GENETIC CHANGES involving oncogenes and tumor suppressor genes contribute to the deregulated expansion of malignant cells. While some of these changes result in increased proliferation, others contribute to an increase in cell numbers by inhibiting apoptosis (programmed cell death).1 Because cytotoxic drugs or irradiation result in cell killing by apoptosis, the genetic changes underlying malignancy often reduce the ability of these agents to destroy malignant cells.1 2 The elucidation of the pathways involved in the regulation of apoptosis in normal and malignant hematopoietic cells is therefore likely to contribute to the development of improved therepeutic stategies in the treatment of leukemia and lymphoma. This review first summarizes recent advances in the understanding of the control of apoptosis. Examples of how this control is altered in leukemic cells is then described.
1. MORPHOLOGICAL AND BIOCHEMICAL FEATURES OF APOPTOSIS
Apoptosis is a tightly regulated form of physiological cell death which is dependent on the expression of cell-intrinsic suicide machinery.3 Prominent morphological changes include cell shrinkage, condensation of the nuclear chromatin, fragmentation of the nucleus, and cleavage of chromosomal DNA at internucleosomal sites, resulting in the generation of a characteristic ladder pattern of DNA fragments on electrophoresis. Blebbing of the cell surface results in the release of membrane-bound apoptotic bodies.3Phosphatidylserine, which is normally located on the inner face of the plasma membrane, becomes exposed on the outer surface and provides a recognition signal for engulfment by phagocytes.4 5 Thus, apoptosis results in the rapid and efficient removal of superfluous or damaged cells.
2. GENETIC STUDIES IN CAENORHABDITIS ELEGANS PROVIDE A FRAMEWORK FOR UNDERSTANDING PATHWAYS OF APOPTOSIS REGULATION
Genetic studies in the nematode C elegans have resulted in the identification of a set of genes involved in the regulation of apoptosis.6 The ced-3 gene encodes a cysteine protease, which is homologous to members of the caspase protease family that execute the apoptotic program in mammalian cells (see section 3.1). The ced-4 gene product is required for the activation of CED-3. This activation step is blocked by the CED-9 protein, which is homologous to mammalian BCL-2. BCL-2 can substitute for CED-9 in blocking apoptosis in C elegans7,8 whereas overexpression of CED-4 induces apoptosis in mammalian cells,9 suggesting a high degree of conservation of the mechanisms of apoptosis regulation. Therefore, the C elegansmodel has been of value in the identification of the proteins that control apoptosis in human cells (see sections 3 and 4).
3. SIGNAL TRANSDUCTION PROCESSES IN THE REGULATION OF APOPTOSIS
The induction of apoptosis may conveniently be divided into three stages: (1) the interaction of the inducing signal with the cell, (2) biochemical transduction of the death signal, and (3) the execution of apoptosis. Because different extracellular signals and signal transduction pathways converge on a final common pathway during the execution phase, this terminal stage of apoptosis will be summarized first.
3.1. The caspase family of proteases mediates the terminal stages of apoptotic cell death.
The terminal stages of apoptosis involve the activation of a related family of proteases, the caspases.10,11 These enzymes possess an essential cysteine residue within their active sites and cleave substrates adjacent to aspartate residues. The cDNAs encoding 10 caspases have been cloned.10
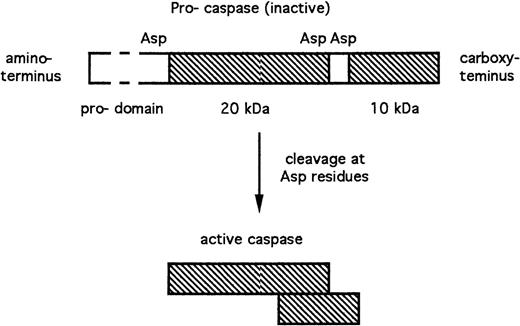
Caspases are expressed as inactive pro-enzymes. Cleavage of these pro-caspases adjacent to aspartate residues results in the generation of active subunits of approximately 10 and 20 kD (Fig 1). These subunits dimerize, with the resultant generation of a complete active site.10 The N-terminal pro-domains (Fig 1) of some pro-caspases, which are removed during activation, nevertheless play important roles in mediating regulatory interactions between caspases and other proteins. The requirement for cleavage adjacent to aspartate residues during caspase activation, together with the aspartate specificity of caspases, raises the possibility that cascades of caspase activation events may be involved in apoptosis regulation (see sections 3.3 and 4.3).
Structure and activation of caspases. The peptide sequences that contribute to the enzymatically active caspase are shown in cross-hatch. Asp, aspartate residues at which the inactive pro-caspase is proteolytically cleaved with the resultant generation of the active subunits.
Structure and activation of caspases. The peptide sequences that contribute to the enzymatically active caspase are shown in cross-hatch. Asp, aspartate residues at which the inactive pro-caspase is proteolytically cleaved with the resultant generation of the active subunits.
Caspases 3, 6, and 7 are terminal members of caspase cascades and recognize critical cellular substrates, whose cleavage contributes to the morphological and functional changes associated with apoptosis.10 Caspase 3 substrates include poly (ADP-ribose) polymerase,12,13 an enzyme involved in regulation of DNA repair and gelsolin, a cytoskeletal protein.14 Caspase 3 activation also results in DNA cleavage via inactivation of an inhibitor of DNA fragmentation factor, the endonuclease responsible for internucleosomal cleavage of chromatin.15 Caspase 6 substrates include the nuclear structural protein, lamin.16Thus, the cleavage of a relatively restricted set of critical caspase substrates contributes to the apoptotic demise of cells via disassembly of structural components, cleavage of the genetic material, and prevention of DNA repair.
3.2. Specific protease inhibitors block cell death by targetting terminal caspases.
In vitro studies suggest that the inhibitor of apoptosis (IAP) family of proteins may modulate cell death via abrogation of caspase activity.
These proteins are similar to the baculovirus-encoded caspase inhibitor p35. IAP1 and 2, XIAP (X-linked IAP) and survivin contain one to three BIR (baculovirus IAP repeat) motifs that are essential to their function.17 IAPs 1 and 2 and XIAP specifically target caspases 3 and 7, which function at the distal end of proteolytic cascades. Therefore, IAP expression may serve to reprieve cells otherwise committed to apoptotic death. However, the Drosophila IAPs (DIAP-1 and DIAP-2) block apoptosis by direct binding via the BIR motifs to noncaspase death-inducing proteins encoded by thereaper, hid, and grim genes.18Therefore, it is possible that the mammalian IAPs may also inhibit cell death by mechanisms other than binding to caspases.
The key question regarding cell death regulation concerns the mechanisms by which caspase activation steps are triggered by apoptotic signals. Different extracellular signals interact with the caspase system in different ways.
3.3. Ligation of FAS or the tumor necrosis factor (TNF) receptor results in the direct activation of caspases.
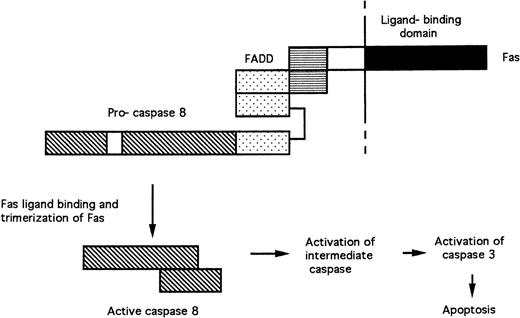
FAS and the TNF receptor are structurally related transmembrane receptor proteins. Their extracellular domains bind FAS ligand and TNF, respectively, resulting in the formation of receptor trimers. The cytoplasmic domain of FAS contains a “death domain” (Fig 2), whose elimination results in the abrogation of cell killing.19 The death domain of FAS recruits the death domain of the FADD (Fas-associated death domain) protein following receptor trimerization.20 FADD also contains a death-effector domain, which mediates interaction with similar amino acid sequences in the pro-domain of pro-caspase 8.21 22 Trimerization of the FAS/FADD/pro-caspase 8 complex after ligand binding results in the cleavage of the pro-caspase and generation of active caspase 8. Caspase 8 then cleaves pro-caspase 3, probably via activation of an unidentified intermediate caspase (Fig 2).
Signal transduction by FAS. Protein-protein interactions between the death domains (horizontal stripes) of FAS and FADD and the death-effector domains (stippled) of FADD and pro-caspase 8 are shown. Trimerization of receptor complexes after binding of FAS to FAS ligand results in the cleavage of pro-caspase 8 and the release of active caspase 8.
Signal transduction by FAS. Protein-protein interactions between the death domains (horizontal stripes) of FAS and FADD and the death-effector domains (stippled) of FADD and pro-caspase 8 are shown. Trimerization of receptor complexes after binding of FAS to FAS ligand results in the cleavage of pro-caspase 8 and the release of active caspase 8.
Activation of apoptosis after TNF receptor ligation follows a similar pattern. However, the TNF receptor does not bind FADD directly, but does so via a linking protein, TRADD (TNF receptor-associated death domain).19
3.4. Apoptosis induction by the perforin/granzyme system.
Killing of target cells by cytotoxic T lymphocytes plays a major role in defense against malignant and virus-infected cells, and contributes to transplant rejection and autoimmune disease. Killing is preceded by the release of the contents of cytotoxic T-cell granules, which contain perforin and the serine proteases granzymes A and B. Perforin forms a pore in the plasma membrane of the target cell, thereby allowing entry of granzyme B into the cytosol.23 Granzyme B cleaves and activates caspase 3 in cell-free systems.24,25 However, the primary target of granzyme B in intact cells is likely to be caspase 10, whose activation results in the subsequent activation of caspase 3.26 In cell-free systems, the addition of granzyme B initiates cleavage of several apoptosis-specific substrates and also induces chromatin condensation. Abrogation of these events by selective inhibitors suggests that activation of caspase 3 (and possibly of caspase 7) may be important mediators of apoptosis induction by cytotoxic T-cell–derived granzyme B.26 However, genetic studies have shown that the granules of cytotoxic T cells contain additional cytotoxic components in addition to perforin and granzymes A and B.27
4. THE BCL-2 PROTEIN FAMILY
4.1. The BCL-2 protein family plays a central role in the regulation of apoptosis.
The 26-kD BCL-2 protein protects cells from the induction of apoptosis by diverse stimuli, including the withdrawal of survival factors, heat shock, and treatment with DNA damaging agents.28-30 BCL-2 is the prototype of a family of related proteins. Other anti-apoptotic family members include BCL-XL, BCL-w, MCL-1, and A1. In contrast, the BAX, BAK, and BAD proteins are examples of pro-apoptotic BCL-2 family members whose overexpression promotes cell killing.30 The conserved BH1 (BCL-2 homology 1) and BH2 domains of the anti-apoptotic proteins form a hydrophobic cleft which binds the BH3 domains of pro-apoptotic family members, at least in vitro.31 32
The susceptibility of cells to apoptosis is determined in part by the relative concentrations of pro- and anti-apoptotic BCL-2 family members. The antagonistic actions of these two groups of proteins have been attributed to their ability to form heterodimers.33However, at least some of the dimerization properties of the BCL-2 family may be artefacts induced by detergents in vitro.34Furthermore, genetic studies suggest that BCL-2 and BAX function independently of one another in the regulation of apoptosis.35 Deletion of the BH4 domain of BCL-2 impairs its ability to block apoptosis without affecting its dimerization with anti-apoptotic family members.9 Deletion of the BH3 domain of BAX, which is required for its dimerization, does not impair the ability of the protein to increase the sensitivity of cells to cytotoxic agents.36 Therefore, it is plausible that the actions of pro- and anti-apoptotic members of the BCL-2 family may determine the sensitivity of cells to apoptosis induction via binding to a common target rather than to dimer formation.35 36
BCL-2 targets to the outer mitochondrial membrane, the nuclear envelope, and the endoplasmic reticulum via its C-terminal hydrophobic domain.30,37,38 However, some studies suggest that BAX shows a largely diffuse sub-cellular localization, translocating rapidly to the mitochondria (and possibly other organelles) after the induction of an apoptotic signal.39
The BCL-2 family regulates apoptosis induction via control of the activation of caspases, apparently by a mechanism involving the release of mitochondrial cytochrome c.11 However, it is unclear whether cytochrome c release is a component of the primary apoptosis induction pathway or a means of amplifying the death signal, as summarized later (see sections 4.3 and 4.4). The mechanism of cytochrome c–dependent caspase activation is discussed next.
4.2. Cytochrome c triggers caspase 3 cleavage via activation of caspase 9.
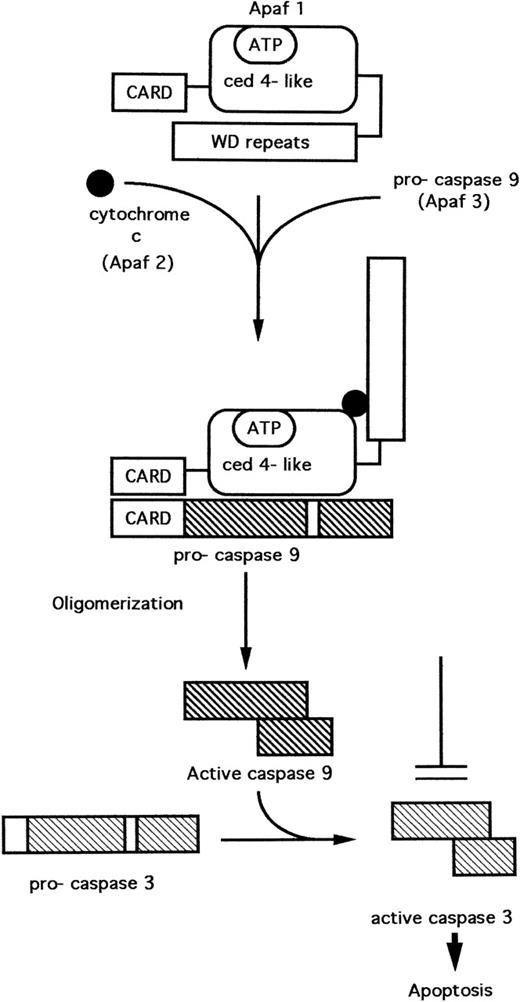
Three proteins have been purified from the cytosol of HeLa cells which, when recombined in the presence of adenosine triphosphate (ATP) (or deoxy ATP), were necessary and sufficient for the cleavage and activation of pro-caspase 3. These proteins, originally designated as Apaf 1, 2, and 3 (Apaf = apoptoticprotease activating factor) have been characterized.40,41 Apaf 1 contains a central domain with homology to C elegans CED-4.40 The amino-terminal domain of Apaf 1 is homologous to the CARDs (caspaserecruitment domains) of some caspases (Fig 3). The carboxy terminus consists of several WD repeats, which mediate interactions between certain regulatory proteins. Apaf 2 was found to be identical to cytochrome c,40 while Apaf 3 is identical to caspase 9.41
A model11,41,42 for the cytochrome c–dependent activation of caspase 3 is depicted in Fig 3. In the absence of cytochrome c, the WD repeat domain prevents interaction of Apaf-1 with pro-caspase 9. Cytochrome c binds Apaf 1, inducing a conformational change that results in the interaction of Apaf 1 and caspase 9, mediated by the CARD pro-domains present on both these proteins. Apaf-1 induces the cleavage and activation of pro-caspase 9 via a mechanism involving oligomerization of Apaf-1, thus facilitating autocatalytic cleavage of the pro-caspase.42 Activated caspase 9 now cleaves and activates caspase 3.
4.3. The BCL-2 protein family apparently regulates the release of cytochrome c from mitochondria.
Cytochrome c is released from mitochondria during apoptosis induced by diverse stimuli.11 Overexpression of BCL-2 or BCL-XL inhibit cytochrome c release induced by etoposide, actinomycin D, oxidative stress, Fas ligation, or interleukin-3 (IL-3) withdrawal.43-45 The BAX protein, on the other hand, triggers redistribution of cytochrome c in the absence of apoptotic stimuli.46 Thus, BCL-2 and BCL-XL may prevent apoposis by inhibiting cytochrome c release while BAX favors cell death by promoting its relocation to the cytosol. However, it is unclear whether these actions of the BCL-2 family result from the direct actions of these proteins on mitochondria, which then initiate caspase activation or whether cytochrome c release is secondary to BCL-2 family-regulated caspase activation and plays a subsequent role in amplification of the apoptotic signal.47
Some evidence suggests a direct role for the BCL-2 family in regulating cytochrome c release. The structure of BCL-XL resembles that of pore-forming bacterial toxins.31 BCL-2, BCL-XL, and BAX form ion channels in vitro.48-50 Because pores formed by BAX protein may show high conductance values under some conditions,50 it is possible that these channels allow the exit of cytochrome c. Anti-apoptotic proteins including BCL-2 may block release by interfering with pore formation by BAX.50 However, channel formation by BCL-2 family proteins has only been shown in synthetic membranes and, in some cases, at nonphysiological pH.48-50Therefore, it has not been established that these proteins can form channels in cellular membranes under physiological conditions.
An alternative hypothesis suggests that BCL-2 family proteins regulate the electrical potential gradient (Δψm) across the inner mitochondrial membrane and thereby regulate mitochodrial volume. The opening of “megachannels” in the inner membrane allows the passage of molecules of less than 1.5 kD, resulting in the dissipation of Δψm. Apoptosis induced by stimuli including antineoplastic drugs and glucocorticoids is apparently preceded by disruption of the gradient. Both the loss of Δψm and subsequent apoptosis induction are blocked by the megachannel antagonist bongkrekic acid or by overexpression of BCL-2 or BCL-XL.51,52 Therefore, changes in the permeability of the inner mitochondrial membrane may be a central coordinating event in the induction of apoptosis, which is inhibited by anti-apoptotic BCL-2 family members.51 The reported ability of BCL-2 to modulate ion fluxes across the inner mitochondrial membrane53 is compatible with this hypothesis. The precise mechanistic relationship between loss of Δψm and cytochrome c release has not been established. However, because loss of Δψm results in mitochondrial swelling, Δψm loss may cause cytochrome c release as a result of outer membrane rupture.45
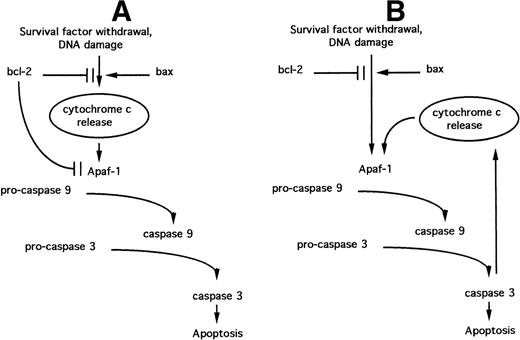
It is now apparent that the BCL-2 family regulates steps in apoptotic signaling distal to cytochrome c release. BCL-2 overexpression blocks apoptosis in cells containing high concentrations of cytosolic cytochrome c, induced either by transient expression of BAX54 or by direct microinjection.55 These observations may be accounted for by the direct action of some anti-apoptotic BCL-2 family members on caspase activation, because BCL-XL can bind Apaf-1 directly and block its ability to activate pro-caspase 9.56 Such a function for BCL-2 family proteins parallels the role of the C elegans BCL-2 homologue CED-9.57 A model that combines the putative ability of BCL-2 family members to regulate both cytochrome c release and the Apaf-1–mediated activation of pro-caspase 9 (the “Swiss army knife” model47) is depicted in Fig 4A.
Models relating the roles of BCL-2 family proteins, cytochrome c release, and Apaf-1–dependent caspase activation during the induction of apoptosis. (A) “Swiss army knife” model; (B) “death cycle” model. See text for details.
Models relating the roles of BCL-2 family proteins, cytochrome c release, and Apaf-1–dependent caspase activation during the induction of apoptosis. (A) “Swiss army knife” model; (B) “death cycle” model. See text for details.
4.4. Caspase action on mitochondria amplifies the initial apoptotic signal via a positive feedback loop.
Caspases can themselves trigger cytochrome c release, because selective inhibitors of these proteases can abrogate release in response to some stimuli.45 Furthermore, death signals including ligation of Fas, which directly activate caspases (section 4.2), may nevertheless be amplified via caspase-mediated cytochrome c release.45Recombinant caspases disrupt Δψm and induce cytochrome c release when added to isolated mitochondria.58 The mechanism of this action of caspases is unclear.
The operation of a positive feedback loop raises the possibility that anti-apoptotic BCL-2 family members may not play a direct role in the modulation of cytochrome c release. The ability of these proteins to directly inhibit Apaf-1 function56 could instead modulate cytochrome c release indirectly via the caspase-dependent feedback loop. This “death cycle” model47 implies that cytochrome c release is not a component of the apoptosis-initiating pathway but serves in the amplification of an initial signal generated via the regulation of Apaf-1 (Fig 4B).
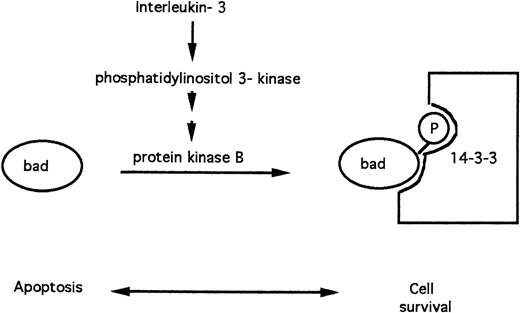
4.5. Survival factors regulate apoptosis via phosphorylation of the BAD protein.
The BAD protein is a pro-apoptotic member of the BCL-2 family.30,59 FL5.12 lymphoid cells depend on IL-3 for their survival in vitro. In cells cultured in the presence of IL-3, the BAD protein is phosphorylated on serine residues. Phosphorylated BAD is sequestered via binding to the 14-3-3 protein and is, therefore, unable to promote apoptosis.59 The BAD protein rapidly becomes dephosphorylated in the absence of IL-3, dissociates from 14-3-3, and triggers apoptosis (Fig 5).
Apoptosis modulation by IL-3 via phosphorylation of the pro-apoptotic BAD protein.
Apoptosis modulation by IL-3 via phosphorylation of the pro-apoptotic BAD protein.
Recent evidence has implicated protein kinase B as the protein kinase that mediates BAD phosphorylation in response to IL-360 and other survival factors61 (Fig 4). IL-3 triggers activation of phosphatidylinositol 3-kinase, a lipid kinase that initiates the eventual generation of phosphatidylinositol 3,4-bisphosphate, an allosteric activator of protein kinase B.62
5. THE p53 PROTEIN
The p53 protein plays an important role in the coupling of DNA damage to cell-cycle arrest and to the induction of apoptosis. In cells with undamaged DNA, p53 protein levels are maintained at a low level as a result of rapid turnover. An increase in stability after the induction of DNA damage results in an increased level of p53.63 The protein product of the ataxia telangiectasia (atm) gene participates in a pathway that links the detection of DNA damage to the upregulation of p53.64 However, the carboxy terminus of the p53 protein itself can bind to damaged DNA,65 suggesting that both p53 and the putative damage detector may colocalize at the site of DNA damage. The radiation resistance of thymocytes derived from p53 “knockout” mice when compared with wild-type thymocytes66,67 emphasizes the importance of p53-dependent mechanisms in the induction of apoptosis after DNA damage induction. Upregulation of p53 also results in cell-cycle arrest. The pathways involved in this facet of p53 action have recently been reviewed.63
5.1. Transcriptional activation by p53.
A tetramer of p53 molecules functions as a transcription factor that binds to consensus sequences in the 5′ untranslated regions of specific target genes.63 The upstream region of the BAX gene contains p53 consensus binding sites.68 Enforced p53 expression augments BAX expression, which is followed by apoptosis induction.69 Genetic studies on apoptosis induction in adriamycin-treated mouse fibroblasts suggest that BAX is an important (but not the only) effector of p53-mediated apoptosis.70However, thymocytes isolated from p53 “knockout” mice expressing elevated levels of BAX are as resistant to etoposide-induced apoptosis as are thymocytes from p53 “knockout” mice expressing normal levels of bax.71 Therefore, mechanisms other than BAX induction may mediate p53-dependent apoptosis, at least in some cell types.
Polyak et al72 have identified 12 mRNA species which were rapidly induced after adenovirus-mediated transfer of the p53 gene into p53 null colorectal carcinoma cells. Thesep53-induced genes (PIGs) either encode proteins that catalyze redox reactions and consequently generate reactive oxygen species (ROS) or whose expression is augmented by ROS generation. Indeed, introduction of p53 into p53 null cells results in a burst of ROS generation that is followed by apoptosis induction. Inhibitors of ROS generation do not interfere with induction of PIG genes by p53 but do abrogate apoptosis induction, suggesting that the upregulated expression of PIG genes and the subsequent generation of ROS play a critical role in the induction of apoptosis by p53 at least in some cell types.72 ROS can themselves trigger cytochrome c release from mitochondria,44 possibly via modulation of ion transport within these organelles,53 73 suggesting an additional mechanism for the induction of apoptosis by p53.
Transient expression of p53 results in ROS generation in cells that are susceptible to p53-mediated apoptosis but not in resistant cells, compatible with the hypothesis that ROS are downstream mediators of p53-induced apoptosis.74 However, the role of ROS in the regulation of apoptosis remains controversial, because in some studies the induction of cell death is not abrogated at very low oxygen tension.75 76
5.2. Transcriptional repression by p53.
In addition to its transactivating properties, p53 represses transcription from several promoters that lack p53 binding sites. BCL-2 can relieve this transcriptional repression and also protect cells from apoptosis, suggesting that inhibition of transcription of specific but as yet unidentified genes may contribute to the ability of p53 to induce apoptosis.77 However, p53 is also able to induce apoptosis via pathways that are not dependent on the regulation of gene expression.78 Therefore, induction of apoptosis by p53 can occur by diverse pathways depending on the cellular context. It is also clear that some apoptotic pathways do not involve p53. For example, thymocytes from p53 knockout mice are resistant to etoposide and radiation but not to glucocorticoids.67 Furthermore, HL60 cells, which have lost both p53 alleles, are extremely sensitive to apoptosis induction by drugs that induce DNA strand-breaks.79 The p53 dependence of apoptotic pathways is also tissue dependent, because radiation-induced apoptosis is compromised in the thymus of p53 knockout mice, but not in the lung.80
5.3. p53 loss results in resistance to cytotoxic regimes.
When transplanted into imunodeficient mice, fibrosarcomas expressing functional p53 show a high proportion of apoptotic cells and regress after treatment with adriamycin or γ radiation. In contrast, tumors lacking p53 show few apoptoses and are resistant to adriamyin or radiation.81 Therefore, inactivation of p53 can result in the resistance of tumors to DNA damaging agents. The elevation of BAX expression in response to radiation is only detected in human leukemia cell lines that express p53 and that die by apoptosis in response to DNA damage. By contrast, p53-negative lines do not elevate BAX levels and are also resistant to radiation-induced apoptosis.82Therefore, p53-mediated elevation of BAX contributes to the killing of some tumor cells by cytotoxic regimes.77
5.4. The p53-related p73 gene product.
The p73 gene encodes a protein that is closely related to p53.83 Overexpresion of p73 results in the induction of some genes that are also targets of p53, and also induces apoptosis. However, p73 expression is apparently not augmented after the induction of DNA damage.83 Therefore, there is at present no evidence implicating p73 in DNA damage-induced cell killing.
6. THE PHYSIOLOGICAL ROLES OF APOPTOSIS IN THE HEMATOPOIETIC AND LYMPHOID SYSTEMS
During hematopoiesis, the survival of progenitor cells is regulated both positively and negatively by a complex, interacting network of cytokines and adhesion molecules.84 Noncycling primitive CD34+ human hematopoietic progenitors require the continuous presence of IL-3 or granulocyte-macrophage colony-stimulating factor (GM-CSF) for survival in vitro. In contrast, other cytokines including IL-6 and IL-11 trigger proliferation of these progenitors.85 Stem cell factor, Flt ligand, and IL-3 suppress apoptosis in single-cell assays designed to test the direct actions of cytokines on primitive progenitors. Thrombopoietin is more effective in preventing apoptosis than any of these cytokines.86 Cytokines show target cell selectivity in preventing apoptosis. For example, stem cell factor selectively promotes survival of primitive hematopoietic cells, whereas IL-3 blocks cell death in more committed progenitors.87 Flt ligand is selective for progenitors committed to the myeloid lineage.88
Other cytokines promote the apoptotic death of both primitive and committed progenitors.84 The flt3 ligand-mediated survival of primitive progenitors is counteracted by both transforming growth factor-β (TGF-β) and TNF-α.89 Interferon-γ (IFN-γ) suppresses the survival of long-term culture-initiating cells. The action of IFN-γ is more potent when this cytokine is secreted by stromal cells in culture than when added to the medium, stressing the importance of the hematopoietic microenvironement in modulating survival.90 Induction of apoptosis by both IFN-γ and TNF-α may be mediated in part by increasing expression of FAS on the surface of hematopoietic progenitors.91Subsequent ligation of this death receptor then triggers cell killing.
Primitive bone marrow B-lymphoid progenitors require direct contact with bone marrow stromal cells for survival.92 These survival-promoting interactions are dependent on interactions between the β1 integrins VLA-4 and VLA-5 expressed on the B-cell surface and fibronectin generated by the fibroblasts.93 It is unclear whether interactions between these adhesion molecules directly generate survival signals or whether the close juxtaposition of the lymphoid progenitors to fibroblasts enhances the actions of unidentified survival factor generated by the fibroblasts.
Apoptotic death of progenitor cells following deprivation of survival factors is an active rather than a passive process. Hematopoietic cells from p53 “knockout” mice are more resistant to the induction of apoptosis after factor withdrawal than are corresponding cells from control animals.94 Murine 32Dc13 myeloid precursor cells depend on IL-3 for survival in vitro. Withdrawal of IL-3 results in apoptotic death, which is dependent on the expression of wild-type p53,95 suggesting that the activation of a cell-intrinsic pathway involving p53 is a prerequisite for cell killing after removal of survival factors.
Cytokines modulate both the basal survival of some leukemia cell lines and also compromise their killing by cytotoxic treatments.84 G-CSF, GM-CSF, IL-3, IL-6, or IFN-γ protect murine myeloid leukemia cell lines from apoptotic death induced by cytotoxic drugs.96,97 Apoptosis induced by the introduction of wild-type p53 into a p53-negative murine AML cell line is abrogated by IL-6,98 suggesting that cell killing on deprivation of this cytokine proceeds via a p53-mediated pathway.
The generation of the recognition repertoires of T and B lymphocytes is dependent on the apoptotic deletion of cells with inappropriate specificities.99 Killing of cells after ligation of FAS or the receptor proceeds via induction of apoptosis.19 Activation of T lymphocytes after encounters with cognate antigen/major histocompatibility complexes (MHC) results in activation and concomitant upregulation of FAS ligand. Subsequent interactions between FAS ligand and FAS induces apoptotic death of the activated T cells, thereby downregulating the immune response. Elimination of autoreactive B lymphocytes is also mediated by the FAS system.19 Triggerring of specific cytotoxic T cells by viral antigens displayed at the surface of target cells induces expression of FAS ligand. Interaction of the ligand with FAS expressed by the target cell initiates apoptosis.19 Cytotoxic T lymphocytes also kill target cells via the perforin/granzyme system (section 3.4).
7. THE INFLUENCE OF GENETIC AND MICROENVIRONMENTAL FACTORS ON APOPTOSIS INDUCTION IN LEUKEMIA CELLS
Treatment of leukemia cell lines with cytotoxic drugs results in the release of cytochrome c43-45 and the activation of caspases.100,101 Caspases are also activated after cytotoxic treatment of freshly isolated B chronic lymphocytic leukaemia (B-CLL) cells.102 However, the mechanisms that couple DNA damage to more downstream regulatory events are largely unclear. Evidence of a largely circumstantial nature suggests that some of the mechanisms of apoptosis control described in sections 3, 4, and 5 are deregulated in leukemia cells, thus contributing to their abnormal expansion and, in some cases, to drug and radiation resistance. Deregulation of apoptosis results from translocations involving genes that encode cell death–regulating proteins. However, microenvironmental factors also impinge on both the basal survival of leukemia cells and their killing by cytotoxic regimes. Some of this evidence is summarized next.
8. ALTERED EXPRESSION OF BCL-2 IN LEUKEMIA AND LYMPHOMA
8.1. Translocation of the BCL-2 gene in non-Hodgkin’s lymphoma (NHL).
The t(14;18) chromosomal translocation associated with NHL results in the juxtaposition of the BCL-2 gene to the Ig heavy chain (IgH) locus.103 Translocation results in enhanced levels of BCL-2 mRNA, which may be partially attributable to the presence of a powerful transcriptional enhancer in the IgH locus.98 The efficiency of splicing of BCL-2 exons is also increased as a result of their fusion to Ig gene introns in t(14;18) cells. The resulting increase in cellular levels of spliced BCL-2 open reading frames also contributes to the upregulation of BCL-2 protein levels in NHL cells.104 Transgenic mice carrying a BCL-2 gene expressed via the IgH gene enhancer overexpress BCL-2 specifically in B-lymphoid cells. These mice accumulate abnormal numbers of small, nonproliferating B cells that show extended survival in vitro.105,106 BCL-2 overexpression alone is insufficient for lymphomagenesis. However, doubly transgenic mice in which overexpression of both BCL-2 and c-MYC is targetted to B-lymphoid cells rapidly develop tumors originating from primitive lymphoid-committed lymphoid cells, suggesting that a second genetic event is necessary for the malignant transformation of lymphocytes overexpressing BCL-2.106
Enforced overexpression of the BCL-2 or BCL-XL gene in leukemia cell lines confers increased resistance to cytotoxic drugs.107-109 However, low-grade NHL patients with increased BCL-2 expression respond well to chemotherapy, although complete remissions are rare.110 Although it is likely that overexpression of BCL-2 impairs apoptosis induction in follicular lymphoma cells, additional microenvironemental signals are required to maintain cell viability. NHL cells remain viable for 1 to 2 days in culture and then die rapidly.111 Cell death is preceded by the downregulation of BCL-XL expression, although BCL-2 expression is maintained. Both the decrease in BCL-XL and cell death are prevented by ligation of CD40, suggesting that continuous signaling by this cell-surface molecule is required to maintain viability of the lymphoma cells via upregulation of BCL-XL expression.111 BCL-2 antisense oligodeoxynucleotides have been used in the treatment of nine patients with relapsed NHL. A reduction in tumor mass was observed in two patients and a decrease in circulating tumor cells in two others. In two of five samples that were studied by flow cytometry, a decrease in BCL-2 protein levels was detected after treatment.112
NHL cells express variable levels of cell-surface FAS, but are resistant to killing after FAS ligation. Therefore, loss of sensitivity to this apoptotic pathway may contribute to the expansion of the lymphoma cells by allowing their escape from normal immune regulatory mechanisms.113
8.2. BCL-2 expression in CLL.
CLL cells show an extended life span in vivo. They proliferate very slowly, suggesting that a failure to die by apoptosis contributes to the accumulation of malignant cells in this disease.114Translocations of the BCL-2 gene to Ig loci are detected in less than 2% of CLL cases.115 Nevertheless, CLL cells from some patients express high levels of BCL-2 protein compared with BAX.116,117 In vitro, malignant cells isolated from 30% of CLL cases survive for several weeks in the absence of added cytokines.118 However, malignant cells from the remaining 70% of patients undergo rapid apoptosis in culture, but are protected by the addition of cytokines including IL-4 and IFN-α or -γ.118-122 IL-4 and IFN-α may promote CLL cell survival by preventing loss of expression of BCL-2 in vitro.118-120Interaction with bone marrow stroma, which is mediated by the β1 and β2 integrins, also maintains viability of CLL cells.123 124 Normal B cells do not adhere to stroma and are not protected from apoptosis. Therefore, upregulation of integrins on the surface of CLL cells relative to normal B cells may contribute to their extended life span in vivo, via the activation of unknown intracellular pathways.
The ratio of BCL-2 to BAX correlates inversely with the sensitivity of B-CLL cells to cytotoxic drugs in vitro.116,117 However, in vitro sensitivity to fludarabine failed to correlate with the achievement of clinical response.125 In a limited study of 58 CLL patients, high levels of the anti-apoptotic BCL-2 family member MCL-1 were found to correlate significantly with a failure to achieve complete remission.125
8.3. BCL-2 expression by acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL) cells.
Genetic changes that directly result in augmented expression of BCL-2 have not been described in AML. However, those AML patients in which greater than 20% of blasts express detectable BCL-2 levels show shorter survival and lower rates of achievement of complete remission compared with patients whose malignant cells express low BCL-2 levels.126 Immunohistochemical staining of BCL-2 is more intense in malignant cells from AML patients who fail to achieve remission than in those who respond to chemotherapy.127Malignant cells from patients showing high BCL-2 expression are drug resistant in vitro.128 Therefore, high BCL-2 expression, resulting from unknown mechanisms, may confer drug resistance on AML cells. However, other mechanisms may also confer protection even in the absence of high BCL-2 expression, because some AML isolates with low BCL-2 expression are also drug-resistant in vitro.128Elevated expression of MCL-1 at relapse suggests that cytotoxic regimes may result in the selection of AML clones expressing high levels of this anti-apoptotic BCL-2 family protein.129
Incubation of AML blasts with antisense oligonucleotides designed to decrease BCL-2 expression increases their sensitivity to cytosine arabinoside in vitro,130 emphasizing the potential importance of this protein in conferring drug resistance to these cells.
The expression of BCL-2 by the malignant cells of ALL patients at presentation is highly variable. However, BCL-2 expression does not correlate either with the ability of the ALL cells to survive in vitro or with the response of the patients to intensive chemotherapy.131 Therefore, it is likely that other factors play important roles in modulating the survival of ALL cells. Interactions between B-lineage ALL cells and stromal fibroblasts, mediated by interactions between β1 integrins and fibronectin, promote the survival of the leukemia cells.92 93 The intracellular pathways of survival promotion triggerred by these interactions are, however, unknown.
9. DELETION AND MUTATION OF THE p53 TUMOR SUPPRESSOR GENE IN LEUKEMIA AND LYMPHOMA
Deletion and/or mutation of p53 alleles results in the generation of tumors with impaired expression of functional p53 protein.63 The distribution of p53 mutations in human leukemia, which has been extensively reviewed 132, will be briefly summarized here. In general, p53 alterations are more frequent in aggressive disease and are associated with drug resistance and poor survival.
9.1. p53 mutations in myelodysplastic syndromes, CML, and CLL.
p53 changes are seen in 4% of myelodysplastic syndromes and are more frequent in advanced stages.132 p53 mutations are rare in the chronic phase of CML but are more frequent in blast crisis.133 p53 mutations are detected in 10% to 15% of CLL and are associated with poor response to therapy and shorter survival.134-136 Mutations are more frequent (about 40%) in Richter’s immunoblastic transformation.137 However, sensitivity of B-CLL cells to camptothecin analogs or fludarabine in vitro did not correlate with the presence of p53 mutations.116
9.2. p53 mutations in lymphoma and ALL.
A high proportion (30%) of Burkitt’s lymphoma and 55% of its leukemic counterpart, L3 ALL, harbor p53 mutations in addition to translocation and overexpression of the MYC oncogene.137The transformation of follicular lymphoma to diffuse aggressive disease correlates with p53 mutation and decreased survival in 25% to 30% of cases.138 p53 mutation is associated with loss of the short arm of chromosome 17, which carries the p53 gene, in Ph1-positive ALL.139 Overall, p53 loss is observed in 13% of ALL.132 However, the incidence of these changes is much lower in pediatric ALL (2%) and may correlate with the excellent response of these childhood ALL to cytotoxic drugs.140
9.3 p53 mutations in AML.
Genetic changes involving p53 are rare in AML, but are more frequent in cases with deletion of chromosome 17p.141 Again, loss of functional p53 in AML is associated with low rates of complete remission and with decreased survival.135 The blast cells from the majority AML patients require the addition of exogenous cytokines for both survival and growth in vitro.142,143Factor-dependent blasts die rapidly when deprived of GM-CSF, but are protected from apoptosis by antisense oligonucleotides that downregulate p53 expression, suggesting that the killing of these cells after factor deprivation is p53-dependent.143 GM-CSF and IL-3 also protect blast cells from 70% of AML patients from apoptosis induction by doxorubicin.144
In summary, p53 loss is relatively rare in leukemia. However, small sub-populations of leukemia cells may harbor these genetic changes, resulting in their relative resistance to cytotoxic regimes. p53-negative sub-populations may, therefore, survive drug treatment and initiate relapsed disease showing a more aggressive phenotype and increased drug resistance.
9.4. Adenoviruses lacking the E1B gene selectively kill tumor cells lacking functional p53.
Adenovirus infection of human cells requires expression of the viral E1B gene. The product of this gene binds to cell-encoded p53, thereby permitting viral replication and eventual killing of the host cells. Mutant adenoviruses lacking the E1B gene are, therefore, unable to proliferate in normal human cells, but are able to do so in tumor cells lacking functional p53.145 Human cervical carcinomas carried as xenografts in immunodeficient mice regress following direct injection of E1B-negative adenovirus. Primary infection of only 2% of the tumor cells may be sufficient to induce regression, due to the infectious nature of the adenovirus.145 It remains to be established whether the strategy outlined above will result in the selective killing of human leukemia cells.
9.5. Genetic changes involving the atm gene.
Ataxia telangiectasia patients show a high incidence of lymphoid, but not of myeloid, malignancies.146 Sixty percent of patients with T-prolymphocytic leukemia show homozygous loss of atmgenes within the tumor cells.147 The malignant cells of approximately 35% of B-CLL patients harbour atm mutations and show decreased expression of the ATM protein. This subset of patients was characterized by a more aggressive form of disease compared to patients with normal ATM expression. In some cases, heterozygous mutations are present in all somatic cells, suggesting a genetic predisposition to the disease.148 149 The role of the ATM-encoded protein in regulating apoptosis via the p53 pathway (section 5) suggests that loss of its expression in some B-CLL may contribute to the resistance to apoptosis characteristic of this malignancy.
10. THE CHIMERIC BCR/ABL ONCOGENE IN PHILADELPHIA (Ph1) CHROMOSOME-POSITIVE CHRONIC MYELOID LEUKEMIA (CML) AND ALL
The Ph1 translocation [t(9;22)], is associated with CML and results in the fusion of the bcr and abl genes. The fusion gene encodes a 210-kD oncoprotein (p210bcr/abl) with enhanced protein tyrosine kinase activity compared with the normal abl-encoded protein.150 A variant Ph1 translocation associated with ALL encodes a 185-kD BCR/ABL oncoprotein (p185bcr/abl) with increased transforming potential compared with p210bcr/abl.151
The normal abl-encoded protein is involved in the induction of apoptosis in some cell types.152 By contrast, expression of p210bcr/abl protects cells from killing by radiation, cytotoxic drugs, and ligation of FAS.153-155p210bcr/abl protects cells from killing by cytotoxic drugs by preventing the release of cytochrome c156 and activation of caspase 3.156 157
Both p210bcr/abl and p185bcr/abl activate phosphatidylinositol 3′-kinase,158,159 and this pathway is essential for transformation by these oncoproteins.160 Kinase activation is mediated via binding of a complex containing the CRKL and CBL adaptor proteins to a proline-rich domain of both chimeric oncoproteins.150 161 Because activation of phosphatidylinositol 3-kinase results in phosphorylation of the pro-apoptotic BAD protein via protein kinase B (section 4.5), the anti-apoptotic actions of BCR/ABL oncoproteins may be mediated at least in part by this route.
Drugs that selectively inhibit the bcr/abl-encoded protein kinases may be of value in the treatment of CML and Ph1-positive ALL. Herbimycin A162 and CGP 57148163,164 selectively inhibit the expansion of cells and cell lines expressing bcr/abl oncoproteins. The actions of herbimycin A on Ph1-positive cell lines is markedly enhanced by combination with etoposide or γ radiation.165Antisense oligonucleotides that suppress expression of bcr/abloncoproteins may also enhance apoptosis induction by drugs or radiation.153 However, the actions of at least somebcr/abl antisense oligonucleotides on CML cell lines may be nonspecific.166 These nonspecific cytotoxic effects may be attributable to the release of deoxyribonucleotides as a consequence of exonucleolytic degradation of the oligonucleotides.167
11. CHROMOSOMAL TRANSLOCATIONS INVOLVING TRANSCRIPTION FACTOR GENES RESULT IN THE DYSREGULATION OF APOPTOSIS
Chromosomal translocations associated with specific sub-types of acute leukemia and lymphoma result in the rearrangement of a variety of transcription factor genes. Some of these translocations result in enhanced expression of the transcription factor due to the juxtaposition of its gene next to Ig or T-cell antigen receptor loci, which contain powerful transcriptional enhancer elements. Other translocations involve the breakage and rejoining within introns of two transcription factor genes. The resulting hybrid genes encode chimeric transcription factors with novel properties which contribute to leukemogenesis.168 Some of these chimeric or aberrantly expressed proteins may contribute to malignant transformation via the suppression of apoptosis. Selected examples are described here.
11.1. Translocations resulting in inhibition of apoptosis.
The t(9;14) translocation, which is associated with 50% of lymphoplasmacytoid lymphoma, juxtaposes the paired box-containing genepax-5 to the IgH locus, where transcription from the pax-5promoters is augmented due to the proximity of the powerful Eμ enhancer.169,170 The PAX-5 protein represses transcription of the p53 gene.171 Therefore, decreased expression of p53 in cells bearing the t(9;14) translocation may contribute to malignant transformation via reducing expression of p53.
The chimeric PML-RARα transcription factor is generated as a consequence of juxtaposition of the retinoic acid receptor α gene (rarα; chromosome 17) and the pml gene (chromosome 15) in acute promyelocytic leukemia. Ectopic expression of this protein in myeloid cell lines diminishes apoptotic cell death. However, a reduced capacity to differentiate may also contribute to malignant transformation by the PML-RARα protein.172
The t(17;19) translocation, which is associated with pre-B cell leukemia, results in the fusion of the genes encoding the transcription factors E2A and HLF (hepatic leukemia factor). Human leukemia cells expressing the chimeric E2A-HLF protein undergo rapid apoptotic death after ectopic expression of a dominant negative inhibitor of E2A-HLF function. In addition, ectopic expression of E2A-HLF in nonmalignant pro-B lymphocytes abrogates apoptosis induction induced by IL-3 withdrawal or by p53 expression.173 Therefore, the oncogenic action of E2A-HLF may be related to its ability to prevent apoptosis. The similarity of the DNA binding/dimerization domains of HLF to the CES-2 (cell death specification-2) protein of C elegans suggests that E2A-HLF may block apoptosis via inducing transcription of a gene whose protein product blocks an early step in the apoptotic pathway.173
The t(10;14) translocation is associated with some cases of T-cell leukemia. The resulting juxtaposition of the hox 11 gene to the IgH locus results in its overexpression. Disruption of the hox 11 gene in mice results in the apoptotic death of spleen cells, again suggesting that oncogenic transformation by deregulated hox 11 expression is the result of protection from apoptosis.174
Overexpression of the tal1(scl) gene as a result of the t(1;14) translocation is a frequent event in T-ALL. Ectopic expression of TAL1 in an immature human T-lymphoid cell line does not perturb cell-cycle control but results in a marked resistance to cytotoxic drugs and to FAS ligation.175 This anti-apoptotic action is dependent on the DNA-binding domain of TAL1, suggesting that induction of expression of an unknown gene(s) underlies the resistance of TAL1 overexpressing cells to cell killing.
11.2. Translocations resulting in induction of apoptosis.
By contrast, some transcription factors involved in chromosomal translocations induce apoptotic cell death. Expression of the MYC gene is deregulated as a result of juxtaposition to the IgH locus in Burkitt’s lymphoma and L3 ALL cells bearing the t(8;14) translocation.176 The ability of the MYC gene product to trigger cell-cycle transit contributes to malignant transformation induced by its overexpression. However, the MYC protein also induces apoptosis when ectopically expressed at high levels in fibroblasts. Although p53 is required for MYC-induced apoptosis,177 p53 “knockout” mice develop normally.178 Therefore, it is probable that levels of MYC protein generated during normal physiological responses do not induce apoptosis.
A net increase in cell number after enforced induction of MYC in murine fibroblasts requires that the apoptotic pathway be blocked by survival-inducing cytokines.179 Therefore, oncogenic transformation by deregulated MYC may require additional genetic events that abrogate apoptosis induction and may explain the high proportion of Burkitt’s lymphoma and L3 ALL cases bearing p53 gene lesions.137 The ability of overexpressed BCL-2 to collaborate with MYC in promoting the generation of lymphoid tumors in doubly transgenic mice is also consistent with the concept that oncogenenic transformation by MYC is dependent on the suppression of apoptosis.106
The e2a-pbx1 fusion gene results from the t(1;19) translocation associated with B-cell precursor ALL. The protein product of this fusion gene rapidly induces apoptosis in B-cell progenitors. The dependence of apoptosis induction on the DNA-binding homeodomain of the PBX1 moiety suggests that cell killing is dependent on transcriptional activation of an unknown gene(s).180 Apoptosis induced by E2A-PBX1 expression in cell lines is blocked by BCL-2 expression. Therefore, oncogenic transformation by this chimeric gene may also depend on additional genetic changes that block apoptosis induction.180
PERSPECTIVES
Knowledge of the complex biochemical pathways involved in the regulation of apoptosis in hematopoietic cells is advancing rapidly. Here we have focused on aspects of apoptosis regulation with particular relevance to the hematopoietic system. The BCL-2 protein family, the release of mitochondrial cytochrome c, p53-mediated transcriptional control, FAS, and the TNF receptor are involved in the control of apoptosis induction at least in some hematopoietic cells. Diverse regulatory mechanisms converge on a final common pathway involving activation of the caspase family of proteases. Additional interactions involving the BCL-2 family may also be important in apoptosis regulation and have been reviewed elsewhere.181
Elevated expression of BCL-2, loss of functional p53, constitutive activativation of protein tyrosine kinases, or the generation of chimeric, oncogenic transcription factors as a result of chromosomal translocations abrogate apoptosis induction and antagonize the actions of cytotoxic drugs or of radiation on leukemia cells. Therefore, strategies designed to bypass blocks in the detection of apoptotic signals or in the signal transduction phase may be of value in overcoming resistance to cytotoxic regimes.2 However, apoptosis is a complex physiological process dependent on the integrated functioning of a large number of gene products. Therefore, any therapeutic stratagem that is dependent solely on the induction of apoptosis will lead to the rapid evolution of clones resistant to killing. Microenvironemental factors also influence the outcome of cytotoxic treatments by modulating pathways of apoptosis control. Therefore, manipulation of the cytokine levels of leukemia or lymphoma patients may also impact on the efficacy of chemotherapy or radiation.182
Elucidation of the precise mechanisms involved in the apoptotic killing of leukemia cells (as opposed to cell lines) and of the strategies by which malignant cells escape killing by cytotoxic agents are major topics for future research. It is anticipated that an understanding of these facets of leukemia and lymphoma cell biology will lead to the design of effective strategies for the treatment of hematopoietic malignancies that are resistant to conventional treatment.
REFERENCES
NOTES ADDED IN PROOF
(1) New members of the cell-surface death receptor (DR) family have been described. DR3 triggers apoptosis consequent to binding APO 3 ligand.183 TNF-related apoptosis-inducing ligand (TRAIL) initiates cell death following binding to DR4 or DR5. The actions of DR4 and 5 are limited by decoy receptors which bind TRAIL but are unable to transduce apoptotic signals.183 (2) Binding of the MDM2 protein to p53 targets the latter for degradation. Phosphorylation of p53 by the ATM protein kinase results in its dissociation from MDM2 and consequent stabilization.184,185(3) Phosphorylation and inactivation of caspase 9186 and of the forkhead family transcription factor FKHRL1187 by protein kinase B contribute to the suppression of apoptosis by pro-survival cytokines. (4) The bcl10 gene involved in the t(1;14)(p22;q23) translocation of mucosa-associated lymphoid tissue (MALT) lymphoma has been cloned. Translocation results in expression of a truncated protein which lacks the pro-apoptotic activity of the wild-type BCL10 protein.188 (5) Mitochondria of cells primed for apoptosis can release death-inducing proteins other than cytochrome c. These include caspases 2 and 9 and a flavoprotein, apoptosis-inducing factor (AIF).189
Author notes
Address reprint requests to R. Gitendra Wickremasinghe, PhD, Department of Hematology, Royal Free and University College Medical School, Rowland Hill St, London NW3 2PF, UK.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal