μg (lanes 1 and 4), 0.5 μg (lanes 2 and 5), and 0.05 μg (lanes 3 and 6), respectively.
Semiquantitative RT-PCR for the expression of various integrins in CMCs 4 weeks after the initiation of culture. RNAs obtained from +/+ CMCs (lanes 1 through 3) or from mi/miCMCs (lanes 4 through 6) were reverse-transcribed and PCR-amplified with 3, 4, 5, v, β1, β7, or β-actin primer. PCR products were electrophoresed in 2.0% agarose gel containing ethidium bromide. Amounts of RNA used for the reverse transcription were 5.0 μg (lanes 1 and 4), 0.5 μg (lanes 2 and 5), and 0.05 μg (lanes 3 and 6), respectively.
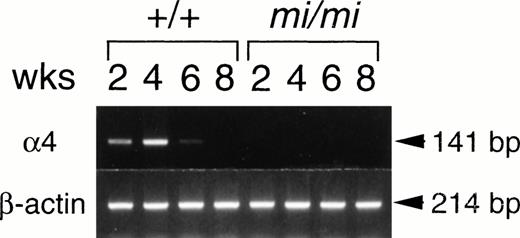
The expression of the α4 subunit gene was investigated at various times after the initiation of the culture. More than 95% of cells were alcian blue-positive throughout the experimental period and were considered to be mast cells (Table 1). The α4 expression was slight 2 weeks after culturing +/+ spleen cells and was clearly observed at 4 weeks (Fig 2). The α4 gene expression started to decrease after 6 weeks of culture and was not detectable after 8 weeks of culture. The expression of α4 subunit was barely detectable in mi/mi CMCs throughout the observation period (Fig 2).
Expression of integrin 4 subunit mRNA in CMCs at 2, 4, 6, and 8 weeks after initiation of the culture. RNAs (5.0 μg) obtained from +/+ or mi/mi CMCs were reverse-transcribed and PCR-amplified with 4 or β-actin primer. PCR products were electrophoresed in 2.0% agarose gel containing ethidium bromide.
Expression of integrin 4 subunit mRNA in CMCs at 2, 4, 6, and 8 weeks after initiation of the culture. RNAs (5.0 μg) obtained from +/+ or mi/mi CMCs were reverse-transcribed and PCR-amplified with 4 or β-actin primer. PCR products were electrophoresed in 2.0% agarose gel containing ethidium bromide.
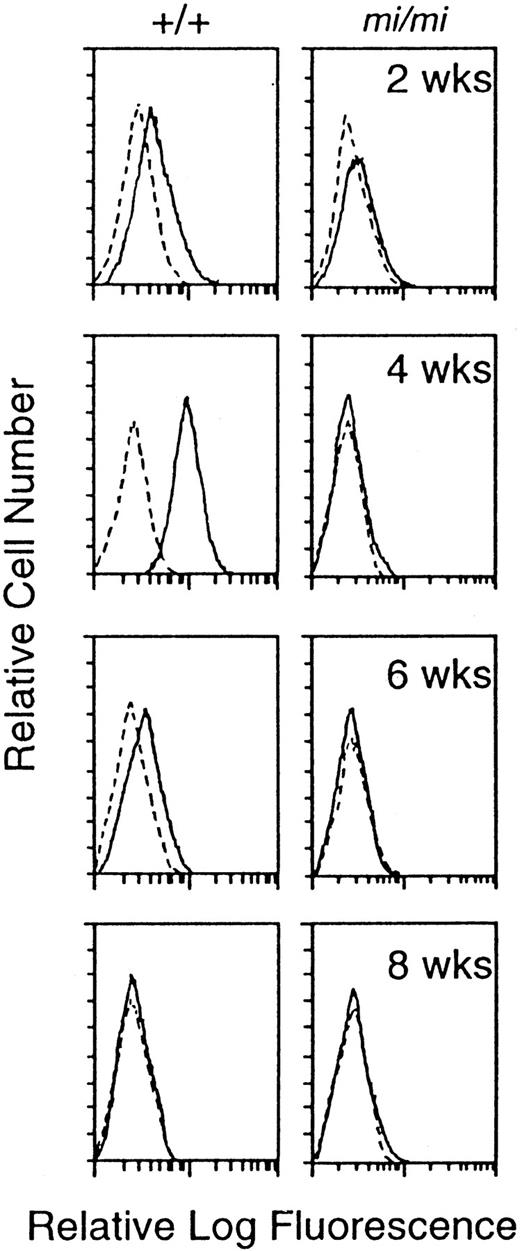
We compared the surface expression of integrin α4 protein between +/+ and mi/mi CMCs by flow cytometry. The expression of α4 subunit was detected in +/+ CMCs 4 weeks after the initiation of culture, and the expression level started to decrease after 6 weeks of the culture and was not detectable after 8 weeks (Fig 3). In mi/mi CMCs, the surface expression of integrin α4 subunit was not detectable throughout the observation period (Fig 3).
Surface expression of integrin 4 subunit protein at 2, 4, 6, and 8 weeks after initiation of the culture. Cells were incubated with either rat anti-4 MoAb (solid line) or control rat IgG (dotted line).
Surface expression of integrin 4 subunit protein at 2, 4, 6, and 8 weeks after initiation of the culture. Cells were incubated with either rat anti-4 MoAb (solid line) or control rat IgG (dotted line).
Because the addition of rmSCF normalized the deficient mRNA expression of MMCP-5 by mi/mi CMCs,9 we examined the effect of SCF-c-kit signals on mRNA expression of the α4 subunit. First, we examined the expression of α4 protein on the surface ofW/Wv CMCs that lack normal SCF-c-kitsignals.29,41 42 Despite the defect of the c-kitreceptor tyrosine kinase, W/Wv CMCs expressed the α4 protein normally (Fig 4). Second, rmSCF was added to the culture medium of CMCs. The α4 expression was not influenced by the addition of rmSCF in all +/+, mi/mi, andW/Wv CMCs (Fig 4).
Effect of the addition of rmSCF on the expression of 4 protein on the surface of +/+, mi/mi, orW/Wv CMCs. Three weeks after initiation of the culture, various CMCs were incubated with 10% PWM-SCM and 50 ng/mL rmSCF for 0, 1, or 2 weeks. The surface expression of integrin 4 subunit was examined by flow cytometry. Cells were incubated with either rat anti-4 MoAb (solid line) or control rat IgG (dotted line).
Effect of the addition of rmSCF on the expression of 4 protein on the surface of +/+, mi/mi, orW/Wv CMCs. Three weeks after initiation of the culture, various CMCs were incubated with 10% PWM-SCM and 50 ng/mL rmSCF for 0, 1, or 2 weeks. The surface expression of integrin 4 subunit was examined by flow cytometry. Cells were incubated with either rat anti-4 MoAb (solid line) or control rat IgG (dotted line).
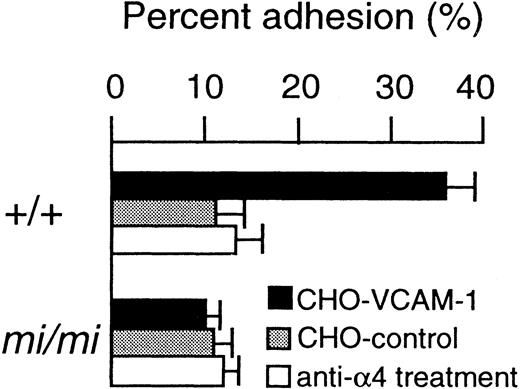
VCAM-1 was the ligand for the integrin α4β1.27 The expression vector containing the sense VCAM-1 cDNA was transfected into CHO cells (hereafter called CHO-VCAM-1). The expression vector for an antisense VCAM-1 cDNA was also transfected into CHO cells as a negative control (hereafter called CHO-control). The proportion of +/+ CMCs adhering to CHO-VCAM-1 cells was significantly higher than that of +/+ CMCs adhering to CHO-control cells (Fig 5). The adhesion of +/+ CMCs to CHO-VCAM-1 cells was inhibited by the treatment of +/+ CMCs with anti-α4 MoAb (Fig 5). No adhesion ofmi/mi CMCs to CHO-VCAM-1 cells was detectable. Themi/mi CMCs did not adhere to CHO-control cells, either (Fig 5).
Adhesion of CMCs to VCAM-1. CMCs were used 4 weeks after the initiation of culture, because the expression of 4 subunit reached the maximum level at that time. The adhesion of CMCs to CHO cells transfected with either sense (CHO-VCAM-1) or antisense VCAM-1 cDNA (CHO-control) was determined. The adhesion of CMCs pretreated with anti-4 MoAb was also determined. Bars indicate the standard error of three assays.
Adhesion of CMCs to VCAM-1. CMCs were used 4 weeks after the initiation of culture, because the expression of 4 subunit reached the maximum level at that time. The adhesion of CMCs to CHO cells transfected with either sense (CHO-VCAM-1) or antisense VCAM-1 cDNA (CHO-control) was determined. The adhesion of CMCs pretreated with anti-4 MoAb was also determined. Bars indicate the standard error of three assays.
Transactivation of integrin α4 subunit gene by +-MITF.
We cloned 949 bases of the 5′-upstream region of the integrin α4 gene to examine the regulation mechanism by +-MITF. The reporter plasmid that contained the luciferase gene under the control of the integrin α4 promoter starting from nt −949 (hereafter called −949 reporter plasmid) was constructed. We also constructed the deleted reporter plasmid that contained the α4 promoter starting from nt −819, −516, −434, or −199 to examine the elements that mediate the transactivation (hereafter called −819, −516, −434, or −199 reporter plasmid, respectively). The reporter plasmids were transfected into the mast cell line, IC-2 cells, which expressed both +-MITF and integrin α4 subunit mRNAs. When the −949 or −819 reporter plasmid was introduced, the IC-2 cells showed only a low level of luciferase activity (Fig 6A). The luciferase activity increased eightfold when the −516 or −434 reporter plasmid was transfected. The deletion of the promoter to −199 reduced the luciferase activity to one fourth the value obtained with the −434 reporter plasmid (Fig 6A). The bHLH-Zip proteins recognize the CANNTG motif (any nucleotides are compatible with the position N).43 There was only one CANNTG motif in the region between nt −434 and −199, ie, CACTTG between nt −294 and nt −289. We mutated the CACTTG motif to CTCTAG in the reporter plasmid starting from nt -434. The mutation decreased the luciferase activity to a level comparable to that obtained by the −199 reporter plasmid.
(A) Luciferase activity under the control of normal, deleted, or mutated 4 promoter in IC-2 cells. (B) The effect of overexpression of +- or mi-MITF on the luciferase activity in IC-2 cells. The luciferase gene under the control of the normal, deleted, or mutated 4 promoter was cotransfected with +- ormi-MITF or with the expression vector. The data represent the mean ± SE of five experiments. In some cases, the standard error was too small to be shown by bars.
(A) Luciferase activity under the control of normal, deleted, or mutated 4 promoter in IC-2 cells. (B) The effect of overexpression of +- or mi-MITF on the luciferase activity in IC-2 cells. The luciferase gene under the control of the normal, deleted, or mutated 4 promoter was cotransfected with +- ormi-MITF or with the expression vector. The data represent the mean ± SE of five experiments. In some cases, the standard error was too small to be shown by bars.
We next transfected the -434 reporter plasmid into IC-2 cells overexpressing +- or mi-MITF cDNA. IC-2 cells overexpressing expression vector alone were used as a control. The luciferase activity in IC-2 cells overexpressing +-MITF was comparable to that of IC-2 cells overexpressing the vector alone (Fig 6B). The luciferase activity was significantly decreased by the overexpression of mi-MITF (Fig 6B). The intact CACTTG motif was necessary for such a negative effect of mi-MITF, because either its deletion or mutation abolished the effect of overexpressing mi-MITF.
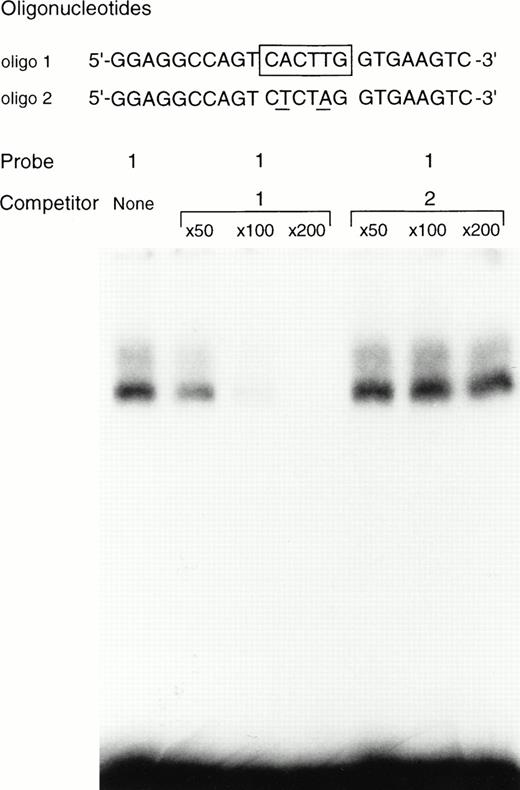
The binding of +-MITF to the oligonucleotide containing the CACTTG motif was examined. We performed EGMSA by using the nuclear extract of +/+ CMCs, but a specific DNA/protein complex was barely detectable. We then used the purified GST-+-MITF fusion protein. When the oligonucleotide containing the CACTTG motif was used as a probe, the specific binding of +-MITF was observed (Fig 7). The excess amount of the nonlabeled oligonucleotide containing the CACTTG motif abolished the binding of +-MITF to the CACTTG motif, whereas the excess amount of unlabeled oligonucleotide mutated in the CACTTG motif (ie, CTCTAG) did not affect the binding (Fig 7).
EGMSA using GST-+-MITF fusion protein. The labeled 5’-GGAGGCCAGTCACTTGGTGAAGTC (oligo 1) was used as a probe (hexameric motif is shown by underlined and boxed by a solid line in the figure). The sequence of the oligonucleotide mutated in the CACTTG motif (to CTCTAG) is also shown as oligo 2. The mutated nucleotides are underlined. The excess amount of nonlabeled oligo 1 or oligo 2 was added as a competitor.
EGMSA using GST-+-MITF fusion protein. The labeled 5’-GGAGGCCAGTCACTTGGTGAAGTC (oligo 1) was used as a probe (hexameric motif is shown by underlined and boxed by a solid line in the figure). The sequence of the oligonucleotide mutated in the CACTTG motif (to CTCTAG) is also shown as oligo 2. The mutated nucleotides are underlined. The excess amount of nonlabeled oligo 1 or oligo 2 was added as a competitor.
The effect of mi-MITF on the binding of +-MITF was examined (Fig 8). The amount of DNA/protein complex was slightly reduced by the addition of an equivalent amount of GST-mi-MITF to the GST-+-MITF. The addition of GST-mi-MITF at twice the amount of GST-+-MITF clearly inhibited the DNA binding of +-MITF. The addition of GST-mi-MITF at four times the amount of GST-+-MITF completely abolished the binding of +-MITF. The GST-mi-MITF itself did not bind to DNA, as reported previously.5
Effect of GST-mi-MITF on the binding of GST-+-MITF. The oligo 1 shown in Fig 7 was used as a probe. Various amounts of GST-mi-MITF were added to the GST-+-MITF, and EGMSA was performed.
Effect of GST-mi-MITF on the binding of GST-+-MITF. The oligo 1 shown in Fig 7 was used as a probe. Various amounts of GST-mi-MITF were added to the GST-+-MITF, and EGMSA was performed.
DISCUSSION
The expression of the integrin α4 subunit gene was deficient inmi/mi CMCs. The adhesion to CHO-VCAM-1 was detectable in +/+ CMCs but not in mi/mi CMCs, indicating that the expression of integrin α4 subunit at a functional level did not occur inmi/mi CMCs. We surmised that the mi/mi mouse was a mutant with deficient expression of integrin α4 subunit in mast cells. The overexpression of +-MITF but not mi-MITF transactivated the α4 promoter containing the CACTTG motif. The mutation or deletion of the CACTTG motif significantly decreased the transactivation ability of +-MITF. These results indicated that the CACTTG motif played an important role for the transactivation by +-MITF. The specific binding of +-MITF to the CACTTG motif suggested that the +-MITF directly transactivated the integrin α4 subunit gene.
The overexpression of mi-MITF reduced the luciferase activity in IC-2 cells. Hemesath et al3 reported the mechanism of dominant negative behavior of mi-MITF; the mi-MITF did not bind DNA, but it dimerized with proteins such as TFE3 and thereby inhibited the DNA binding of its normal partners. This was consistent with the present result. Apparently the overexpression ofmi-MITF in IC-2 cells decreased the luciferase activity by inhibiting the binding of endogenous +-MITF to the CACTTG motif.
Meirsman et al39 analyzed the mouse integrin α4 promoter and 5′-untranslated region in a lymphocytic leukemia cell line. The region around the CACTTG motif was not examined, but they demonstrated the presence of a silencer between nt −936 and −735 and an enhancer between nt +33 and +221. This finding is consistent with our present result. The transfection of the −949 or −819 reporter plasmid but not of the −516 reporter plasmid showed a low level luciferase activity, suggesting that some negative factors for α4 gene transcription bound upstream from nt −517. All reporter constructs used in the present study contained the region between nt +33 and +221. Even when the CACTTG motif was deleted or mutated, the reporter activity was not completely abolished. Endogenous transcription factors in IC-2 cells may associate the region between nt +33 and +221 and may transactivate the α4 gene in collaboration with +-MITF.
We previously demonstrated that the addition of rmSCF changed the gene expression in mi/mi CMCs, although their reaction to rmSCF was deficient due to the low c-kit expression.9 The expression of the MMCP-5 gene was impaired in mi/mi CMCs, but rmSCF increased its mRNA level. In contrast, the expression of MMCP-6, whose mRNA level was also reduced in mi/mi CMCs, was not increased by the addition of rmSCF. We examined whether rmSCF affected the expression of the α4 gene in mi/mi CMCs. The addition of rmSCF did not increase the α4 expression in mi/mi CMCs. The level of α4 expression was also not reduced inW/Wv CMCs, indicating that SCF-c-kitsignals were not necessary for the α4 expression. The present result is consistent with the report of DuCharme et al23that the addition of rmSCF did not increase the α4 expression in +/+ CMCs.
The function of the integrin α4 subunit was extensively studied in lymphocytes. The interaction of α4 subunit with VCAM-1 mediated the migration of lymphocytes into sites of inflammation.44,45Because the number of mast cells increased in some inflammatory sites,46 the progenitors of mast cells were apparently recruited from peripheral blood to the inflammatory sites. In fact, we previously reported that the concentration of mast cell progenitors decreased in peripheral blood but increased at the site of the parasite (Nippostrongylus brasiliensis) infection.47Recently, Palacanda et al28 reported the adhesion of two rat mast cell lines RBL-1 and RCMC-1 to VCAM-1 and suggested the involvement of α4 subunit-VCAM-1 interaction in the augmentation of mast cells at inflammatory sites. In the present study, we demonstrated that +/+ CMCs but not mi/mi CMCs bound VCAM- 1. Further analysis using mi/mi mice will clarify the role of α4 subunit-VCAM-1 interaction in the increase of mast cells at inflammatory sites.
The integrin α4 subunit was important not only for the migration to inflammatory sites but also for the differentiation of lymphocytes. Arroyo et al48 generated chimeric mice by injecting α4 gene-targeted embryonic stem cells into recombination activating gene-1 (RAG-1)–targeted blastocysts. The lymphocytes in this chimeric mouse were α4-deficient, because lymphocytes derived from RAG-1 gene-targeted blastocysts did not survive. In the chimeric mice, development of B cells was apparently blocked before the pro-B–cell stage, and no T cells were detected in the thymus, showing that the α4 subunit was essential for differentiation of both B and T cells. Although mast cell number in tissues of the chimeric mice was not reported, there is a possibility that the development of mast cells may also be impaired in the chimeric mice.
Smith and Weis27 demonstrated that CMCs but not peritoneal mast cells expressed the α4 subunit. They suggested that the α4 subunit was important for the migration of mast cells from blood to tissues. The present result is consistent with their idea. Mast cell progenitors of mi/mi mice may show some difficulties in invading tissues due to the lack of the α4 subunit. Mast cell progenitors have not been identified in the peripheral blood of adult mice. Such identification would promote further analysis of the migration of mast cell progenitors. The mi/mi mice should be useful in evaluating the physiological roles of the integrin α4 subunit.
ACKNOWLEDGMENT
The authors thank Dr S. Nomura and Dr N. Matsuura of Osaka University for valuable discussions, Dr K. Nakajima of Osaka University for pSPLuc, Dr S. Nagata of Osaka University for pEF-BOS, and Kirin Brewery Co Ltd for rmSCF.
Supported by grants from the Ministry of Education, Science and Culture and a grant from the Mochida Memorial Foundation for Medical and Pharmaceutical Research.
Address reprint requests to Eiichi Morii, MD, Department of Pathology, Osaka University Medical School, Yamada-oka 2-2, Suita 565-0871, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.