Abstract
The FAC protein encoded by the Fanconi anemia (FA) complementation group C gene is thought to function in the cytoplasm at a step before DNA repair. Because FA cells are susceptible to mitomycin C, we considered the possibility that FAC might interact with enzymes involved in the bioreductive activation of this drug. Here we report that FAC binds to NADPH cytochrome-P450 reductase (RED), a microsomal membrane protein involved in electron transfer, in both transfected COS-1 and normal murine liver cells. FAC-RED interaction requires the amino-terminal region of FAC and the cytosolic, membrane-proximal domain of the reductase. The latter contains a known binding site for flavin mononucleotide (FMN). Addition of FMN to cytosolic lysates disrupts FAC-reductase complexes, while flavin dinucleotide, which binds to a distinct carboxy-terminal domain, fails to alter FAC-RED complexes at concentrations similar to FMN. FAC is also functionally coupled to this enzyme as its expression in COS-1 cells suppresses the ability of RED to reduce cytochrome c in the presence of NADPH. We propose that FAC plays a fundamental role in vivo by attenuating the activity of RED, thereby regulating a major detoxification pathway in mammalian cells.
© 1998 by The American Society of Hematology.
THE AUTOSOMAL RECESSIVE disease Fanconi anemia (FA) can lead to birth defects, bone marrow failure, and myeloid leukemia.1,2 Although the disorder is genetically heterogeneous, there are several shared features that include chromosome breakage, enhanced sensitivity to mitomycin C (MMC) and to related bifunctional alkylating agents (also called crosslinkers), delays in the G2 phase of the cell cycle, and predisposition to apoptosis.2 The hypersensitivity to crosslinking agents has served as the basis for assigning FA cells to at least eight different complementation groups3-5 and for cloning the disease genes in two groups.6,7 The genes for FA groups A (FAA),8 C (FAC),3and D (FAD)9 have been mapped to different chromosomal loci, suggesting that mutations in several distinct genes can give rise to a similar disease phenotype.4 The genes for complementation groups A7,10 and C6 are present in single copies and encode unique proteins, which are expressed at low levels in most tissues. The ≈163-kD protein encoded by the FAA gene contains a nuclear localization signal (NLS), but otherwise it is devoid of any sequence motifs that may suggest a biological function,7,10 save for limited homology to a class of peroxidases.11 Although an initial study with an epitope-tagged form of FAA showed that the chimeric protein localizes to the cytoplasm,12 more recent subcellular localization studies have shown that a significant fraction of FAA is nuclear.13,14 Moreover, forced localization of FAA to the cytoplasm was shown to abolish its ability to correct the hypersensitivity of FA group C cells to MMC.14 FAA has no homology to FAC, and their biochemical relationship, if any, is not apparent from their sequences. The FAC protein consists of 558 amino acids with a predicted molecular mass of ≈63 kD.6 Recent sequence analysis suggests that FAC may possess a catalase domain.11 If confirmed, these data would seem to indicate that both FAA and FAC may participate in cellular detoxification processes. Several studies have shown that FAC localizes primarily to the cytoplasm of mammalian cells under both steady-state and stress conditions, and about one third associates with internal membranes.15-17 Transfection studies have shown that FAC prevents the formation of interstrand DNA crosslinks induced by MMC, but it has little or no effect on the turnover or repair of such lesions.18 These data have led us to suggest that FAC may act as a sensor of crosslinkers or other reactive metabolites. Two additional studies have reported that a fraction of FAC also localizes in the nucleus,13,19 but the functional consequences of this observation are not clear. Although historically the pathogenesis of FA has been attributed to a fundamental deficiency in DNA repair,20 we believe that the preponderance of data on the group C subset argues against this model.
The distinctive sensitivity of FA cells to crosslinkers has led us to consider the possibility that FAC modulates the toxicity of these agents directly or indirectly. MMC and diepoxybutane (DEB) are among the most popular agents in this group. MMC is an antineoplastic drug that requires metabolic activation to unmask its cytotoxic function.21 The reduction of MMC by cellular enzymes generates highly reactive species that can generate interstrand crosslinks in double-stranded DNA. In turn, reactive oxygen metabolites can degrade DNA and contribute to the cytotoxicity of MMC. The relative contribution of these pathways to the pathogenesis of FA is not clear. However, it is noteworthy that the chromosomal instability can be attenuated by low oxygen tension and exacerbated by normal or high concentrations of oxygen.22-24
One approach to deciphering the function of FAC may be through the identification of its binding partners, which include at least three ubiquitous cytoplasmic proteins.17,25 Because FA cells are highly sensitive to MMC, we investigated whether FAC interacts with enzymes involved in the bioreductive activation of this drug.26 A key enzyme in this pathway is NADPH:cytochrome c (P-450) reductase (RED; EC 1.6.2.4), a 77-kD integral microsomal enzyme that can transfer electrons from NADPH to an isozyme of the cytochrome P450 family26-33 as well as to cytochrome c. Tethered by a short hydrophobic sequence to the microsomal membrane, RED extends into the cytosol and contains binding sites for several prosthetic groups, including flavin mononucleotide (FMN), flavin dinucleotide (FAD), and NADPH. Electrons donated by NADPH are initially transferred internally from FAD to FMN, then externally to one of the cytochromes P450 in microsomes. An outcome of this chain of events is the oxidative metabolism of various drugs, xenobiotics, and endogenous substrates, such as steroids and fatty acids.
A potential interaction between FAC and RED seemed attractive for several reasons. First, during attempts to identify FAC-associated proteins, cytoplasmic proteins in the 69- to 90-kD range were found to bind to glutathione-S-transferase (GST)-FAC, but not to GST.25 Second, similar to the phenotype of FA cells, RED overexpression in a non-FA cell line was shown to induce MMC hypersensitivity,33 and acquired resistance to MMC correlated with reduced activity of RED.34 Here we show that FAC binds to the cytosolic domain of RED, which can be inhibited in vitro by FMN. In vivo, FAC suppresses the catalytic function of RED. These data suggest a model in which an important component of the defect in FA group C cells involves the uncoupling of FAC-RED interaction. Without appropriate attenuation of RED activity by FAC, reactive species (eg, of MMC or oxygen metabolites) could accumulate and affect cell viability.
MATERIALS AND METHODS
Expression plasmids.
Full-length human FAC and RED cDNAs as well as cDNAs encoding human cytochrome P4501A1, NADPH:Quinone Oxidoreductase1 (NQO1), NADPH:Quinone Oxidoreductase2 (NQO2), BclXL, p34cdc2 kinase, and cyclin B were cloned into either pMT2 (gift of Dr R. Wise, Brigham and Women’s Hospital, Boston, MA) or pcDNA3 (Invitrogen, Carlsbad, CA). Wild-type FAC and a panel of deletion mutants generated by polymerase chain reaction were also subcloned as fusion cDNAs upstream of the human IgG1 heavy-chain cDNA, as before.17 Recombinant GST-FAC expressed in Escherichia coli was prepared as described previously.25
Preparation and analysis of liver cellular extracts.
Livers from three C57BL/6 mice were homogenized in ice-cold homogenization buffer (50 mmol/L Tris-HCl [pH 7.4], 0.25 mol/L sucrose]. Nuclei and unbroken cells were pelleted by centrifugation at 3,000g for 10 minutes, and mitochondria were pelleted by a further centrifugation at 9,000g for 20 minutes. The clarified supernatant was then centrifuged at 100,000g for 60 minutes to yield cytosol (supernatant) and microsome (pellet). The latter fraction was resuspended in homogenization buffer before protein interaction studies. Each fraction was immunoprecipitated with affinity-purified anti-FAC antibodies raised against the GST-FAC recombinant protein, as described,35 or with a control antibody against MxA36 prepared by the same affinity-purification procedure. After incubation of lysates with each antibody in phosphate-buffered saline (PBS) containing 0.1% NP-40 for 1 hour, immune complexes were precipitated with protein A-agarose, washed, and analyzed by immunoblotting. Protein concentrations were determined by the Bradford assay (Bio-Rad, Richmond, CA) corrected for detergent effects.
Transfection and immunoprecipitation (IP).
COS-1 cells were transfected by the diethyl aminoethyl (DEAE)-dextran method. For metabolic labeling, cells were preincubated for 1 hour in Dulbecco’s Modified Essential Medium (DME) lacking cysteine and methionine, followed by incubation in the same medium containing Expre35S35S label (0.2 mCi/mL; DuPont, Wilmington, DE) for 1 hour at 37°C. Monolayers were then washed in PBS and lysed in 0.4 mL lysis buffer (20 mmol/L Tris-HCl [pH 8.0], 50 mmol/L NaCl, 0.1% NP-40, 2 mmol/L EDTA, and protease inhibitors). Supernatants were incubated for 1 hour with either pre-immune serum or affinity-purified anti-FAC antibody in the presence or absence of the indicated competitors or in higher concentrations of NP-40. Immune complexes were then precipitated with protein A-agarose beads (Bio-Rad). After washing in lysis buffer, beads were boiled in Laemmli sample buffer containing reducing agents and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Western analysis of immune complexes.
Lysates of transfected COS-1 cells or mouse liver extracts were analyzed either directly by immunoblotting or after IP with anti-FAC or anti-RED antibodies. Immune complexes precipitated with protein A-agarose beads were resolved by SDS-PAGE and transferred to polyvinyldifluoride membranes (DuPont) by electroblotting. After blocking with 10% nonfat milk and 1% bovine serum albumin (BSA), membranes were reacted sequentially with a primary antibody (either anti-FAC or anti-RED antiserum [Novus Molecular Inc, San Diego, CA]) and horseradish peroxidase–conjugated goat anti-rabbit IgG (GIBCO-BRL, Grand Island, NY), and bands were visualized by chemiluminescence (DuPont).
Yeast two-hybrid analysis.
RED deletion mutants generated by PCR were cloned into the vector pAD-GAL4 (Stratagene, La Jolla, CA) downstream of the GAL4 transcriptional activation domain,37 and full-length human FAC cDNA was cloned into the vector pBD-GAL4 Cam. Both inserts were under the control of the ADH1 promoter. After cotransformation of the yeast host strain YRG-2, a filter color assay was used to assess the transcriptional activation of lacZ (β-galactosidase activity) as an indicator of a physical interaction between AD-RED and BD-FAC, and the intensity of the color reaction was scored in a semi-quantitative manner by visual inspection.
Enzyme assays.
Ten- to 50-μL aliquots of COS-1 cell lysates were incubated with 20 μmol/L NADPH, 0.6 μmol/L cytochrome c, and 50 mmol/L potassium phosphate (pH 7.6) in a final volume of 1 mL, as before.38An increase in absorbance at 550 nm due to the NADPH-dependent reduction of cytochrome c was taken as an index of RED activity. Enzyme activity was calculated using the extinction coefficient of cytochrome c (18.5 cm−1mmol/L−1). For assessment of NQO1 activity, COS-1 cell lysates were incubated in 25 mmol/L Tris-HCl (pH 7.4), 0.18 mg/mL BSA, 5 μmol/L FAD, 0.01% Tween 20, 200 μmol/L NADPH, and 50 μmol/L 2,6-dichlorophenolindophenol, as before,39 and the reaction rate was monitored by a decrease in absorbance at 600 nm.
RESULTS
FAC binds to RED in transfected COS-1 cells.
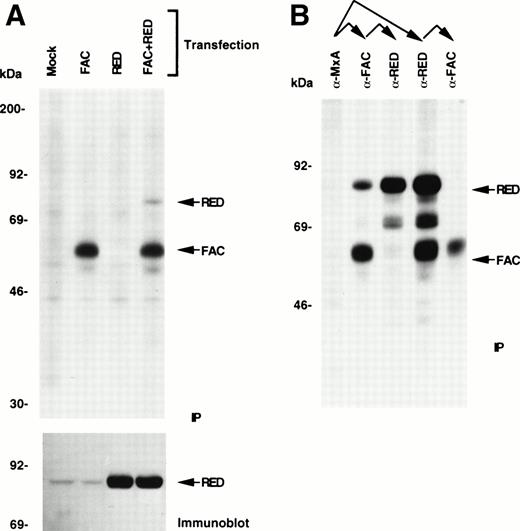
We used several strategies to test the hypothesis that an interaction between FAC and RED takes place in vivo. First, COS-1 cells were transiently transfected with a combination of mammalian expression vectors encoding FAC and RED and analyzed by metabolic 35S labeling and IP. Cytosolic lysates from cells expressing FAC alone showed the expected 63-kD protein when immunoprecipitated with anti-FAC antibodies, while lysates from cotransfected cells showed an additional band of ≈80 kD, consistent with the size of RED (Fig 1A). When the concentration of NP-40 in the lysate was increased from 0.1% to 0.25%, there was a marked reduction (>90%) in the amount of the 80-kD protein that coprecipitated with FAC, but not in the amount of precipitable FAC (data not shown). This result suggests that the association of FAC with the 80-kD protein is detergent-sensitive. The identity of this band was further established by immunoblotting of unlabeled lysates with anti-RED antibodies (Fig 1A). The 80-kD band was expressed at much greater levels in cells transfected with RED cDNA, which reacted specifically with a polyclonal antipeptide-antibody directed against human RED. As before,17,25 35 IP with FAC preimmune serum failed to show either FAC or other associated proteins (data not shown). Coexpression of FAC with two other proteins involved in xenobiotic metabolism (cytochrome P4501A1 and NQO2) as well as with other cytoplasmic proteins thought to be irrelevant for MMC metabolism (including BclXL, p34cdc2 kinase, and cyclin B) showed no evidence of physical interactions between FAC and each of these proteins. However, under the same conditions there was a weak interaction between FAC and NQO1 (data not shown).
Binding of FAC to RED. (A) IP of FAC-RED complexes. COS-1 cells overexpressing FAC, RED, or a combination were radiolabeled with a mixture of 35S-cysteine and methionine, cytoplasmic lysates immunoprecipitated sequentially with anti-FAC antibody and protein A-agarose and analyzed by 10% SDS-PAGE and autoradiography. The same panel of unlabeled lysates was also analyzed by immunoblotting with anti-RED antibody. Twenty times as much lysate was used for binding to RED as that applied directly in the immunoblotting experiment. (B) FAC-bound and unbound forms of RED. Sequential IP of cytoplasmic lysates from COS-1 cells transfected with both FAC and RED shows that a fraction of the total intracellular pool of FAC and RED associate with each other. Relative molecular masses are shown.
Binding of FAC to RED. (A) IP of FAC-RED complexes. COS-1 cells overexpressing FAC, RED, or a combination were radiolabeled with a mixture of 35S-cysteine and methionine, cytoplasmic lysates immunoprecipitated sequentially with anti-FAC antibody and protein A-agarose and analyzed by 10% SDS-PAGE and autoradiography. The same panel of unlabeled lysates was also analyzed by immunoblotting with anti-RED antibody. Twenty times as much lysate was used for binding to RED as that applied directly in the immunoblotting experiment. (B) FAC-bound and unbound forms of RED. Sequential IP of cytoplasmic lysates from COS-1 cells transfected with both FAC and RED shows that a fraction of the total intracellular pool of FAC and RED associate with each other. Relative molecular masses are shown.
The amount of coprecipitable RED was quantified by sequential IP experiments (Fig 1B). IP with an irrelevant antibody, anti-MxA,36 prepared by the same affinity-purification method as that for FAC, failed to precipitate either FAC or RED. However, when the MxA-depleted lysate was incubated with anti-FAC antibody, most if not all of radiolabeled FAC was precipitated along with a small fraction of RED. Finally, using FAC-depleted lysate and anti-RED antibody, the remainder of RED that had presumably remained unbound to FAC was immunoprecipitated. Conversely, initial incubation of the MxA-depleted lysate with anti-RED antibody resulted in the IP of almost all of the radiolabeled RED along with a fraction of FAC. Thus, minor pools of FAC and RED can interact with each other.
Fac-RED complexes in normal liver cells.
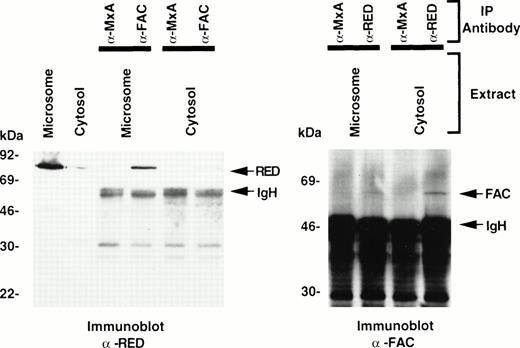
Based on our earlier observation that anti-human FAC antibodies can cross-react with the murine orthologue of FAC, fac,40 and the assumption that FAC-RED interactions may be conserved in other mammals, we attempted to detect fac-RED protein complexes in extracts of non-FA mouse livers. Because RED is primarily microsomal, we prepared cytosolic and microsomal extracts and attempted to detect fac-RED protein complexes by sequential IP and immunoblotting experiments. As expected, the microsomal fraction contained significantly greater amounts of RED than the cytosolic fraction (Fig 2). When each fraction was immunoprecipitated with either anti-FAC antibody or anti-MxA and immunoblots probed with anti-RED antibodies, fac-RED complexes were found in both cytosolic and microsomal extracts. Consistent with the known location of RED in microsomes, fac-RED complexes were significantly more abundant in the microsomal extracts (Fig 2). Conversely, IP with anti-RED antibody and probing of immunoblots with anti-FAC antibody also showed fac-RED complexes. These results demonstrate that fac-RED complexes can be detected in a normal tissue extract, and that the distribution of the complex correlates with the known subcellular location of RED.
Detection of murine fac-RED complexes in liver extracts. Using the indicated antibodies and protein A-agarose, mouse liver cytosolic or microsomal extracts (730 μg) were used to immunoprecipitate fac, and immune complexes were analyzed for the presence of RED by probing the immunoblot with anti-RED antibodies (left). Each subcellular fraction (50-μg aliquots) was also analyzed directly without prior IP. Conversely, immune complexes obtained by IP with anti-RED antibodies were analyzed for the presence of FAC by immunoblotting (right). After SDS-PAGE (10% gel for the left panel, 8% to 20% gradient gel for the right panel), immunoblots were probed with the antibodies indicated in the bottom of the figure.
Detection of murine fac-RED complexes in liver extracts. Using the indicated antibodies and protein A-agarose, mouse liver cytosolic or microsomal extracts (730 μg) were used to immunoprecipitate fac, and immune complexes were analyzed for the presence of RED by probing the immunoblot with anti-RED antibodies (left). Each subcellular fraction (50-μg aliquots) was also analyzed directly without prior IP. Conversely, immune complexes obtained by IP with anti-RED antibodies were analyzed for the presence of FAC by immunoblotting (right). After SDS-PAGE (10% gel for the left panel, 8% to 20% gradient gel for the right panel), immunoblots were probed with the antibodies indicated in the bottom of the figure.
Binding domain localization on FAC.
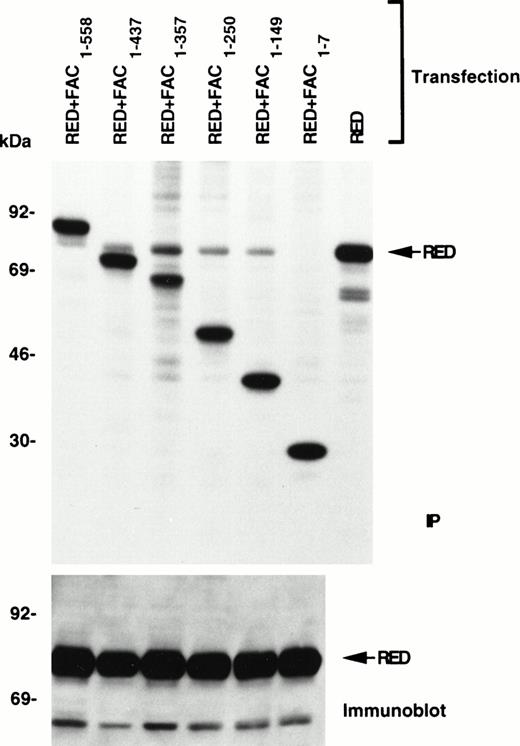
To determine the region of FAC that is necessary for this interaction, we generated a series of carboxy-terminal truncated mutants fused to the constant region of the human IgG1 heavy-chain cDNA, as described previously.17 After coexpression of these constructs with full-length RED in COS-1 cells, single-step IP with protein A-agarose beads showed that residues within the region 8-149 of FAC are necessary for binding to RED (Fig 3). Thus, the amino-terminal domain of FAC is required for interaction with RED.
Localization of the RED-binding domain of FAC to the amino terminal region. Carboxy-terminal truncated fragments of FAC (residues remaining indicated as subscripts) fused to the constant region of the human IgG1 heavy chain were coexpressed with full-length RED in COS-1 cells. Protein interactions were detected by IP of 35S-labeled lysates with protein A-agarose beads, followed by SDS-PAGE and autoradiography. The lower panel shows unlabeled lysates analyzed by immunoblotting with anti-RED antibody.
Localization of the RED-binding domain of FAC to the amino terminal region. Carboxy-terminal truncated fragments of FAC (residues remaining indicated as subscripts) fused to the constant region of the human IgG1 heavy chain were coexpressed with full-length RED in COS-1 cells. Protein interactions were detected by IP of 35S-labeled lysates with protein A-agarose beads, followed by SDS-PAGE and autoradiography. The lower panel shows unlabeled lysates analyzed by immunoblotting with anti-RED antibody.
Binding domain of FAC on RED.
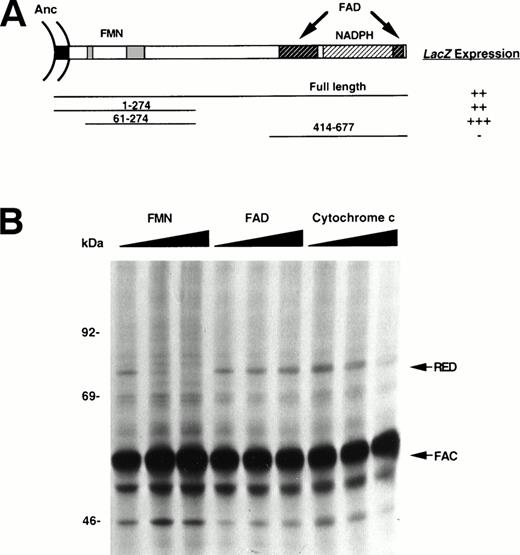
Considerably more is known about the functional organization of RED than of the FAC protein.29-32 The amino-terminal region of RED is homologous to FMN-containing bacterial flavodoxins, and the carboxy-terminus is homologous to FAD-containing ferrodoxin NADP+ reductases. Furthermore, the FMN- and FAD/NADPH-binding domains can be dissected into distinct structural and functional units, which bind to their respective cofactors.26,27 To delineate the FAC-binding domain of RED and, if more than one domain is involved, discern quantitative differences, we performed reciprocal mapping experiments using the yeast two-hybrid system.37 Deletion mutants of RED were fused to the transcriptional activation domain of the GAL4 protein (AD-RED), while FAC was fused to the DNA-binding domain of GAL4 (BD-FAC; Fig 4A). Transformation with AD-RED or BD-FAC alone did not result in transcriptional activation (data not shown). However, transformants expressing either full-length RED or deletion mutants encoding either residues 1-274 (membrane anchor and FMN-binding domain) or 61-274 (containing the FMN-binding domain, but lacking the membrane anchor) turned blue in the presence of BD-FAC in a filter color assay. There was no interaction between BD-FAC and AD-RED constructs lacking the FMN-binding domain. Thus, the cytosolic, membrane-proximal region of RED that is known to bind to FMN also binds FAC. The proximity of FAC to the microsomal membrane is compatible with our previous observation that approximately one third of the total intracellular pool of FAC associates with internal membranes.16 17
Effect of cofactors on FAC-RED interaction. (A) The FMN-binding domain of RED is required for interaction with FAC. Schematic diagram indicating functional domains of RED analyzed for binding to FAC in the yeast two-hybrid system. The intensity of blue color corresponding to β-galactosidase activity was assessed visually and scored as follows: minus, white; double plus, blue; triple plus, dark blue. Anc, membrane anchor. (B) Failure of FAC to bind RED in presence of cofactors. Radiolabeled lysates of COS-1 cells transfected with both FAC and RED were divided into equal volumes, immunoprecipitated sequentially with anti-FAC antibody and protein A-agarose, and analyzed by SDS-PAGE and autoradiography. Increasing amounts (0, 0.1 mmol/L, and 1.0 mmol/L) of FMN, FAD, or cytochrome c were added to otherwise identical lysates during immune complex formation.
Effect of cofactors on FAC-RED interaction. (A) The FMN-binding domain of RED is required for interaction with FAC. Schematic diagram indicating functional domains of RED analyzed for binding to FAC in the yeast two-hybrid system. The intensity of blue color corresponding to β-galactosidase activity was assessed visually and scored as follows: minus, white; double plus, blue; triple plus, dark blue. Anc, membrane anchor. (B) Failure of FAC to bind RED in presence of cofactors. Radiolabeled lysates of COS-1 cells transfected with both FAC and RED were divided into equal volumes, immunoprecipitated sequentially with anti-FAC antibody and protein A-agarose, and analyzed by SDS-PAGE and autoradiography. Increasing amounts (0, 0.1 mmol/L, and 1.0 mmol/L) of FMN, FAD, or cytochrome c were added to otherwise identical lysates during immune complex formation.
Effect of cofactors on FAC-RED interaction.
To assess whether known RED cofactors affect the interaction of FAC with RED, we cotransfected COS-1 cells with expression constructs encoding these cDNAs and immunoprecipitated FAC-RED complexes in the presence or absence of known RED cofactors. The intensity of bands corresponding to RED and FAC on a representative autoradiogram (Fig 4B) were quantified by densitometry (data not shown), and the degree of protein-protein interaction was expressed as the ratio of RED to FAC in control relative to the experimental samples. The inclusion of 0.1 mmol/L FMN in lysates caused a greater than 95% reduction in FAC-RED complex formation. Similar concentrations of FAD did not appear to have any effect. Cytochrome c also partially inhibited this interaction, albeit at a 10-fold higher concentration. Finally, we were unable to show that FMN in the range 0 to 1.0 mmol/L binds directly to recombinant GST-FAC immobilized to glutathione-agarose beads (data not shown). Taken together, these results show that FMN can compete with FAC for interaction with RED.
Suppression of RED activity by FAC.
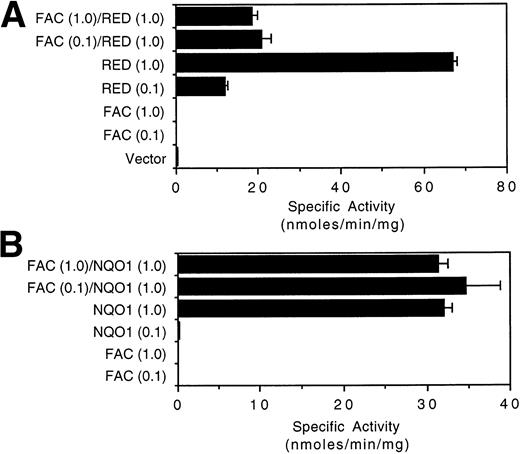
We also determined whether the expression of FAC could affect the catalytic activity of RED in vivo. COS-1 cells transfected with RED expressed dose-dependent levels of reductase activity (Fig 5A). However, cotransfection of COS-1 cells with RED and FAC, but not RED and the empty expression vector, suppressed the activity of RED by 3.2- to 3.6-fold. Interestingly, the extent of suppression was independent of the amount of transfected FAC plasmid DNA over a 10-fold range, and FAC was not able to abolish RED activity completely. By contrast, the catalytic activity of NQO1 was not affected by coexpression of NQO1 with FAC (Fig 5B). These results demonstrate that (1) the catalytic activity of RED can be attenuated by FAC; (2) the final determinant of reductase activity in this cell culture model is the intracellular level of RED, not FAC; and (3) a fraction of RED activity is not subject to regulation by FAC.
FAC suppresses the catalytic activity of RED but not NQO1. Cytosolic lysates of COS-1 cells transfected with the indicated constructs were assayed for (A) RED activity and (B) NQO1 activity as described (Materials and Methods). The indicated amounts of transfected DNA (μg) were standardized with empty vector DNA to a concentration of 1 μg/mL for a final amount of 5 μg. The mean of at least three independent measurements and the standard error of the mean are shown.
FAC suppresses the catalytic activity of RED but not NQO1. Cytosolic lysates of COS-1 cells transfected with the indicated constructs were assayed for (A) RED activity and (B) NQO1 activity as described (Materials and Methods). The indicated amounts of transfected DNA (μg) were standardized with empty vector DNA to a concentration of 1 μg/mL for a final amount of 5 μg. The mean of at least three independent measurements and the standard error of the mean are shown.
DISCUSSION
A critical component of the cytochrome P450 monooxygenase system is the membrane-embedded microsomal enzyme RED, which is essential for the activation of cytochrome P450 enzymes that are involved in the oxidation of many xenobiotics and endogenous compounds. Abnormal metabolism of one or several of these compounds could contribute to the pathogenesis of FA. Here we show that FAC binds to the cytosolic domain of RED (Fig 3) and attenuates its ability to transfer electrons (Fig5). This observation provides the first insight into the molecular function of FAC in the regulation of an important cellular detoxification pathway. Our earlier studies had suggested that FAC interacts with at least three cytoplasmic proteins17,25; RED is one such binding protein.
Both physical and functional data suggest that only a subset of the total intracellular RED interacts with a subset of FAC. The FAC-binding domain on RED corresponds to the known binding site of FMN. To assess whether the effect of this cofactor on FAC-RED interaction is likely to be of any physiological importance, we reasoned that a comparison between FMN and FAD may be instructive (Fig 4). Both cofactors bind to distinct sites on RED. Although their precise intracellular concentrations are uncertain, measurements of FMN and FAD have shown similar contents of cofactor per unit of purified recombinant RED protein (5.5 nmol/mg) and a stoichiometry of 1:1 for FMN/FAD.32 Thus, the inhibition of FAC-RED complexes by FMN, but not by similar concentrations of FAD, may recapitulate normal physiology. Because the usual dissociation constant for FMN is in the range 10−8 to 10−11 mol/L for several FMN-binding enzymes, a large fraction of RED in cells is probably tightly bound to FMN and unable to associate with FAC. Even the remaining fraction can be displaced from FAC by additional FMN (Fig4B). At a functional level, FAC has only a partial effect on the overall activity of RED and cannot suppress it completely despite a large increase in the amount of transfected FAC (Fig 5A). Presumably the limiting component is RED that has remained unbound to FMN. However, this component may also be a member of the cytochrome P450 superfamily that is coupled to RED.41,42 Furthermore, not all of the intracellular FAC is in a complex with RED (Fig 1B). Given the proximity of the FMN- and FAC-binding sites to the microsomal membrane, we postulate that RED interacts chiefly with the smaller pool of FAC that is associated with internal membranes, not the larger cytosolic pool.16,17 FAC is rich in hydrophobic residues,6 and an interaction between FAC and microsomal membranes—which may be expected to be detergent-sensitive—could stabilize its binding to RED.
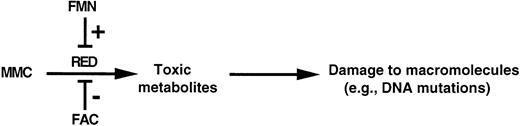
The interaction of these smaller pools through a common binding site for FAC and FMN suggests a dynamic mechanism for the regulation of RED and fine-tuning of the redox state of the cell (Fig 6). FAC and FMN can regulate differentially the activity of RED by binding to its membrane-proximal domain. FAC suppresses the activity of RED; as a corollary, mutations in FAC relieve this suppression and lead to the constitutive activation of RED. Following this proximal derangement, a cascade of biochemical abnormalities could affect the viability of FA group C cells. For example, unopposed RED activity at critical times during development or cell turnover could cause excessive oxidative stress, which could lead to DNA mutations or damage to other macromolecules. Crosslinking by activated MMC may also contribute to the pathogenesis. This presently speculative pathway can be tested in appropriate animal models.
Model of the regulation of RED by FAC. A possible mechanism for this effect is by competition of FAC with FMN for binding to RED and interruption of the electron-transfer chain from NADPH to FMN. In the absence of FAC, unopposed RED activity could generate toxic metabolites (eg, activated MMC, reactive oxygen species, etc), which could damage genomic DNA as well as other macromolecules.
Model of the regulation of RED by FAC. A possible mechanism for this effect is by competition of FAC with FMN for binding to RED and interruption of the electron-transfer chain from NADPH to FMN. In the absence of FAC, unopposed RED activity could generate toxic metabolites (eg, activated MMC, reactive oxygen species, etc), which could damage genomic DNA as well as other macromolecules.
Several aspects of our model are consistent with previous data on RED and the physiological abnormalities observed in FA cells. First, unlike most enzymes involved in bioreductive processes, RED reduces MMC preferentially under aerobic rather than anaerobic conditions.33,34 MMC reduction under aerobic conditions could exacerbate the chromosomal instability of FA cells. Second, the failure to suppress RED activity with increasing levels of FAC is consistent with our earlier demonstration of a threshold effect for FAC: although low levels of FAC protein are both necessary and sufficient to complement FA group C cells, much higher levels do not result in super-resistance to MMC beyond wild-type levels.18 These results had suggested the presence of one or more rate-limiting downstream targets. RED and possibly certain cytochromes P450 may be placed downstream of FAC in this pathway. Third, an increasing body of evidence shows that oxidative damage accounts for a major component of the cellular pathogenesis in FA. There is excess 8-hydroxy-2′-deoxyguanosine, a marker of oxidative damage, in the genomic DNA of FA lymphoblasts treated with hydrogen peroxide43 and in fresh buffy coats from FA patients,44 and oxygen radicals generated by MMC are thought to be chiefly responsible for apoptosis induction in FA group C lymphoblasts.45 A pro-oxidant state created by the dysregulation of RED places the genome, an innocent bystander, at risk for mutations. Fourth, FAC-RED interaction may account for the cytotoxicity of structurally diverse crosslinkers. DEB is thought to act as a direct mutagen and bypass cellular pathways involved in xenobiotic metabolism. However, this view may be premature because certain forms of DEB—eg, stereoisomers or epoxy metabolites—may indeed require metabolic activation to exert clastogenic effects, and P450 enzymes have been shown to be involved in the activation or hydrolysis of DEB-related compounds.46-48 A mechanistic model with FAC-RED as the focal point can potentially account for the cytotoxicity of other crosslinkers implicated in the pathogenesis of FA group C.
RED appears to be one of several FAC-binding proteins. We have recently characterized an intracellular chaperone, GRP94, which interacts with FAC and regulates its intracellular level.49 Others have reported interactions between FAC and FAA13 and between FAC and p34cdc2 kinase50; we have been unable to confirm these data14 (and this report). Nevertheless, FAC may have additional roles, perhaps in other cellular compartments, and distinct domains could mediate these functions.
Supported by grants from the National Institutes of Health (HL52138), the Fanconi Anemia Research Fund, and a Translational Research Award from the Leukemia Society of America.
Address reprint requests to Hagop Youssoufian, MD, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030; e-mail: hagopy@bcm.tmc.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.