Abstract
Marginal zone cell lymphomas of the mucosa-associated lymphoid tissue (MALT) are the most common subtype of lymphoma arising at extranodal sites. The t(11;18)(q21;q21) appears to be the key genetic lesion and is found in approximately 50% of cytogenetically abnormal low-grade MALT lymphomas. We show that the API2 gene, encoding an inhibitor of apoptosis also known as c-IAP2, HIAP1, andMIHC, and a novel gene on 18q21 characterized by several Ig-like C2-type domains, named MLT, are recurrently rearranged in the t(11;18). In both MALT lymphomas analyzed, the breakpoint inAPI2 occurred in the intron separating the exons coding respectively for the baculovirus IAP repeat domains and the caspase recruitment domain. The breakpoints within MLT differed but the open reading frame was conserved in both cases. In one case, the translocation was accompanied by a cryptic deletion involving the 3′ part of API2. As a result, the reciprocal transcript was not present, strongly suggesting that the API2-MLT fusion is involved in the oncogenesis of MALT lymphoma.
RECURRENT TRANSLOCATIONS acquired in a multistep process of transformation leading to the evolution of autonomous cell clones are well recognized in nodal B-cell lymphomas. These translocations characterize distinct subtypes of disease and involve genes controlling cell proliferation and apoptosis.BCL2, which suppresses apoptosis, was cloned from the t(14;18)(q21;q32) found in most cases of follicular B-cell lymphoma, translocations involving the BCL1/CyclinD1 gene on chromosome 11q13 are seen in many cases of mantle cell lymphoma, andc-MYC is rearranged in almost all Burkitt’s lymphomas.1
By contrast, the genetic mechanisms underlying the genesis and disease progression of extranodal marginal zone B-cell lymphomas of mucosa-associated lymphoid tissue (MALT) type, a distinct subtype of B-cell non-Hodgkin’s lymphoma (NHL), are not known.2 MALT lymphomas account for 5% to 10% of all NHLs, and the vast majority of lymphomas arising at extranodal sites. They originate in a setting of chronic inflammation triggered by chronic infection or autoimmune disorders, such as Helicobacter pylori gastritis, Sjögren’s syndrome, and Hashimoto’s thyroiditis.3In vitro experiments have shown that H pylori–specific T cells provide contact dependent help for the growth of the malignant B cells of gastric MALT.4 The etiological link between low-grade gastric MALT lymphomas and H pylori infection has also been shown by the regression of some cases with antibiotic therapy.5,6 The preferential use of Ig variable region genes (VH) associated with autoimmune disorders indicate that some marginal zone B-lymphomas may arise from autoreactive B cells.7,8 Abnormal karyotypic data have been published for only a limited number of MALT lymphomas.9-17 Recurrent abnormalities in these cases include trisomies of chromosomes 3, 7, 12, and 18,11,17,18 the t(1;14)(p22;q32) that has been described in two cases,17 and the t(11;18)(q21;q21) that represents the most frequent structural abnormality and seems to characterize this disease entity.9,12 14
We herein present a detailed molecular genetic characterization of the 11q21 and 18q21 breakpoint regions in two cases of gastrointestinal MALT lymphomas characterized by the t(11;18)(q21;q21) and show that theAPI2 gene also known as c-IAP2,19HIAP1,20 and MIHC,21 an inhibitor of apoptosis, and a novel gene on 18q21, named MLT,are rearranged in this translocation. Our data suggest that truncation of the API2 gene distal to its three baculovirus IAP repeat (BIR) domains and fusion of this truncated gene with the carboxy-terminal region of MLT may lead to increased inhibition of apoptosis and thereby confer a survival benefit to MALT type B-cell lymphomas.
MATERIALS AND METHODS
Tumor specimens.
Two cases of low-grade extranodal gastrointestinal MALT lymphomas displaying the t(11;18)(q21;q21) were selected from the files of the Center for Human Genetics, University of Leuven, Belgium and the Department of Hematology, University of Salamanca, Spain, based on the availability of metaphase spreads and frozen tumor tissue. Case 1 presented with an extended multifocal gastrointestinal MALT lymphoma involving the stomach, the small and large bowel, and the mesenteric lymphnodes. Case 2 was diagnosed with a gastric MALT lymphoma with secondary involvement of the spleen, the bone marrow, and the peripheral blood. Both cases revealed H pylori–associated gastritis and showed the typical morphology and immunophenotype of marginal zone B-cell lymphomas of MALT type2 including the tumor cell characteristics, extension of the marginal zones by tumor cells, follicular colonization, lymphoepithelial lesions, expression of IgM, CD19, CD20, Igκ light chain restriction, and negativity for CD5, CD10, and CD23.
Cytogenetic analysis.
Cytogenetic analysis was performed as described previously11 using tissue of a small bowel biopsy (case 1) and the spleen specimen (case 2). Both cases showed the t(11;18)(q21;q21) as the sole cytogenetic abnormality (case 1: 46,XY,t(11;18)(q21;q21) [17]/46,XY [3]; case 2: 46,XX,t(11;18)(q21;q21) [6]/46,XX [14]). Fluorescence in situ hybridization (FISH) was performed as previously described.22 Chromosomes 11 and 18 were identified by cohybridization with chromosome 11 (pLC11A) and 18 (L1.84) specific α-satellite probes in combination with G-banding using 4,6-diamidino-2-phenylindole-dihydrochlorid (DAPI) counterstain.
Yeast artificial chromosome (YAC) clones.
YAC clones derived from the Centre d’Etude du Polymorphisme Humain (CEPH, Paris, France) human megaYAC library were selected from the YAC contig reported by Chumakov et al23 and data obtained from URL http://www-genome.wi.mit.edu/cgi-bin/contig/yac_infoat the Whitehead Institute for Biomedical Research (Cambridge, MA). In addition, YAC A153A6 hybridizing to the BCL2 gene located at 18q2124 and a probe specific for the MLL gene on 11q23 (Oncor, Gaithersburg, MD) were used. Human YAC inserts were selectively amplified using Alu-polymerase chain reaction (PCR).25 To confirm their cytogenetic position and to determine the relative order of the YAC clones, pairs of differentially labeled YACs were hybridized to normal metaphase spreads obtained from phytohemagglutinin (PHA)-stimulated peripheral blood lymphocytes of a healthy donor.
P1 artificial chromosome (PAC) and plasmid clones.
PAC clones were isolated by screening high-density filters from the Roswell Park Cancer Institute (RPCI; Buffalo, NY) libraries with32P-labeled probes. A walking strategy was used to extend the map. PAC end-fragments were rescued using a vectorette ligation approach.26 The presence of the sequence tagged site (STS) in the relevant PACs was confirmed by PCR and each PAC was analyzed by FISH on normal metaphase spreads. BamHI subclones of PAC 152M5 were generated by ligation of gel-purified fragments in pUC18 (Pharmacia Biotech, Uppsala, Sweden), and transformation into XL10-gold cells (Stratagene, La Jolla, CA). A BamHI restriction map was generated by comparing the sequence of the ends of the BamHI fragments to the sequence of random 1-kb subclones selected for containing the BamHI restriction sites. To generate random subclones of PAC 152M5, DNA was sheared by sonication, the fraction around 1 kb was gel-purified (Qiaquick Gel Extraction; Qiagen, Hilden, Germany), blunted, and ligated in pUC18, and transformed into XL1-blue cells.
Reverse transcriptase (RT)-PCR and cloning.
Total RNA was extracted from tumor-infiltrated gastrointestinal and splenic tissue using the Trizol Reagent (Life Technologies, Inc, Rockville, MD). First-strand cDNA was reverse transcribed from 1 μg of total RNA with Murine Moloney Leukemia Virus reverse transcriptase (Life Technologies, Inc) according to standard procedures using a random hexamer primer. After size fractionation on Microspin S-400 HR columns (Pharmacia Biotech), a poly-A tail was added to the first strand cDNA with dATP and terminal deoxynucleotidyl transferase (Boehringer Mannheim, Mannheim, Germany). Double-stranded cDNA was then generated using standard procedures with primer R2T8 (5′ CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTT 3′). Nested PCR was performed using respectively primers MLTr1 (5′ CCTTCTGCAACTTCATCCAG 3′) and MLTr2 (5′ ATGGATTTGGAGCATCAACG 3′) in combination with primers R2F1 (5′ CCAGTGAGCAGAGTGACG 3′) and R2F2 (5′ GAGGACTCGAGCTCAAGC 3′). Amplification products were cloned in pGEM T-easy (Promega, Madison, WI). The API2-MLT fusion was confirmed by RT-PCR on patient’s RNA using the Titan RT-PCR system (Boehringer Mannheim) with primers API2f1 (5′ CCAAGTGGTTTCCAAGGTGT 3′) and MLTr2 and by sequence analysis of the cloned amplification products.
The MLT consensus cDNA sequence was determined by performing RACE (rapid amplification of cDNA ends) experiments on cDNA of patient 1. Several overlapping clones were analyzed to obtain the 5′ and 3′ sequences of MLT (GenBank Accession No. AF130356).
RESULTS
FISH characterization of the 11q21 and 18q21 translocation.
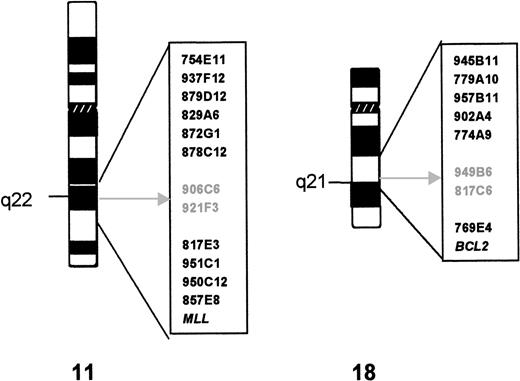
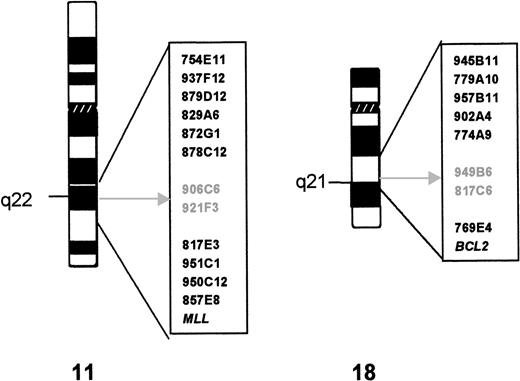
Twelve YACs derived from the chromosomal region 11q21-22.3 and theMLL probe were hybridized to metaphase spreads of case 1. Figure 1 shows their relative position in relation to the t(11;18) breakpoints. The hybridization signals of YACs 906C5 and 921F3 were split by the translocation. Subsequent analysis showed that YACs 906C5 and 921F3 also spanned the translocation breakpoint of case 2.
Cytogenetics and YAC characterization of the t(11;18). The YAC clones used in FISH analysis are indicated to the right of the ideogram of each chromosome. The breakpoint is indicated by an arrow; the YAC clones that yield split signals in both cases are in gray.
Cytogenetics and YAC characterization of the t(11;18). The YAC clones used in FISH analysis are indicated to the right of the ideogram of each chromosome. The breakpoint is indicated by an arrow; the YAC clones that yield split signals in both cases are in gray.
From the 9 YACs assigned to 18q21.1-22, 5 hybridized centromeric and 2 (including A153A6 containing the BCL2 gene) telomeric to the translocation breakpoint (see Fig 1). The hybridization signals of YACs 949B6 and 817C6 were split by the translocation in both cases. YAC 949B6 contains three ordered STSs (cen—D18S887—D18S1055—D18S1129—tel). FISH experiments with PAC clones isolated for these STSs positioned the chromosome 18 breakpoint in case 1 between D18S1055 and D18S1129. A walking strategy initiated from both markers led to the identification of PAC 205G9 and 152M5, which were shown to be split by the t(11;18) in both cases. The breakpoints were further narrowed down by FISH analysis withBamHI fragments subcloned from PAC 152M5 (Fig2A). In case 1, fragment H was split by the translocation, whereas in case 2 the breakpoint could be mapped to fragment D (Fig 3).
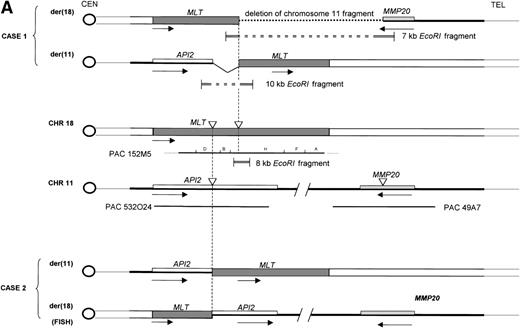
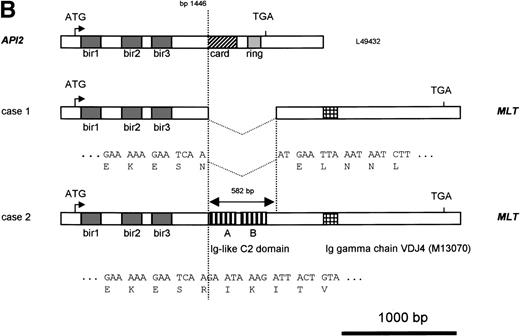
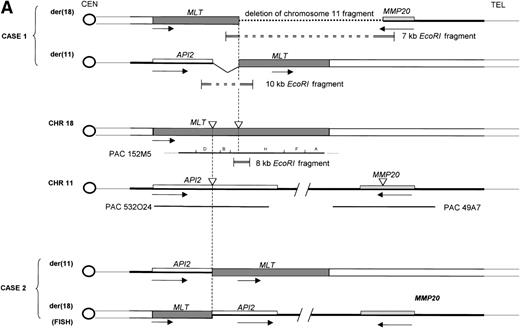
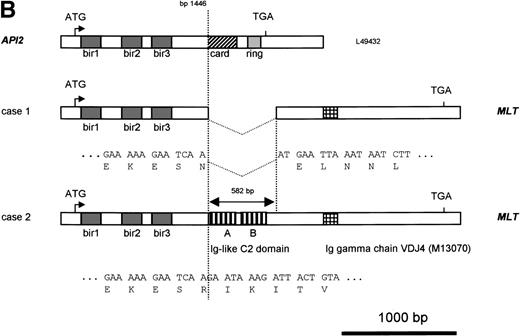
Molecular structure of the t(11;18). The genomic structure of the t(11;18) is shown in (A). In the center the MLT gene is shown as the darkly shaded area on the normal 18q; API2 andMMP20 are shown, respectively, as an open rectangle and a light gray rectangle on the normal 11q (not drawn to scale). Below each gene the PAC isolated for this gene is shown. For PAC 152M5 the position of the different BamHI fragments used for FISH experiments are indicated. On top the rearrangement in case 1 is illustrated: the der(11) fuses the 5′ end of API2 to the 3′ end of MLT,while on the der(18), as a result from the cryptic deletion of chromosome 11, the 5′ end of MLT is fused to the 5′ end ofMMP20. The transcriptional orientation of each gene is indicated by an arrow below each chromosome, showing that on the der(18) MLT and MMP20 do have an opposite transcriptional orientation. The genomic fusion fragments that were cloned from, respectively, the der(11) and the der(18) are indicated by the double lines. Below, the rearrangement of case 2 is shown: the der(11) fuses 5′ API2 to 3′ MLT. The breakpoint in API2 is identical to the one in case 1; the breakpoint in MLT occurred upstream of the breakpoint in case 1 (see 2B). FISH experiments suggest that the der(18) is the balanced reciprocal of the der(11). The localization of all breakpoints is indicated on the normal chromosomes by open triangles. (B) The structure of the different fusion cDNAs. On top the structure of API2 is shown with three aminoterminal BIR domains separated from the carboxyterminal RING domain by a CARD domain. TheAPI2 cDNA is truncated after the third BIR domain and fused in frame to MLT. As a result of the heterogeneity of the genomic breakpoints in case 2, 582 additional nucleotides, encoding two Ig-like C2 domains of MLT, are present in this fusion. An Ig gamma VDJ4-like sequence in MLT is shown by a cross-hatched box. The sequence and the translation of the different junction fragments is shown underneath each cDNA.
Molecular structure of the t(11;18). The genomic structure of the t(11;18) is shown in (A). In the center the MLT gene is shown as the darkly shaded area on the normal 18q; API2 andMMP20 are shown, respectively, as an open rectangle and a light gray rectangle on the normal 11q (not drawn to scale). Below each gene the PAC isolated for this gene is shown. For PAC 152M5 the position of the different BamHI fragments used for FISH experiments are indicated. On top the rearrangement in case 1 is illustrated: the der(11) fuses the 5′ end of API2 to the 3′ end of MLT,while on the der(18), as a result from the cryptic deletion of chromosome 11, the 5′ end of MLT is fused to the 5′ end ofMMP20. The transcriptional orientation of each gene is indicated by an arrow below each chromosome, showing that on the der(18) MLT and MMP20 do have an opposite transcriptional orientation. The genomic fusion fragments that were cloned from, respectively, the der(11) and the der(18) are indicated by the double lines. Below, the rearrangement of case 2 is shown: the der(11) fuses 5′ API2 to 3′ MLT. The breakpoint in API2 is identical to the one in case 1; the breakpoint in MLT occurred upstream of the breakpoint in case 1 (see 2B). FISH experiments suggest that the der(18) is the balanced reciprocal of the der(11). The localization of all breakpoints is indicated on the normal chromosomes by open triangles. (B) The structure of the different fusion cDNAs. On top the structure of API2 is shown with three aminoterminal BIR domains separated from the carboxyterminal RING domain by a CARD domain. TheAPI2 cDNA is truncated after the third BIR domain and fused in frame to MLT. As a result of the heterogeneity of the genomic breakpoints in case 2, 582 additional nucleotides, encoding two Ig-like C2 domains of MLT, are present in this fusion. An Ig gamma VDJ4-like sequence in MLT is shown by a cross-hatched box. The sequence and the translation of the different junction fragments is shown underneath each cDNA.
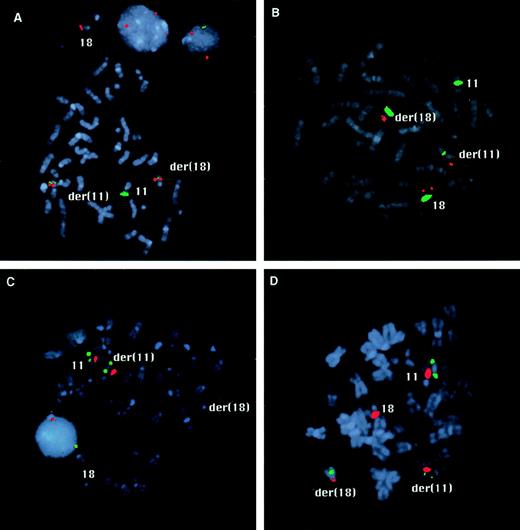
FISH mapping of the chromosome 11 and 18 breakpoints. (A) The hybridization signals of YAC 921F3 (green) and the BamHI fragment H of PAC 152M5 (red) are both split by the translocation in case 1. Signals of both probes are visible on the derivative chromosomes 11 and 18. (B) In case 2, fragment D of PAC 152M5 (red) shows split signals. The centromeric probes for chromosome 11 and 18 appear in green. (C) In case 1, PAC 532O24 (green) is seen on the normal chromosome 11 and on the derivative chromosome 11 (C), whereas this probe is split by the translocation in case 2 (D). The signals of the centromeric probes are shown in red.
FISH mapping of the chromosome 11 and 18 breakpoints. (A) The hybridization signals of YAC 921F3 (green) and the BamHI fragment H of PAC 152M5 (red) are both split by the translocation in case 1. Signals of both probes are visible on the derivative chromosomes 11 and 18. (B) In case 2, fragment D of PAC 152M5 (red) shows split signals. The centromeric probes for chromosome 11 and 18 appear in green. (C) In case 1, PAC 532O24 (green) is seen on the normal chromosome 11 and on the derivative chromosome 11 (C), whereas this probe is split by the translocation in case 2 (D). The signals of the centromeric probes are shown in red.
Cloning of the fusion genes.
To identify genes on chromosome 18q in the vicinity of the breakpoints, the sequences derived from short random subclones from theBamHI fragments D, B, H, and F were compared with the nucleotide databases. Subclone F24, mapping telomeric to the breakpoint, contained a 178-bp fragment identical to a single EST (IMAGE cDNA clone 1420842, GenBank Accession No. AA826328) that resembles a hypothetical Caenorhabditis elegans gene (F22D3.6, GenBank Accession No. U28993). The presence of canonical 5′ and 3′ splice sequences flanking the 178 bp suggested that this represented an exon of a human gene. A second exon with similarity to the same C elegans gene product was predicted by computer analysis of subclone B9, located centromerically to the breakpoint in case 1. RT-PCR experiments confirmed that both exons were part of the same human transcript and indicated the disruption of this gene (named MLTfor ALT-ymphoma associated ranslocation) by the translocation in case 1. Sequence analysis of these RT-PCR products and of 5′ and 3′ RACE products yielded a consensus MLT cDNA sequence of 2,491 bp (GenBank Accession No. AF130356). The first ATG start codon at bp 113 precedes an open reading frame of 2,184 bp, which encodes a protein of 729 amino acids. A transcript of approximately 3,000 bp was detected upon Northern analysis (data not shown), which suggests additional 5′ and/or 3′ MLT sequences.
To identify an eventual chromosome 11 fusion partner for MLT,cDNA transcribed from RNA of case 1 was then used in 5′ RACE experiments with two nested primers (MLTr1 and r2) derived fromMLT sequences telomeric to the breakpoint. The amplification products were cloned and eight clones with an average insert length of 800 bp were sequenced. Five clones contained uniquely MLTsequences. The three remaining clones showed a fusion of MLTsequences to the 5′ part of the API2 gene, an inhibitor of apoptosis mapped to chromosome 11q22. The API2 protein contains three copies of the BIR at its amino-terminus and a caspase recruitment domain or CARD27 followed by a carboxy-terminal zinc binding RING finger domain.28 The chimeric API2-MLTtranscript contains bp 1-1446 of API2 (GenBank Accession No.L49432) fused in frame with bp 786 of the MLT cDNA (GenBank Accession No. AF130356). At the protein level the first 441 amino acids (AA) of API2, containing the three BIR domains, are fused to the carboxy-terminal part of MLT (Fig 2B).
Primers derived from the API2 and MLT cDNA sequences (M&M) were then used to confirm this fusion directly by RT-PCR. An amplification product with the expected size (445 bp) and sequence was obtained for patient 1, confirming the existence of the chimericAPI2-MLT transcript. In contrast, using the same primers and cDNA from patient 2, a 1,000-bp RT-PCR product was obtained, with again an API2-MLT fusion with a continuous open reading frame. The breakpoint in the API2 gene occurred at the same position as described for patient 1. However, the chimeric cDNA contained an additional 582-bp MLT sequence in agreement with the more centromeric localization of the 18q breakpoint in this case as defined by FISH (Fig 2). The consensus cDNA sequences for bothAPI2/MLT fusions are shown in Fig4.
Sequence of the API2-MLT chimeric cDNA. The 5′API2 cDNA sequence (bp 1-1446 according to GenBank Accession No. L49432) is shown in italic, the additional 582 bp of MLT fused to API2 in case 2 are shown in bold. The inframe API2-MLT fusion generates at bp 1446-1448 an AAT (N) for case 1, an AGA (R) in case 2. The three BIR domains of API2 and the two Ig-like C2 domains (Ig-I C2) and the domain similar to a mouse Ig γ chain (VDJ4, GenBank Accession No.M13070) of MLT are underlined. The sequence has been deposited in GenBank under Accession No. AF123094.
Sequence of the API2-MLT chimeric cDNA. The 5′API2 cDNA sequence (bp 1-1446 according to GenBank Accession No. L49432) is shown in italic, the additional 582 bp of MLT fused to API2 in case 2 are shown in bold. The inframe API2-MLT fusion generates at bp 1446-1448 an AAT (N) for case 1, an AGA (R) in case 2. The three BIR domains of API2 and the two Ig-like C2 domains (Ig-I C2) and the domain similar to a mouse Ig γ chain (VDJ4, GenBank Accession No.M13070) of MLT are underlined. The sequence has been deposited in GenBank under Accession No. AF123094.
Absence of a reciprocal MLT-API2 transcript.
To analyze the genomic events leading to the expression of a chimericAPI2-MLT transcript, we cloned the genomic breakpoints of case 1. To this aim, an 8-kb EcoRI fragment spanning the breakpoint was subcloned from fragment H. Southern hybridization with the 5′- and the 3′-end-fragment of this clone detected rearranged EcoRI fragments of, respectively, 7 kb containing the 5′-end of MLTand 10 kb containing the 3′-end of MLT (Fig5B). Long-distance inverse PCR29 was used to amplify the genomic chromosome 11 sequences present in both chimeric fragments, and PAC clones corresponding to these chromosome 11 sequences were isolated. To our surprise, two independent sets of PAC clones were obtained. PAC 532O24 was isolated using chromosome 11 sequences derived from the der(11). This PAC was shown to contain the 5′ end of API2. FISH experiments with this PAC yielded signals on the normal 11 and the der(11) of case 1. PAC 49A7, obtained with chromosome 11 sequences derived from the der(18), however, did not contain API2, and sequencing showed that this clone contained the MMP20 gene instead (Fig 2). FISH experiments with this clone on case 1 resulted in fluorescent signals on the normal 11 and the der(18). Taken together, these data show that in case 1 the t(11;18) is associated with a cryptic deletion of chromosome 11 sequences distal to the breakpoint, resulting in the absence of an MLT-API2 fusion. These data are in agreement with a localization of the MMP gene cluster telomeric to API2. Because the exact distance is not known, we cannot estimate the size of the deletion. Sequencing of the der(18) fusion fragment showed that the MLT gene and the MMP20gene are on opposite strands of the genome, thereby excluding the expression of an MLT-MMP20 transcript. FISH experiments on case 2 detected a signal for the PAC 532O24 on the normal 11, the der(11), and the der(18) consistent with the occurrence of a balanced translocation and a breakpoint in API2. PAC 49A7 yielded fluorescent signals on the normal 11 and the der(18) consistent with the localization of the MMP20 gene distally to API2.
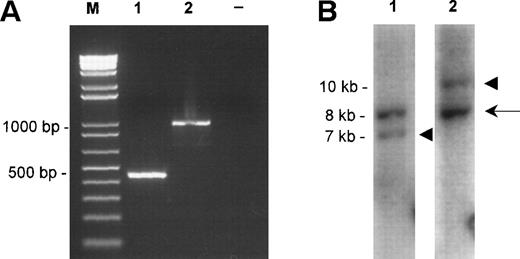
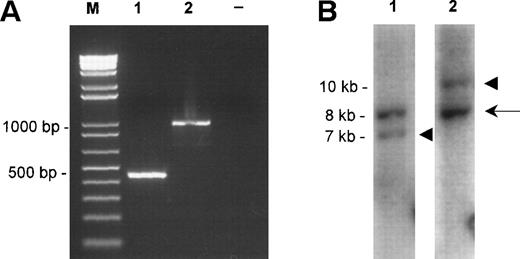
Molecular characterization of the fusions. (A) TheAPI2-MLT products obtained by RT-PCR from case 1 (lane 1) and case 2 (lane 2). Size markers (M) are shown on the left, the negative control (−) is shown on the right. (B) The Southern blot detecting the rearranged EcoRI fragments of case 1. The probes were derived from an 8-kb EcoRI clone from chromosome 18 spanning the breakpoint (see Fig 2A, center). Lane 1 shows hybridization with the probe derived proximally to the breakpoint, lane 2 shows hybridization with the probe derived distally to the breakpoint. The arrow shows the normal 8-kb EcoRI fragment, the arrowheads show the chimeric fragments of, respectively, 7 kb, derived from the der(18) and 10 kb, originating from the der(11).
Molecular characterization of the fusions. (A) TheAPI2-MLT products obtained by RT-PCR from case 1 (lane 1) and case 2 (lane 2). Size markers (M) are shown on the left, the negative control (−) is shown on the right. (B) The Southern blot detecting the rearranged EcoRI fragments of case 1. The probes were derived from an 8-kb EcoRI clone from chromosome 18 spanning the breakpoint (see Fig 2A, center). Lane 1 shows hybridization with the probe derived proximally to the breakpoint, lane 2 shows hybridization with the probe derived distally to the breakpoint. The arrow shows the normal 8-kb EcoRI fragment, the arrowheads show the chimeric fragments of, respectively, 7 kb, derived from the der(18) and 10 kb, originating from the der(11).
DISCUSSION
We show here that the t(11;18)(q21;q21) associated with extranodal marginal zone B-cell lymphomas of the MALT type results in the expression of a chimeric transcript fusing 5′-API2 on chromosome 11 to 3′-MLT on chromosome 18.
Several observations point to the API2-MLT fusion as the oncogenic lesion underlying the t(11;18). The chimeric cDNA was cloned from two independent tumors. In one case the genomic breakpoints were also cloned and the structure of both genes and the localization of the breakpoints is in agreement with the expression of the fusion transcript. The cryptic deletion of the 3′ part of API-2 in case 1 precludes the expression of a reciprocal MLT-API2 transcript in this case. As a result of the deletion, the 5′-end of the MLTgene is fused to the 5′-end of the MMP20 gene on the der(18). Because both genes are present on opposite strands of the DNA, noMLT-MMP20 transcript is expressed. Furthermore, FISH experiments with PACs for, respectively, MLT, API2, andMMP20 clearly suggest that in case 2 a balanced translocation occurred not involving a break in the MMP20 gene, further arguing against any significance of the MLT-MMP20 fusion.
API2 belongs to the family of inhibitor of apoptosis proteins (IAP), which play an evolutionary conserved role in regulating programmed cell death in diverse species.30 The IAPgenes were first identified in baculoviruses in which they demonstrated an ability to suppress the host cell apoptotic response to viral infection.31 Subsequently, five human IAP relatives have been described: NIAP, API1 (also known as cIAP1, HIAP2, MIHB), API2 (cIAP2, HIAP1, MIHC),XIAP-hILP, andsurvivin.19-21,32-35 The common structural features of all IAP family members is a motif termed BIR occurring in one to three copies, a caspase recruitment domain or CARD27 located between the BIR domain(s), and a carboxy-terminal zinc binding RING finger domain28 that is present in all IAPs with the exception of NIAP andsurvivin. The human API1 and API2 proteins were originally identified as proteins that are recruited to the cytosolic domain of the tumor necrosis factor (TNF) receptor II via their association with the TNF-associated factor (TRAF) proteins, TRAF-1 and TRAF-2,19 and have been subsequently shown to suppress different apoptotic pathways by inhibiting distinct caspases, such as caspase-3, caspase-7, and pro-caspase-9.33 36
The function of the novel MLT gene located on chromosome 18q21 is not yet known. Its closest homologue is a hypothetical C elegans gene. The carboxy-terminal part of this gene is characterized by the presence of two Ig-like C2-type domains and a domain similar to the murine Ig γ chain VDJ4 sequence (GenBank Accession No. M13070). The C2 domains are only present in the longer fusion cDNA of case 2 and, thus, probably have no functional significance in the tumor.
The molecular mechanism of action of the API2-MLT fusion remains to be elucidated. We hypothesize that the fusion protein resulting from the t(11;18) may lead to increased inhibition of apoptosis and, thereby, confer a survival advantage to MALT lymphomas and allow antigen-independent proliferation. Indeed, MALT lymphomas have been shown to display low levels of apoptosis37 and to escape from FAS-mediated apoptosis.38 The truncation ofAPI2 after the BIR domains could release their anti-apototic effects from regulation by the CARD and RING domains. Recent studies have shown that the BIR domain–containing regions of API1 andAPI2 are sufficient for inhibition of caspases and suppression of apoptosis.33 The BIR domains of one of theDrosophila homologues (DIAP1) were shown to suppress apoptosis in the Drosophila eye disk, whereas the full-length protein exhibited less activity. Moreover, transgenic flies overexpressing the RING domain alone exhibited increased cell death in the eye, suggesting that the RING domain may act as a negative regulator of cell death suppression in some instances.32 On the other hand, a specific role for the carboxy-terminal MLT domain is suggested by its consistent presence in the fusion and by the recurrency of the t(11;18) in MALT lymphoma. It is possible that the presence of theMLT domain would stabilize the fusion protein, increase its affinity for protein interaction, or influence its subcellular localization, thereby modulating its interactions with other proteins.
The mechanism of gene deregulation by the t(11;18) differs from that seen in most of the B-cell lymphoma–associated translocations, which involve one of the Ig loci on 14q32, 2p12, or 22q11 and lead to deregulated expression of the incoming oncogene due to the proximity of potent B-cell transcriptional enhancers within the Ig loci.39 In this case, the expression of the fusion gene is driven from the promoter of its 5′ partner, API2. The observation that API2 mRNA is highly expressed in adult lymphoid tissues, including spleen, thymus, and peripheral blood lymphocytes, and also in fetal lung and kidney, is in agreement with this.20
At the genomic level the rearrangements appear to be heterogeneous. The breakpoint in MLT occurred in two different introns for both cases. In the API2 gene the breakpoint occurred in the same intron for both cases but it was associated with the deletion of the 3′-end of the gene in only one tumor. The cytogenetic analysis of MALT lymphoma is often hampered by their poor proliferation in vitro. However, we anticipate that the physical maps and the genomic clones generated by this work will allow the development of sensitive interphase FISH assays for this rearrangement. Alternatively, the fusion mRNA or the fusion protein provide new molecular targets for diagnosis. The identification and characterization ofAPI2-MLT–positive neoplasms should indicate whether they represent a distinct clinicopathological subentity.
ACKNOWLEDGMENT
We are grateful to Dr G. Verhoef (Department of Hematology, University of Leuver) and Dr M. Gonzales and Dr T. Flores (Department of Hematology, University of Salamanca) for providing clinical data. We thank Riet Somers and Anja Steyls for expert technical assistance.
J.D. and M.B. contributed equally to the study and should both be regarded as first authors.
Supported by Grants No. G.0153.96 and G.0377.97 of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (F.W.O.) (P.M.; A.H.) and Grant No. 70-2175Dil from the Deutsche Krebshilfe/Dr Mildred Scheel Stiftung für Krebsforschung (J.D.). P.M. is an ‘onderzoeksdirecteur’ and M.B. a ‘Postdoctoraal onderzoeker’ of the ‘Fonds voor Wetenschappelijk Onderzoek—Vlaanderen.’
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Peter Marynen, PhD, Human Genome Laboratory, Center for Human Genetics and Flanders Interuniversity Institute for Biotechnology, Herestraat 49, B-3000 Leuven, Belgium; e-mail: Peter.Marynen@med.KULeuven.ac.be.