Abstract
Treatment of HL-60 human leukemia cells with etoposide induces apoptotic cell death and activation of at least 18 electrophoretically distinct cysteine-dependent aspartate-directed protease (caspase) isoforms, several of which differ only in their isoelectric points. The purpose of the present study was to determine whether activated caspases are phosphorylated. Phosphatase treatment of cytosolic extracts containing active caspases followed by affinity labeling with N-(N-benzyloxycarbonylglutamyl-N-biotinyllysyl)aspartic acid [(2,6-dimethylbenzoyl)oxy] methyl ketone (Z-EK(bio)D-aomk) showed a mobility shift in several of the labeled species, suggesting that phosphorylated forms of these enzymes are present in the extracts. Metabolic labeling with 32P followed by etoposide treatment and subsequent affinity purification of affinity-labeled caspases confirmed that at least three caspase species were phosphorylated. To detect effects of the phosphorylation on enzymatic activity, caspase-mediated cleavage of aspartylglutamylvalinylaspartyl-7-amino-4-trifluoromethylcoumarin (DEVD-AFC) and poly(ADP-ribose) polymerase (PARP) by phosphorylated and dephosphorylated extracts was measured. No significant changes in Km or vmax were detected using DEVD-AFC. In contrast, a slight, but significant enhancement of PARP cleavage was observed in dephosphorylated extracts, suggesting that phosphorylation of active caspases could have an inhibitory effect on enzyme activity. These observations, which provide the first evidence that caspases are phosphoproteins, suggest that caspases may be targets for some of the growing list of protein kinases that are involved in apoptotic events.
© 1998 by The American Society of Hematology.
OVER THE PAST 5 years it has become clear that the initiation and execution of apoptotic cell death involve a complex network of cysteine-dependent proteases termed caspases.1-3 Caspases are expressed in healthy cells as inactive zymogens. Early in apoptosis, autocatalytic activation of certain procaspases triggers a protease cascade that leads to activation of downstream caspases and other enzymes that mediate apoptotic biochemical changes.4-6 How this protease cascade is regulated is the subject of intensive study. Localization of regulatory procaspases to certain subcellular compartments appears to be one key aspect of this regulation.3 It is not currently known whether posttranslational modifications such as protein phosphorylation might also play a role in modulating the activity of the mature caspases and/or regulating the caspase cascade.
Several lines of evidence have previously suggested that apoptosis can be regulated by protein kinases and phosphatases.7-9 It has been shown, for example, that Bruton’s tyrosine kinase (BTK) is required for the apoptotic response to ionizing radiation in chicken DT40 lymphoma cells.10 Two other kinases whose activity is upregulated during apoptosis, PCKδ11 and ASK1,12 have been implicated in activation of the Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) pathway. This pathway, which is stimulated during Fas- and ceramide-mediated apoptosis,13,14 as well as apoptosis that occurs after growth factor withdrawal15 or anticancer drug treatment,16,17 results in c-Jun–dependent transcriptional activation, although it is clear that apoptosis can occur in the absence of transcription as well.18,19 Recent studies have also implicated the closely related Zip20 and DAP21 kinases in cell death, although the exact role of these kinases during apoptosis remains to be determined.
Examination of death receptor signaling pathways has provided even stronger evidence linking kinases with the induction of apoptosis. The serine/threonine kinase RIP22 appears to play an important role in the initiation of tumor necrosis factor-α (TNF-α)–induced cell death by mediating interactions between the type I TNF-α receptor (TNFR1)23 and an adapter protein RAIDD,24 thereby facilitating recruitment of procaspase-2 to the TNFR1 signaling complex. More recently, RIP2/ CARDIAK,25,26 a RIP-related kinase that can bind to the intracellular domains of Fas/CD95 and TNFR1,26 was found to bind to and promote activation of procaspase-1 (but not caspases −2, −4, −9) in vitro,25providing the first example of a direct interaction between a kinase and a procaspase. However, it is not clear whether RIP2/CARDIAK phosphorylates procaspase-1, as the binding between the two molecules requires the so-called caspase recruitment domain27 rather than the kinase domain of RIP2.25 Forced overexpression of RIP2 was shown to induce apoptosis,26 but it remains to be determined whether procaspase-1 (or any other procaspase) is a pertinent substrate when RIP2 kinase initiates apoptotic signaling.
In addition to acting as positive regulators of apoptosis, kinases can also be anti-apoptotic. The Bcr/Abl kinase, for example, delays the onset of apoptosis in granulocytes from patients with chronic myelogenous leukemia,28 possibly by delaying engagement of the apoptotic machinery.29-31 Likewise, activation of a kinase cascade involving phosphatadylinositol-3 kinase and Akt (protein kinase B) has been observed to inhibit apoptosis that ordinarily occurs after withdrawal of interleukin-2 (IL-2), IL-3 or integrin-mediated signals.32-34
The biochemical basis for the effects of many of these kinases on apoptotic events is incompletely understood. Several members of the Bcl-2 family of apoptotic regulators are apparently regulated at least in part by phosphorylation.35 Serine phosphorylation of Bcl-2 has been reported to be either anti-apoptotic36,37 or proapoptotic,38 depending on the site that is phosphorylated. For example, paclitaxel-induced phosphorylation of Bcl-2 results in the disruption of Bcl-2/Bax interactions, the release of Bax to the cytosol, and activation of the apoptotic pathway.39 Similarly, signaling through the phosphatidylinositol 3-kinase/Akt pathway leads to phosphorylation of the proapoptotic Bcl-2 family member BAD, disrupting its interactions with anti-apoptotic Bcl-2 family members and altering its ability to induce apoptosis.40-43 However, these observations do not rule out the possibility that other components of the apoptotic machinery might also be regulated by phosphorylation.
Phosphatases have also been implicated in control of the apoptotic pathway. Association of the tyrosine phosphatase FAP-1 with the carboxyl terminal domain of the death receptor CD95/Fas has been reported to inhibit Fas-mediated apoptosis,44 raising the possibility that phosphorylation of Fas or some other component may be required for activation of this pathway. Conversely, drug-induced apoptosis in human leukemia cells is accompanied by dephosphorylation of the retinoblastoma susceptibility protein (Rb).45,46 In at least one cell line, serine/threonine phosphatase inhibitors can prevent both the drug-induced dephosphorylation of Rb and apoptosis,47 suggesting that dephosphorylation of Rb or some other cellular component is required in this system for engagement of the apoptotic machinery.
Recent studies using affinity labeling and two-dimensional gel electrophoresis to identify the caspase species activated during apoptosis showed that a number of labeled polypeptides differed only in their isoelectric points.48 49 This observation raised the possibility that caspases might be regulated by posttranslational modifications. In view of the growing evidence that kinases and phosphatases are involved in the regulation of apoptosis, the objective of the present study was to determine whether active caspases are phosphorylated in vivo and to perform an initial assessment of the consequences of this modification on enzymatic activity.
MATERIALS AND METHODS
Materials.
Reagents were obtained from the following suppliers: sodium orthovanadate, dialyzed fetal bovine serum (FBS), and d-biotin from Sigma (St Louis, MO); λ phosphatase from New England Biolabs (Beverly, MA); ortho[32P]phosphate from Amersham (Arlington Heights, IL); okadaic acid and DEVD-fmk from Calbiochem (Cambridge, MA); Nonidet P40 from BioRad (Richmond, CA); sodium deoxycholate from BDH (Poole, UK); immunopure immobilized avidin from Pierce (Rockford, IL); Sephadex G-25 medium and Immobiline drystrips (pH 4-7L, 11 cm) from Pharmacia (Uppsala, Sweden); and other reagents as previously specified.48
Cell culture.
HL-60 cells were cultured in RPMI 1640 medium containing 10% (vol/vol) heat-inactivated FBS and gentamicin (50 μg/mL) at concentrations of less than 106 cells/mL to insure logarithmic growth. For metabolic labeling, cells (5 × 105 cells/mL) were centrifuged and resuspended in phosphate-free RPMI 1640 medium containing 10% (vol/vol) dialyzed FBS and 10 μCi/mL ortho[32P]phosphate. Cells were labeled for 24 hours at 37°C before induction of apoptosis.
Induction of apoptosis and preparation of cytosolic extracts.
Etoposide was added to HL-60 cells at a concentration of 68 μmol/L, a concentration shown to induce apoptotic cell death in previous studies.29,48 After 5 hours, cytosol was prepared from washed cells as previously described,48 with the exception that okadaic acid was included at a final concentration of 20 nmol/L.
Preparation of concentrated extracts from HL-60 cells.
After drug treatment, all steps were performed at 4°C (modified from Lazebnik et al50). Cells were sedimented at 300g for 5 minutes, and washed with incomplete MIG buffer (50 mmol/L piperazine-N,N′-bis[2-ethanesulfonic acid] [PIPES, pH 7.0], 50 mmol/L KCl, 10 mmol/L EDTA, 10 mmol/L sodium orthovanadate). Cells were washed again in complete MIG buffer, consisting of incomplete buffer supplemented just before use with 20 nmol/L okadaic acid, 20 μmol/L cytochalasin B, 1 mmol/L dithiothreitol (DTT), 100 μmol/L α-phenylmethylsulfonyl fluoride, and 1 μg/mL chymostatin, leupeptin, antipain, and pepstatin A. The cell pellet was resuspended in 1/3 vol of complete MIG buffer and transferred to a 2-mL microcentrifuge tube. Cells were disrupted by sonication at 4°C for 30 seconds at 100 W using a Misonix Model XL2010 sonicator equipped with a 1/8” microtip probe (Branson Ultrasonics, Danbury, CT). The cell lysate was centrifuged at 250,000gmax for 2 hours, yielding a clear extract. Protein concentration of the extract, measured by the Bradford assay,51 was estimated to be 40 mg/mL.
Phosphatase treatment of HL-60 cytosolic extracts.
Aliquots containing 300 μg of cytosolic protein were supplemented with 8 U/mL of λ phosphatase in the presence or absence of 10 mmol/L sodium orthovanadate and incubated at 30°C for 30 minutes. At the end of the incubation, the reaction was terminated by addition of sodium orthovanadate to a final concentration of 10 mmol/L to each tube. Samples were frozen at −80°C until analyzed by affinity labeling with Z-EK(bio)D-aomk followed by immunoblotting (see below) or assayed for DEVD-AFC cleavage activity as previously described.29 48
Affinity labeling and inhibitor competition experiments.
Aliquots containing 5.2 mg of cytosolic protein from etoposide-treated HL-60 cells were incubated for 5 minutes at 37°C with the tetrapeptide inhibitor DEVD-fmk or the same volume of dilulent (DMSO). Z-EK(bio)D-aomk was then added to a final concentration of 1 μmol/L and the incubation was continued for 15 minutes at 37°C.
Purification of caspases from radiolabeled extracts.
After radiolabeled extracts were reacted with Z-EK(bio)D-aomk, unreacted affinity label was removed by passing the lysates through a Sephadex G25 gel filtration column (1.5 mL of packed resin). The flow through was diluted with an equal volume of denaturation buffer (0.5% wt/vol sodium dodecyl sulfate [SDS], 20 mmol/L PIPES-KOH, pH 7.0, 20% [vol/vol] β-mercaptoethanol) and boiled for 5 minutes. Four volumes of correction buffer (1% [vol/vol] Nonidet P40, 1% [wt/vol] sodium deoxycholate, 2 mmol/L EDTA, and 10 mmol/L PIPES-KOH, pH 7.0) was added. The corrected lysate (900 μL) was incubated with 60 μL of immunopure immobilized avidin for 1 hour with agitation at 4°C. The resin was washed three times with 1 mL of wash buffer (0.05% [wt/vol] SDS, 10 mmol/L PIPES, 4% β-mercaptoethanol, 0.8% [vol/vol] Nonidet P40, 0.8% [wt/vol] sodium deoxycholate, and 1.6 mmol/L EDTA), and once with 1 mL of final wash buffer (10 mmol/L PIPES, 10% [vol/vol] β-mercaptoethanol, 2 mmol/L EDTA). The bound proteins were eluted by boiling the resin for 5 minutes in elution buffer (50 mmol/L Tris-HCl [pH 8.6], 15% [wt/vol] sucrose, 2 mmol/L EDTA, 3% [wt/vol] SDS, and 2 mmol/L d-biotin).
Western blot analysis.
Samples containing purified radiolabeled caspases were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 16% (wt/vol) acrylamide gels and transferred to nitrocellulose. Radioactivity was quantitated using a Storm 840 Phosphorimager (Molecular Dynamics, Sunnyvale, CA), and digital data were processed using the ImageQuant software package. The same nitrocellulose membrane was later probed with peroxidase-labeled streptavidin and visualized by enhanced chemiluminescence.29 48
Alternatively, samples treated with λ phosphatase and labeled with Z-EK(bio)D-aomk were precipitated using methanol/chloroform and resuspended in 100 μL of urea solubilization buffer (8 mol/L urea, 4% [vol/vol] Nonidet P-40, 2% [vol/vol] β-mercaptoethanol, 20% [vol/vol] BioLyte 3-10 carrier ampholytes). Two-dimensional gel electrophoresis was performed in a Multiphor II Unit using immobilized pH gradients as previously described.52 Samples were focused on an immobilized linear pH gradient isoelectric focusing gel (110 mm, pH 4 to 7) for 30,500 volt-hours, transferred to the top of a 16% (wt/vol) SDS-polyacrylamide gel, resolved at 15 mA for 6 hours, transferred to nitrocellulose, and probed with streptavidin as described above.
Analysis of poly(ADP-ribose) polymerase (PARP) cleavage activity.
Extracts treated with phosphatase were incubated with 125 ng of purified bovine PARP at 37°C for various amounts of time. Proteins were resolved by SDS-PAGE on 7.5% (wt/vol) acrylamide gels and transferred to nitrocellulose. PARP was detected using C-II-10 monoclonal antibody and visualized by enhanced fluorescence (ECF). Quantitation on the phosphorimager was performed as described above.
RESULTS
Detection of active caspases in extracts from apoptotic HL-60 cells.
Our previous studies showed that at least eight electrophoretically distinct caspase species, including multiple isoforms of caspase-3 and caspase-6, were detectable in cytosol prepared from apoptotic HL-60 cells.48 To more completely characterize the caspases activated during HL-60 cell apoptosis, whole-cell extracts were prepared from sonicated cells and reacted with Z-EK(bio)D-aomk, an affinity labeling reagent that covalently labels the large subunits of all caspases previously tested.48 After labeling, the extracts were subjected to affinity chromatography on avidin agarose. Polypeptides eluted from the column with free biotin were resolved by two-dimensional gel electrophoresis using isoelectric focusing with an immobilized pH gradient in the first dimension and SDS-PAGE in the second. The region of the gel containing polypeptides in the 15- to 25-kD molecular-weight range (the known molecular weights of the caspase large subunits53) is shown in Fig 1. Using this methodology, 18 distinct Z-EK(bio)D-aomk-reactive species were observed (Fig 1).
High-resolution, high-sensitivity detection of active caspases in apoptotic HL-60 cell extracts by two-dimensional gel electrophoresis. (A) Mobilities of Z-EK(bio)D-aomk reactive caspases on two-dimensional PAGE. Caspases were partly purified from extracts of apoptotic HL-60 cells (see Materials and Methods), labeled with Z-EK(bio)D-aomk, resolved by 2-dimensional PAGE using an immobilized pH gradient, transferred to nitrocellulose membrane, reacted with peroxidase-conjugated streptavidin, and detected by ECL. (B) Indexing of active caspases purified from HL-60 cytosol. Shaded circles correspond to species previously shown to comigrate with known caspases. Spots A1, C1, C3 comigrate with Caspase-3, while spot B1 comigrates with Caspase 6.48 The migration of relevant molecular-weight markers is shown at the left of (A) and (B). (C) Isoelectric points of the detected caspases. (D) Control experiments performed with ortho[32P]phosphate labeled extracts showed the presence of Z-EK(bio)D-aomk reactive species in extracts prepared from etoposide treated cells (lanes 2, 2′), but not in extracts from control healthy cells (lanes 1, 1′).
High-resolution, high-sensitivity detection of active caspases in apoptotic HL-60 cell extracts by two-dimensional gel electrophoresis. (A) Mobilities of Z-EK(bio)D-aomk reactive caspases on two-dimensional PAGE. Caspases were partly purified from extracts of apoptotic HL-60 cells (see Materials and Methods), labeled with Z-EK(bio)D-aomk, resolved by 2-dimensional PAGE using an immobilized pH gradient, transferred to nitrocellulose membrane, reacted with peroxidase-conjugated streptavidin, and detected by ECL. (B) Indexing of active caspases purified from HL-60 cytosol. Shaded circles correspond to species previously shown to comigrate with known caspases. Spots A1, C1, C3 comigrate with Caspase-3, while spot B1 comigrates with Caspase 6.48 The migration of relevant molecular-weight markers is shown at the left of (A) and (B). (C) Isoelectric points of the detected caspases. (D) Control experiments performed with ortho[32P]phosphate labeled extracts showed the presence of Z-EK(bio)D-aomk reactive species in extracts prepared from etoposide treated cells (lanes 2, 2′), but not in extracts from control healthy cells (lanes 1, 1′).
A number of observations indicate that these Z-EK(bio)D-aomk-labeled species are active caspases: (1) Z-EK(bio)D-aomk efficiently labels cloned caspases.48 (2) Extracts made by an identical method from nonapoptotic control cells, which lack active caspases,48 lack 15- to 25-kD polypeptides that react with Z-EK(bio)D-aomk (Fig 1D). (3) Treatment with the irreversible caspase inhibitors YVAD-cmk, DEVD-fmk, or Z-VAD-fmk inhibits labeling with Z-EK(bio)D-aomk (eg, see Fig 3B).48,54 (4) Several of the labeled species shown in Fig 1 comigrate in two-dimensional gels with cloned human caspases expressed in Sf9 cells. Spots A1, C1, and C3 comigrate with caspase-3; and spot B1 comigrates with caspase-6.48 (5) Direct sequencing of two-dimensional gel spots obtained after a similar purification yielded sequences corresponding to active caspase-3 and caspase-6.49Collectively, these observations indicate that the Z-EK(bio)D-aomk-reactive species detected in Fig 1A are active caspases. Morover, the results of this experiment indicate that the pattern of caspase activation in this cell line is substantially more complex than previously suspected based on conventional two-dimensional gel analysis of cytosol alone.
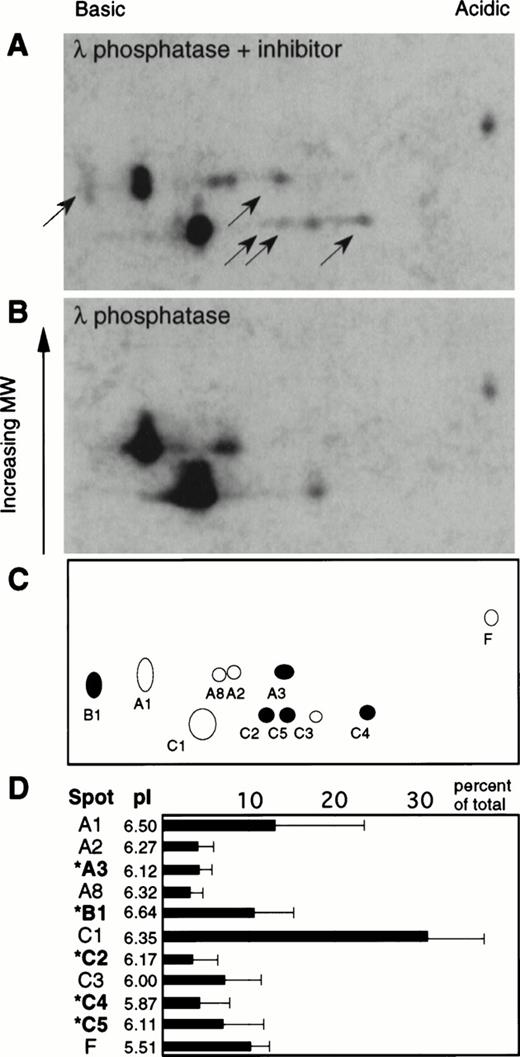
Effect of phosphatase treatment on the pattern of affinity labeled caspases.
Several of the labeled caspase species detected in Fig 1 differed from one another only in their isoelectric points. To investigate the possibility that some of these species might represent phosphorylated forms, cytosol was prepared from etoposide-treated HL-60 cells in the presence of the phosphatase inhibitor okadaic acid, reacted with Z-EK(bio)D-aomk, and treated with λ phosphatase (8 U/mL, 30 minutes at 30°C) in the presence or absence of sodium orthovanadate as a phosphatase inhibitor. Cytosol was used for this analysis to decrease the number of reactive species, thereby simplifying the analysis. In the absence of inhibitor, λ phosphatase induced a substantial alteration of the gel pattern (Fig 2), suggesting that at least some of the species are phosphorylated forms of the large subunits of active caspases. Quantitation of the active caspases detected by enhanced chemiluminescence (ECL) showed that the five species whose mobility shifted upon phosphatase treatment corresponded to approximately 30% of the covalently bound affinity label or, to a first approximation because labeling is stoichiometric, 30% of the active caspases detected by this protocol. In additional experiments (not shown), similar results were obtained when potato acid phosphatase was substituted for λ phosphatase. Moreover, similar changes were observed when the order of treatment with Z-EK(bio)D-aomk and λ phosphatase was reversed.
Effect of phosphatase treatment on the two-dimensional gel pattern of active caspases. Z-EK(bio)D-aomk treated cytosol from etoposide-treated HL-60 cells was incubated with 400 U λ phosphatase in the presence (A) or absence (B) of the inhibitor sodium orthovanadate, then subjected to analysis by two-dimensional PAGE. Black arrows point to caspases that disappear upon phosphatase treatment. (C) Indexing of the active caspases present in (A). Filled circles correspond to caspases that disappear upon phosphatase treatment. (D) Bar chart illustrating the relative abundance of the various species shown in (A) (average of three independent experiments, with the standard deviation indicated). Also shown is the nomenclature of the various active caspases detected in this experiment together with their corresponding isoelectric points. Species indicated by an asterisk (*) correspond to caspases that disappear upon dephosphorylation.
Effect of phosphatase treatment on the two-dimensional gel pattern of active caspases. Z-EK(bio)D-aomk treated cytosol from etoposide-treated HL-60 cells was incubated with 400 U λ phosphatase in the presence (A) or absence (B) of the inhibitor sodium orthovanadate, then subjected to analysis by two-dimensional PAGE. Black arrows point to caspases that disappear upon phosphatase treatment. (C) Indexing of the active caspases present in (A). Filled circles correspond to caspases that disappear upon phosphatase treatment. (D) Bar chart illustrating the relative abundance of the various species shown in (A) (average of three independent experiments, with the standard deviation indicated). Also shown is the nomenclature of the various active caspases detected in this experiment together with their corresponding isoelectric points. Species indicated by an asterisk (*) correspond to caspases that disappear upon dephosphorylation.
Purification and detection of active caspases in etoposide-treated ortho[32P]phosphate-labeled HL-60 cells.
To confirm that the caspases whose mobility shifted after treatment of extracts with λ phosphatase were in fact phosphorylated, HL-60 cells were labeled in vivo with ortho[32P]phosphate and treated with etoposide to induce apoptosis (Fig3A). The active caspases present in extracts prepared from these metabolically labeled cells were derivatized with Z-EK(bio)D-aomk and purified using avidin agarose. To distinguish purified [32P]-labeled caspases from other labeled cellular proteins that also bound to the avidin-agarose, control extracts were pre-incubated with the covalent caspase inhibitor DEVD-fmk before derivatization with Z-EK(bio)D-aomk. This had no effect on background binding of extract proteins to the avidin-agarose, but completely abolished the Z-EK(bio)D-aomk–dependent binding of [32P]-labeled activated caspases (Fig 3B, lanes 2′, 6′). Purified [32P]-labeled proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and exposed for 7 days to a phosphorimager plate. The same membrane was later incubated with peroxidase-coupled streptavidin and visualized by enhanced chemiluminescence. The mobility of proteins detected by ECL, previously shown to correspond to active caspases,48,49,54 and the labeled species detected using the phosphorimager were then compared (Fig 3B). Quantitation of the phosphorimager counts corresponding to the rectangular areas A5, A5′, and A6 shown in Fig 3B enabled us to construct the profiles shown in Fig 3C. This analysis showed that phosphorylated species 5 and 8 comigrated on one-dimensional gels with active caspases, previously designated IRP1 and IRP4.54Furthermore, recovery of species 5 and 8 was significantly diminished after pretreatment of extracts with DEVD-fmk, consistent with the notion that both species are indeed active caspases. One additional labeled species (no. 2) was also substantially decreased after DEVD-fmk pretreatment (see Fig 3C, merged). Although this species did not comigrate with one of the known biotinylated caspases labeled with Z-EK(biotin)D-aomk, it may correspond to a minor caspase species that is labeled to relatively high specific activity with 32P under this protocol. When combined with the two-dimensional analysis of phosphatase-treated caspases, these data suggest that several active caspases are indeed phosphorylated in HL-60 cells.
Detection of 32P-labeled active caspases in extracts from apoptotic HL-60 cells. (A) Experimental protocol. HL-60 cells labeled with ortho[32P]phosphate were exposed to etoposide to induce apoptosis. Concentrated extracts from these cells were reacted with Z-EK(bio)D-aomk (±pretreatment with DEVD-fmk) and bound to monomeric avidin-agarose. The input, flow-through, and eluted fraction (with 0.1% SDS) were resolved on a 16% SDS-polyacrylamide gel and then analyzed (B) by autoradiography of the 32P (lanes 1 through 6) or by ECL (lanes 1′ through 6′). Four percent of the total extract was loaded for lanes 1, 2, 3, 4, 1′, 2′, 3′, 4′. Eighty percent of the purified protein fraction was loaded for lanes 5, 6, 5′, 6′. Lanes 2, 4, 6, 2′, 4′, 6′ correspond to cytosol preincubated with DEVD-fmk before Z-EK(bio)D-aomk labeling. The rectangular areas, A5, A5′, A6 were subsequently subjected to quantitative analysis using the phosphorimager. (C) Quantitation of the phosphorimager data (counts per minute) of areas A5, A6, and densitometry (arbitrary units) for area A5′. The box designated “MERGED” corresponds to a superimposition of A5 and A6, showing the relative position of the ECL bands in A5′ as black rectangles on the x-axis.
Detection of 32P-labeled active caspases in extracts from apoptotic HL-60 cells. (A) Experimental protocol. HL-60 cells labeled with ortho[32P]phosphate were exposed to etoposide to induce apoptosis. Concentrated extracts from these cells were reacted with Z-EK(bio)D-aomk (±pretreatment with DEVD-fmk) and bound to monomeric avidin-agarose. The input, flow-through, and eluted fraction (with 0.1% SDS) were resolved on a 16% SDS-polyacrylamide gel and then analyzed (B) by autoradiography of the 32P (lanes 1 through 6) or by ECL (lanes 1′ through 6′). Four percent of the total extract was loaded for lanes 1, 2, 3, 4, 1′, 2′, 3′, 4′. Eighty percent of the purified protein fraction was loaded for lanes 5, 6, 5′, 6′. Lanes 2, 4, 6, 2′, 4′, 6′ correspond to cytosol preincubated with DEVD-fmk before Z-EK(bio)D-aomk labeling. The rectangular areas, A5, A5′, A6 were subsequently subjected to quantitative analysis using the phosphorimager. (C) Quantitation of the phosphorimager data (counts per minute) of areas A5, A6, and densitometry (arbitrary units) for area A5′. The box designated “MERGED” corresponds to a superimposition of A5 and A6, showing the relative position of the ECL bands in A5′ as black rectangles on the x-axis.
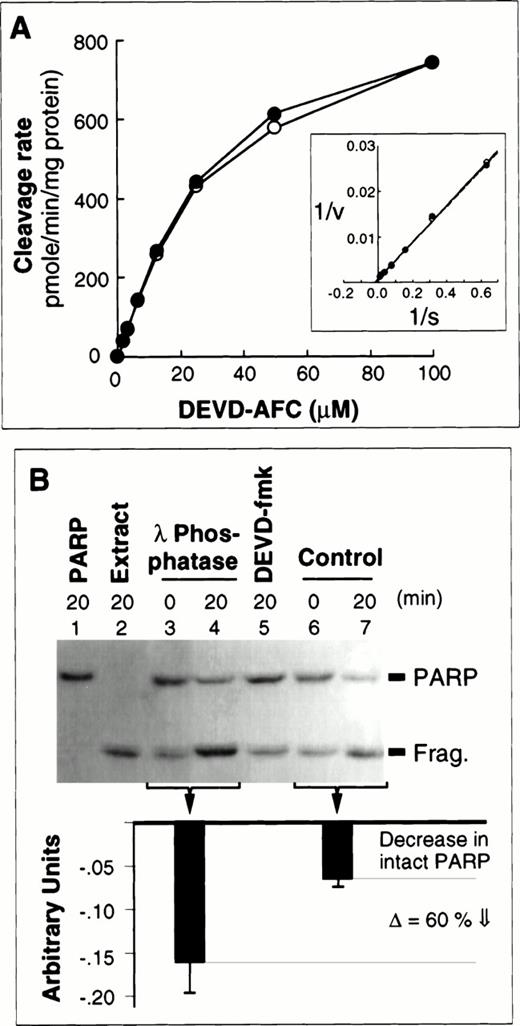
Effects of dephosphorylation of active caspases on cleavage of peptide and native protein substrates.
The results presented in Fig 2 show that the intensity of one polypeptide previously shown to comigrate in two-dimensional gels with caspase-3 (spot C1)48 increased upon phosphatase treatment of extracts. This suggests that a portion of the activated caspase-3 is phosphorylated in HL-60 cells undergoing apoptosis. Analysis of the three-dimensional structure of caspase-3, based on the reported atomic coordinates,55 showed several putative phosphorylation sites, including a protein kinase C (PKC) consensus phosphorylation site, on the p20 subunit in close proximity to the substrate binding pocket. PKCδ and PKCθ have been reported to be cleaved and activated during apoptosis in a caspase-dependent manner.11,56,57 More recently, activation of PKCα has also been reported to occur before58 or after59caspase activation in HL-60 cells. Although the precise role of PKC isoforms during apoptosis clearly remains to be clarified, these observations led us to ask whether activity of caspase-3–like enzymes was altered in phosphatase-treated samples. A priori, we expected any such effects to be small, because the great bulk of the active caspase-3 in these extracts (spots A1 and C1) is apparently not phosphorylated and shows no mobility shift following λ phosphatase treatment (Fig 2). In fact, when caspase activity was measured using the tetrapeptide substrate DEVD-AFC, phosphatase treatment failed to alter either the vmax or Km (≈20 μmol/L) for this substrate (Fig 4A). However, recent experiments have suggested that tetrapeptide substrates might not accurately reflect all aspects of caspase-mediated cleavage.60 To investigate whether phosphorylation might influence the interaction of active caspases with physiological protein substrates, we compared the ability of control and dephosphorylated extracts to cleave the canonical caspase substrate PARP (Fig 4B). These experiments showed that full-length PARP is cleaved more effectively in the dephosphorylated extracts than in control extracts. Quantitation performed on results obtained from three independent experiments indicated that PARP cleaving activity of dephosphorylated extracts is approximately 60% higher than that of control extracts (Fig 4B, bottom).
DEVD-AFC and PARP cleavage activity assays using dephosphorylated and control extracts. (A) Measurements using a fluorogenic assay showed no differences between the ability of dephosphorylated (open symbols) and control extracts (closed symbols) to cleave the synthetic substrate DEVD-AFC. Inset, data replotted by the method of Lineweaver and Burke. (B) Dephosphorylated extracts exhibit enhanced ability to cleave PARP. (Top) PARP cleavage was examined by enhanced chemofluorescence. Lanes 1 and 2 correspond to PARP and extract incubated alone, respectively. (This extract contains the cleaved PARP fragment.) Lanes 3 and 4, cleavage of PARP by dephosphorylated extract. Lane 5, PARP was incubated in the presence of control extract following pretreatment with DEVD-fmk. (There is no significant decrease in intact PARP—the fragment is from the extract as in lane 2.) Lanes 6 and 7, cleavage of PARP by control extract. (Bottom) Quantitation of the PARP cleavage results (average of three independent experiments, with the standard deviations indicated). Downward black bars correspond to the relative decrease (relative to the starting level) in the amount of full-length PARP remaining after incubation for 20 minutes in phosphatase-treated and control extracts. Both the decrease in intact PARP and corresponding increase in the cleaved fragment are enhanced in phosphatase-treated extracts.
DEVD-AFC and PARP cleavage activity assays using dephosphorylated and control extracts. (A) Measurements using a fluorogenic assay showed no differences between the ability of dephosphorylated (open symbols) and control extracts (closed symbols) to cleave the synthetic substrate DEVD-AFC. Inset, data replotted by the method of Lineweaver and Burke. (B) Dephosphorylated extracts exhibit enhanced ability to cleave PARP. (Top) PARP cleavage was examined by enhanced chemofluorescence. Lanes 1 and 2 correspond to PARP and extract incubated alone, respectively. (This extract contains the cleaved PARP fragment.) Lanes 3 and 4, cleavage of PARP by dephosphorylated extract. Lane 5, PARP was incubated in the presence of control extract following pretreatment with DEVD-fmk. (There is no significant decrease in intact PARP—the fragment is from the extract as in lane 2.) Lanes 6 and 7, cleavage of PARP by control extract. (Bottom) Quantitation of the PARP cleavage results (average of three independent experiments, with the standard deviations indicated). Downward black bars correspond to the relative decrease (relative to the starting level) in the amount of full-length PARP remaining after incubation for 20 minutes in phosphatase-treated and control extracts. Both the decrease in intact PARP and corresponding increase in the cleaved fragment are enhanced in phosphatase-treated extracts.
DISCUSSION
In the present study, we used two complementary techniques to evaluate the possibility that caspases are phosphoproteins. Results of these experiments not only provide the first evidence that some activated caspase species contain covalently bound phosphate, but also provide an indication that phosphorylation might inhibit the activity of caspase species against certain substrates. These observations have potentially important implications for current understanding of the manner in which apoptosis is regulated.
Previous studies have shown that multiple caspase species differing only in isoelectric point are detectable in the cytosol of apoptotic leukemia cells.48,49 We have extended those studies by demonstrating that phosphatase treatment of cytosol results in disappearance of some species and increases in others (Fig 2). To confirm this result using independent techniques, caspases were affinity purified from 32P-labeled cells after induction of apoptosis (Fig 3). At least two caspase species were found to contain covalently bound 32P. Coupled with the previous reports of charge heterogeneity among activated caspases,48 49 these results provide strong evidence that activated caspases are phosphoproteins.
Among the affinity-labeled caspase species that disappear from the two-dimensional gels with phosphatase treatment are B1, which comigrates with recombinant caspase-6, and four currently unidentified species. These changes were accompanied by increased amounts of C1, which comigrates with caspase-3. It is unlikely that any of the unidentified species corresponds to either caspase-2 or caspase-1, the only two caspases shown thus far to associate with protein kinases (the former with RIP indirectly via the adapter RAIDD24 and the latter directly with RIP2/CARDIAK25). Immunoblotting and activity measurements previously failed to detect any significant maturation of procaspase-1 or -2 in HL-60 cells undergoing etoposide-induced apoptosis.48
Despite the fact that only about 30% of the active caspase species appear to be phosphorylated in these extracts (Fig 2), a reproducible increase in PARP cleavage was observed after dephosphorylation of cytosol (Fig 4B). This observation could mean that phosphorylation of certain caspase species has a significant inhibitory effect on their ability to cleave protein substrates. Because our detection of active caspases depends on covalent labeling with the substrate analog Z-EK(bio)D-aomk, we cannot at present rule out the possibility that phosphorylated caspases are labeled less efficiently by this reagent. If this were the case, then the data in Fig 2 might have underestimated the level of caspase phosphorylation in apoptotic HL-60 cells. On the other hand, although it is clear that the λ phosphatase acts directly on certain caspases, as judged by the changes in their mobility in two-dimensional gels, it is also difficult to rule out the possibility that the enhanced PARP cleavage observed after phosphatase treatment is due to dephosphorylation of another factor(s) that somehow modulates PARP cleavage in these extracts. It will be important in the future to map the phosphorylation sites on the caspases and identify the relevant kinases. These experiments await the availability of high affinity antibodies that can cleanly immunoprecipitate defined caspases from apoptotic cell extracts.
In view of the evidence that phosphorylation plays an important role in the apoptotic process (see Introduction), it is possible that phosphorylation also affects other aspects of caspase function. For example, phosphorylation has been shown to be an important factor regulating the nuclear transport of certain polypeptides.61-63 We have recently shown that C2 and C4, two of the species whose mobility in two-dimensional gels is sensitive to phosphatase treatment, are present in cytosol but absent from nuclei of apoptotic HL-60 cells,29 raising the possibility that phosphorylation might affect caspase targeting within apoptotic cells. In MDA-MB-468 breast carcinoma cells, on the other hand, three of the caspase species shown here to be phosphorylated (B1, C4, and C5) were found in the nuclei (T.J.K. and L.M.M., unpublished observations, January 1998). Thus, if phosphorylation is involved in the regulation of caspase nuclear import, the phenomenon might vary depending on the cell type.
It is also possible that phosphorylation is important in regulating procaspases. Although the methods used in the present study detect only the active caspases, preliminary experiments have shown the existence of multiple charge isoforms of procaspase-2 and procaspase-3 in untreated cells (T.J.K. and S.H.K., unpublished observations, May 1998). These observations raise the possibility that changes in phosphorylation state might be involved in caspase activation and/or caspase interactions with regulatory molecules.
Although the role(s) of caspase phosphorylation remain to be more fully elucidated, the present report opens the way to the investigation of a new level of caspase regulation in vivo. It will be important in future experiments to identify both the phosphorylated species and the relevant kinases in order to begin to understand the role of caspase phosphorylation in the apoptotic response.
Supported by a Principal Research Fellowship from the Wellcome Trust (W.C.E.) and Public Health Service Grant No. CA69008 (S.H.K. and W.C.E.). L.M.M. was supported by a studentship from the Programa Gulbenkian de Doutoramento em Biologia e Medicina. S.H.K. was a Scholar of the Leukemia Society of America. W.C.E. is a Principal Research Fellow of the Wellcome Trust.
Address reprint requests to William C. Earnshaw, PhD, Institute of Cell & Molecular Biology, University of Edinburgh, Edinburgh, EH9 3JR, Scotland, UK; e-mail: bill.earnshaw@ed.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 1. High-resolution, high-sensitivity detection of active caspases in apoptotic HL-60 cell extracts by two-dimensional gel electrophoresis. (A) Mobilities of Z-EK(bio)D-aomk reactive caspases on two-dimensional PAGE. Caspases were partly purified from extracts of apoptotic HL-60 cells (see Materials and Methods), labeled with Z-EK(bio)D-aomk, resolved by 2-dimensional PAGE using an immobilized pH gradient, transferred to nitrocellulose membrane, reacted with peroxidase-conjugated streptavidin, and detected by ECL. (B) Indexing of active caspases purified from HL-60 cytosol. Shaded circles correspond to species previously shown to comigrate with known caspases. Spots A1, C1, C3 comigrate with Caspase-3, while spot B1 comigrates with Caspase 6.48 The migration of relevant molecular-weight markers is shown at the left of (A) and (B). (C) Isoelectric points of the detected caspases. (D) Control experiments performed with ortho[32P]phosphate labeled extracts showed the presence of Z-EK(bio)D-aomk reactive species in extracts prepared from etoposide treated cells (lanes 2, 2′), but not in extracts from control healthy cells (lanes 1, 1′).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/9/10.1182_blood.v92.9.3042/5/m_blod42155001w.jpeg?Expires=1769092294&Signature=PPbi1Hv~YLp-9Nv489jry3k3T0jqIKrmcWXo0gES0NDrHnWyDvTuYrL-KyznoWH2qlVuTVcp-SUJZy8-TWLTHSp0XTCo-e9F0G8w5tYcNslD3~KJCJve75rVXMDosAkHNwrFByc4oVl6bDD6aS-NPeIM9jYThYGS0xU3GnvzwxxjiM4eJI0zdYgqzfp22GotZVwZHvBG88BJKzBAnCKQU5C0lswR9hHduPa2RA~SSrOLLeb1JYXK7Zhnee8emRDKclyIWQSltYaf6yqD4Eh8n~c4bVBgch90mhuNKnQzMMIdP17KcCKO3KDQF4TPFix-oYOS1vPABCP2c5zLTldsTQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Detection of 32P-labeled active caspases in extracts from apoptotic HL-60 cells. (A) Experimental protocol. HL-60 cells labeled with ortho[32P]phosphate were exposed to etoposide to induce apoptosis. Concentrated extracts from these cells were reacted with Z-EK(bio)D-aomk (±pretreatment with DEVD-fmk) and bound to monomeric avidin-agarose. The input, flow-through, and eluted fraction (with 0.1% SDS) were resolved on a 16% SDS-polyacrylamide gel and then analyzed (B) by autoradiography of the 32P (lanes 1 through 6) or by ECL (lanes 1′ through 6′). Four percent of the total extract was loaded for lanes 1, 2, 3, 4, 1′, 2′, 3′, 4′. Eighty percent of the purified protein fraction was loaded for lanes 5, 6, 5′, 6′. Lanes 2, 4, 6, 2′, 4′, 6′ correspond to cytosol preincubated with DEVD-fmk before Z-EK(bio)D-aomk labeling. The rectangular areas, A5, A5′, A6 were subsequently subjected to quantitative analysis using the phosphorimager. (C) Quantitation of the phosphorimager data (counts per minute) of areas A5, A6, and densitometry (arbitrary units) for area A5′. The box designated “MERGED” corresponds to a superimposition of A5 and A6, showing the relative position of the ECL bands in A5′ as black rectangles on the x-axis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/9/10.1182_blood.v92.9.3042/5/m_blod42155003w.jpeg?Expires=1769092294&Signature=eWY7isSb3m2DZDtIlc~6MHpC024pWnteO29Vpkf-htWaxe2cDv9J-HC99pKcfQWt~1pq0ip4ARfLKAIBhy98sWXm9fK3EXx7ucTZC2lFqTy8qHdQbGl5j6Keg~b0Obr9lZXU2JuK~BsPL4lOGn4~c4CT1g6ZJ6cyMk02wZUe2gDc5NHzK28rij9J6u-LabCSPx2yVAEadpYyadliTK~QXfbNYnBSj4862OoucXTk3xY19vt-qeN7FRmcBa8-S7b7SXYP9vZE9shvRjUuO5LCce34pDWVss2z7Sb5erxZ7vlGlfC00XrxfyWGlNOImHzqg3Ug~h0-JHOrUPAsTVIWHg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal