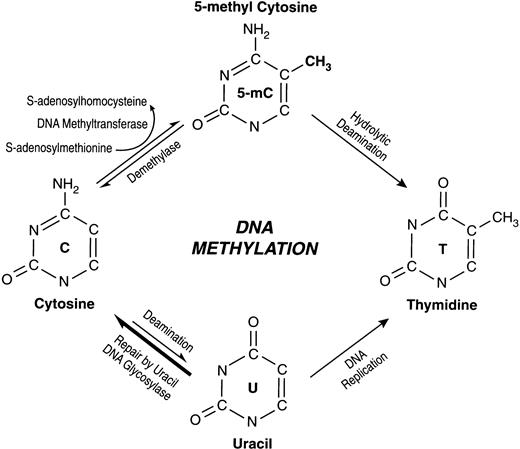
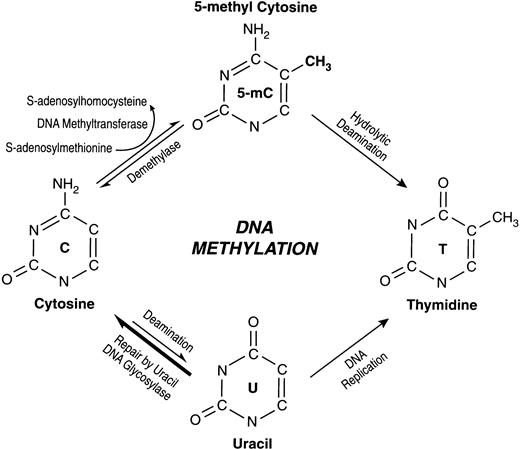
SINCE ITS FIRST recognition in 1948, the fifth base of human DNA, 5-methylcytosine (5-mC) has generated much interest and considerable controversy during attempts to understand its significance (for review, see Weissbach1). DNA methylation in eukaryotes involves addition of a methyl group to the carbon 5 position of the cytosine ring (Fig 1). This reaction is catalyzed by DNA methyltransferase in the context of the sequence 5′-CG-3′, which is also referred to as a CpG dinucleotide. It is the most common eukaryotic DNA modification and is one of the many epigenetic (alteration in gene expression without a change in nucleotide sequence) phenomena. Although extensive in plants and mammals, the absence of detectable DNA methylation in some eukaryotes such as Drosophila2 and Saccharomyces cerevisiae3 has raised doubts about its significance in normal development and tissue-specific gene expression. However, recent studies showing abnormal development and embryonic lethality in transgenic mice expressing decreased but not completely absent DNA Methyltransferase (MTase) activity after DNA-MTase gene knockout4 lends support to a critical role for DNA methylation in developmental gene regulation. Others have proposed that the control of intragenomic parasites is the primary function of DNA methylation in mammalian cells.5
Schematic representation of the biochemical pathways for cytosine methylation, demethylation, and mutagenesis of cytosine and 5-mC.
Schematic representation of the biochemical pathways for cytosine methylation, demethylation, and mutagenesis of cytosine and 5-mC.
In this review, we will discuss the fundamental aspects of DNA methylation and its role in transcription repression, neoplasia, and transgene silencing and during development. In the interest of brevity, the role of DNA methylation in genomic imprinting and X chromosome inactivation are not discussed here, but have been reviewed recently elsewhere.6-8
DISTRIBUTION OF METHYLATED CYTOSINES AND CpG ISLANDS
Eukaryotic genomes are not methylated uniformly but contain methylated regions interspersed with unmethylated domains.9 During evolution, the dinucleotide CpG has been progressively eliminated from the genome of higher eukaryotes and is present at only 5% to 10% of its predicted frequency.10-12 Cytosine methylation appears to have played a major role in this process, because most CpG sites lost represent the conversion through deamination of methylcytosines to thymines. Approximately 70% to 80% of the remaining CpG sites contain methylated cytosines in most vertebrates, including humans.10,12 These methylated regions are typical of the bulk chromatin that represents the late replicating DNA with its attendant histone composition and nucleosomal configuration and is relatively inaccessible to transcription factors.13 In contrast to the rest of the genome, smaller regions of DNA, called CpG islands, ranging from 0.5 to 5 kb and occurring on average every 100 kb, have distinctive properties. These regions are unmethylated, GC rich (60% to 70%), have a ratio of CpG to GpC of at least 0.6, and thus do not show any suppression of the frequency of the dinucleotide CpG.10,14 Chromatin containing CpG islands is generally heavily acetylated, lacks histone H1, and includes a nucleosome-free region.13 This so called open chromatin configuration may allow, or be a consequence of, the interaction of transcription factors with gene promoters.14
The patterns of DNA methylation reflect two types of gene 5′ regulatory regions in the genome. Approximately half of all genes in mouse and humans (ie, 40,000 to 50,000 genes) contain CpG islands.10 These are mainly housekeeping genes that have a broad tissue pattern of expression, but approximately 40% of genes with a tissue-restricted pattern of expression are also represented.14 Promoter region CpG islands are usually unmethylated in all normal tissues, regardless of the transcriptional activity of the gene. The main exceptions include nontranscribed genes on the inactive X-chromosome and imprinted autosomal genes where one of the parental alleles may be methylated.15 Tissue-specific genes without CpG islands are variably methylated, often in a tissue-specific pattern, and usually methylation is inversely correlated with the transcriptional status of the genes (for review, see Bird16 and Cedar17).
DNA METHYLTRANSFERASES
The enzymes that transfer methyl groups to the cytosine ring, cytosine 5-methyltransferases, or DNA methyltransferases (DNA-MTase) have been characterized in a number of eukaryotes.18 The target site for DNA-MTase in DNA is the dinucleotide palindrome CG (commonly referred to as CpG, with p denoting the phosphate group). The first eukaryotic DNA-MTase gene cloned from mouse almost a decade ago19 is now referred to as Dnmt1. This gene is highly conserved among eukaryotes. Dnmt1 orthologs have been identified in various species, including humans(DNMT1).20 Interestingly, Dnmt1 orthologs have not been found in organisms lacking DNA methylation, such asSaccharomyces cerevisiae, Caenorhabditis elegans, andDrosophila melanogaster.
Mammalian Dnmt1 methyltransferase has a high affinity for hemimethylated substrates but is also capable of performing de novo methylation of unmethylated substrates in vitro. The de novo activity of mammalian Dnmt1 methyltransferase has been shown to be stimulated by aberrant DNA structures21 and 5-mC residues in single-stranded22 or double-stranded DNA substrates.23 After it was found that the human and mouse Dnmt1 genes contain additional 5′ transcribed sequences with protein-encoding potential,24 it became apparent that previous biochemical and cell culture assays had relied on an incomplete Dnmt1 cDNA sequence that codes for a truncated protein. Pradhan et al25 demonstrated that full-length Dnmt1 MTase synthesized in Baculovirus was equally active on unmethylated and hemimethylated substrates, in part by virtue of an inhibitory effect of the additional N-terminal amino acids on activity with hemimethylated DNA. These results are evidence that theDnmt1 MTase protein has inherent de novo methylating activity that may be altered by protein-protein interactions and enhanced by aberrant structures or 5-mC residues in the substrate DNA.
Evidence for an additional mammalian DNA-MTase capable of de novo methylation came from the experiments of Lei et al.26 These investigators generated a null mutation of the Dnmt1 gene through homologous recombination in mouse embryonic stem cells.Dnmt1 null embryonic stem cells were viable and contained low but stable levels of methylcytosine and methyltransferase activity. Interestingly, integrated provirus DNA in Moloney murine leukemia virus (MoMuLV)-infected homozygous Dnmt1 knockout ES cells exhibited de novo methylation to a similar extent as in wild-type cells. Although a second DNA-MTase has recently been identified in mouse (Dnmt2)27 and in humans(DNMT2),28 it is not yet clear if it is the long sought after de novo methyltransferase present in Dnmt1deficient ES cells.
TECHNIQUES TO STUDY DNA METHYLATION
Early techniques to study site-specific DNA methylation relied primarily on the inability of methylation-sensitive type II restriction enzymes to cleave sequences containing one or more methylated CpG sites, combined with Southern hybridization.29-31 This method requires large amounts of high molecular weight DNA, detects methylation only if more than a few percent of alleles are methylated, and only provides information about those CpG sites found within the recognition sequence of methylation-sensitive restriction enzymes. Singer-Sam et al32 improved the sensitivity of methylation detection by combining the use of methylation-sensitive restriction enzymes and polymerase chain reaction (PCR). After cleaving the DNA with methylation-sensitive restriction endonucleases, eg, HpaII, PCR amplification with specific primers flanking the restriction site will only occur if DNA cleavage has been prevented by methylation. However, this method, like Southern-based approaches, can only monitor CpG methylation in methylation-sensitive restriction sites. Also, a false-positive result may be obtained due to incomplete restriction enzyme digestion of cellular DNA.
Genomic sequencing protocols that have previously been used to study DNA methylation use Maxam and Gilbert chemical cleavage reactions performed on genomic DNA33 with linker-mediated PCR (LMPCR) to enhance the signal.34 These methods are based upon the fact that 5-mC is not cleaved during the standard Maxam and Gilbert cytosine cleavage reaction.35 Thus, 5-mC is identified in a sequencing gel by the lack of a band that corresponds to a cleavage product of a cytosine degradation reaction. This assay is technically demanding and is subject to both false-positive and false-negative results.
Frommer et al36 introduced a procedure based on bisulfite-induced oxidative deamination of genomic DNA under conditions in which cytosine is converted to uracil and 5-mC remains unchanged. The target sequence is then amplified by PCR using strand-specific primers. Upon sequencing of the amplified DNA, all uracil and thymine residues become detectable as thymine and only 5-mC residues amplify as cytosines. This method, as shown in Fig 2, is presently the method of choice for the detailed analysis of 5-mC in any given genomic target sequence.
(A) Illustration of the steps involved in determining the methylation status of cytosines in a known DNA sequence by the bisulfite conversion method. (B) In vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are only seen in adult erythroid cells. Positions indicated are relative to the transcription start site. Cytosines that are not associated with CpG dinucleotides (sequence shown in [C]) have all been converted to thymidines in both 5-day and adult erythroid cells. (C) Graphic representation of the in vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are clearly seen in (A) (data not shown for CpG dinucleotide at position −15). Primers R and F indicate the sequence of rho globin promoter used for designating internal primers and for sequencing.
(A) Illustration of the steps involved in determining the methylation status of cytosines in a known DNA sequence by the bisulfite conversion method. (B) In vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are only seen in adult erythroid cells. Positions indicated are relative to the transcription start site. Cytosines that are not associated with CpG dinucleotides (sequence shown in [C]) have all been converted to thymidines in both 5-day and adult erythroid cells. (C) Graphic representation of the in vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are clearly seen in (A) (data not shown for CpG dinucleotide at position −15). Primers R and F indicate the sequence of rho globin promoter used for designating internal primers and for sequencing.
A number of rapid methods to detect 5-mC have been developed based on the bisulfite deamination reaction in combination with PCR amplification. These are suitable for examining limited numbers of CpG dinucleotides that are either found within or immediately adjacent to the PCR primer sequences37,38 or within a restriction enzyme recognition sequence.39
DNA METHYLATION AND TRANSCRIPTIONAL REPRESSION
A role for DNA methylation in the differential regulation of gene expression was hypothesized many years ago.40,41 The potential mechanism was suggested by a number of early observations in which site-specific cytosine methylation within or adjacent to genes was found to correlate with transcriptional repression.31 42-45 Subsequently, this inverse relationship between cytosine methylation and transcription has been observed in a large number of genes, although not universally.
Despite the long held view that DNA methylation might act as a negative regulator of transcription, the precise mechanism involved has remained elusive. Numerous reports have shown the ability of promoter DNA methylation to inhibit transcription of a wide variety of genes in in vitro transfection assays, and in some cases, such methylation corresponds to the inactive state of the gene under study in vivo (reviewed in Bird16 and Razin and Cedar46).
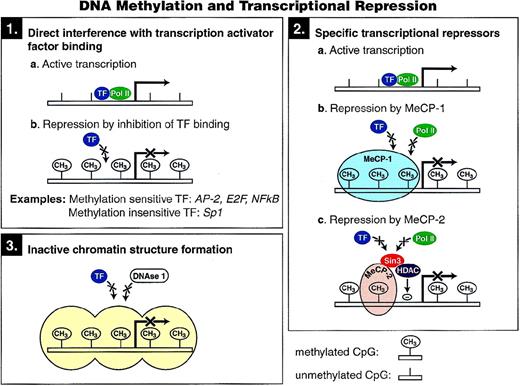
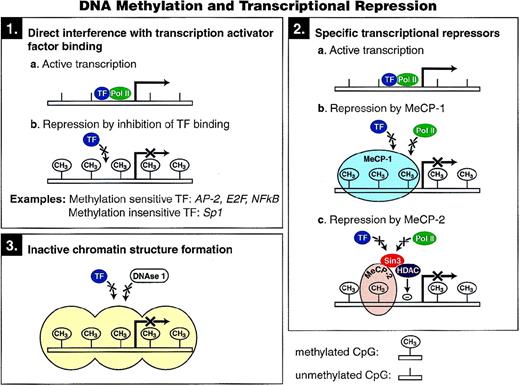
Three possible mechanisms have been proposed to account for transcriptional repression by DNA methylation, and each is shown in Fig 3. The first mechanism involves direct interference with the binding of specific transcription factors to their recognition sites in their respective promoters. Several transcription factors, including AP-2, c-Myc/Myn, the cyclic AMP-dependent activator CREB, E2F, and NF-κB, recognize sequences that contain CpG residues, and binding to each has been shown to be inhibited by methylation. In contrast, other transcription factors (eg, Sp1 and CTF) are not sensitive to methylation of their binding sites,47 and many factors have no CpG dinucleotide residues in their binding sites.
Proposed mechanisms of transcriptional repression mediated by cytosine methylation.
Proposed mechanisms of transcriptional repression mediated by cytosine methylation.
A second potential mechanism for methylation induced silencing is through the direct binding of specific transcriptional repressors to methylated DNA. Two such factors, MeCP-1 and MeCP-2 (methyl cytosine binding proteins 1 and 2), have been identified and shown to bind to methylated CpG residues in any sequence context. Although in vertebrates DNA methylation has been posited to inhibit transcription initiation, methylation has also been shown to block transcription elongation in Neurospora through a mechanism that may be mediated through MeCP-1 and/or MeCP-2.48
MeCP-1 binds to DNA containing multiple symmetrically methylated CpG sites, as opposed to hemimethylated CpGs, and manifests as a large complex on electrophoretic mobility shift assay.49Repression of transcription from densely methylated genes can be mediated by MeCP-1, and cells deficient in MeCP-1 show much reduced repression of methylated genes.50 In a further study, it was demonstrated that sparse methylation could repress transfected genes completely, but the inhibition was fully overcome by the presence in cis of an SV40 enhancer. However, densely methylated genes could not be reactivated by the strong enhancer. It was proposed that sparsely methylated genes form an unstable complex with MeCP-1 that prevents transcription when the promoter is weak. This complex can be disrupted by a strong promoter, thereby allowing the methylated gene to be transcribed.51
We have recently shown that a complex with electrophoretic mobility similar to MeCP-1 forms efficiently with the methylated but not with unmethylated embryonic rho-globin gene promoter sequences, and the complex can be detected using nuclear extracts from the same primary avian erythroid cells in which methylation-mediated transcriptional inhibition was demonstrated.52 These results, in conjunction with the demonstration of a role for methylation in silencing rho-globin gene transcription in vivo in normal adult avian erythroid cells,53 54 suggest a role for MeCP-1 or a similar complex in developmental silencing of embryonic globin genes during normal erythropoiesis.
A component of the MeCP-1 complex, PCM1, has a methyl-CpG binding domain (MBD) and two cysteine-rich domains (CXXC) that are found in animal DNA methyltransferases and in the mammalian HRX proteins, MLL and ALL-1. Although the functional significance of the CXXC domain is not known, there is evidence that it is a part of a transcriptional repression domain.55,56 PCM1 has been shown to repress transcription in vitro in a methylation-dependent manner.57
MeCP-2 is more abundant than MeCP-1 in the cell and is able to bind to DNA containing a single methylated CpG pair.58 MeCP-2, like DNA methyltransferase, is dispensable for the viability of embryonic stem cells, but is essential for embryonic development.59MeCP-2 has two domains: a methyl-CpG binding domain that is essential for chromosomal localization and a transcriptional repressor domain (TRD) that can inhibit transcription from a promoter at a distance, suggesting that MeCP-2 interacts with the transcriptional machinery or the initiation complex.60 Recently, a region of MeCP-2 that localizes with the TRD was shown to associate with a corepressor complex containing the transcriptional repressor mSin3A and histone deacetylases. Transcriptional repression in vivo was relieved by the deacetylase inhibitor trichostatin A, suggesting that two global mechanisms of gene regulation, DNA methylation and histone deacetylation, can be linked by MeCP-2.61,62 However, in some instances DNA methylation has been shown to play a dominant role over histone deacetyloses in transcriptional repression.53,54 It has been proposed that MeCP-2 might contribute to the assembly of a more stable repressive chromatin structure.63 However, MeCP-2 can repress transcription of naked DNA in a cell-free in vitro assay,60 suggesting that chromatin formation is not necessary for its repressive action.
A third mechanism by which methylation may mediate transcriptional repression is by altering chromatin structure. Keshet et al64 transfected mouse L cells with M13 plasmid constructs containing the human β-globin gene, as well as several other eukaryotic genes, after enzymatic methylation. Unmethylated DNA sequences, after integration and stable propagation in cell culture, were all detected in active chromatin, as measured by DNAse 1-sensitivity, in contrast to DNA sequences that were methylated in vitro before transfection that were contained in DNAse1-resistant, transcriptionally inactive chromatin. Experiments using microinjection of certain methylated and nonmethylated gene templates into nuclei have shown that methylation inhibits transcription only after chromatin is assembled.65 Even a strong transcriptional activator, GAL4-VP16, cannot counteract the effect of chromatin once it has assumed the inactive state induced by DNA methylation.65Therefore, in addition to stabilizing the inactive state, methylation also prevents activation by blocking the access of transcription factors.63 66 Whether this chromatin effect of methylation is mediated solely by MeCP-2–associated histone deacetylase remains to be determined.
One important issue regarding DNA methylation and transcriptional silencing has been whether methylation is a primary control mechanism or a secondary effect of gene activity. In the case of some genes with sparse CpG nucleotides, gain of methylation occurs after transcriptional silencing67; in other cases, loss of methylation occurs after transcriptional activation.68 At the same time, in other systems it has been shown that despite optimum nuclear conditions for transcription, including DNAseI-sensitive chromatin, transcription can be tightly repressed by CpG methylation.52 53 It appears that methylation, particularly of CpG-rich genes, may serve as a locking off mechanism that may follow or precede other events that turn a gene off, but that once in place can prevent activation despite an optimum nuclear environment for transcription.
DNA DEMETHYLATION DURING DEVELOPMENT AND TISSUE-SPECIFIC DIFFERENTIATION
A critical aspect of the overall regulatory role of DNA methylation is the process of demethylation. Using methylation-sensitive restriction enzymes to monitor the general level of DNA methylation, it was shown that, during early development, a dramatic reduction in methylation levels occurs in the preimplantation embryo.69 This is followed by a wave of de novo methylation involving most CpG residues but leaving the CpG islands unmethylated at the time of implantation.70 After implantation, most of the genomic DNA is methylated, whereas tissue-specific genes undergo demethylation in their tissues of expression.46 Thus, a means for both global and tissue- or gene-specific removal of methylcytosines from DNA must exist.
The mechanism underlying the process of demethylation has not been fully elucidated. In some cases, demethylation could be a passive process, ie, inhibition of methylation after DNA replication.45 Despite the tight coordination of DNA methylation and replication that has been demonstrated in mammalian cells,71 the mechanism of methylation inhibition in the presence of DNA replication remains to be defined. The existence of an active demethylation process not involving DNA replication in mammalian cells has been supported by a number of observations (reviewed in Szyf72). In transient transfection assays using myoblast cells, a methylated α-actin gene was shown to have transcriptional activity similar to unmethylated template secondary to an active demethylation process.73 Using transient transfection assays, Szyf et al74 showed that an in vitro methylated SK-plasmid bearing no sequence homology to eukaryotic genes became fully demethylated between 1 and 2 days after tranfection into the mouse embryonal carcinoma cell line P19. This demethylating activity was not sequence-selective and was independent of DNA replication.74 During B-cell development, both the heavy and light chain Ig genes undergo demethylation.75 Specificcis-acting elements, including both the intronic and 3′ enhancers in the case of the κ light chain gene, have been shown to direct demethylation.75 76 Further analysis of the intronic enhancers suggested the involvement of the NF-κB family of proteins as trans-acting factors in inducing demethylation. Bothcis-acting elements and trans-acting factors, therefore, appear to direct the demethylation machinery to its target locus.
The biochemical mechanism underlying the process of demethylation remains unclear. However, extracts from chicken embryos have been demonstrated to have sequence nonspecific demethylating activity in vitro.77 This enzymatic activity was limited to demethylation of hemimethylated DNA and involved removal of the methylated cytosine and subsequent repair of the resulting apyrimidinic acid residue.78 In the case of rat myoblasts, demethylation activity is performed by an active component whose activity is to excise methylated nucleotides from the DNA template and replace them with an unmodified form.79 Interestingly, in the presence of a myoblast extract, demethylation of the α-actin but not of the adenine phosphoribosyl transferase (APRT) CpG island fragment was observed. Although this demethylating activity was initially felt to be dependent on an RNA component, subsequent investigation of a more purified active fraction demonstrated complete resistance of the activity to RNAse treatment.80
It has long been supposed that direct removal of the methyl group by breaking the carbon-to-carbon bond of 5-mC would be so energetically unfavorable as to be an unlikely event. However, a gene encoding an enzyme capable of directly removing the methyl group from methylated CpG-containing DNA in cell free reactions as well as in transfected cells has been reported recently.81 If the corresponding endogenous gene product can be shown to have the same activity, this will provide a remarkable new avenue for studying the control of demethylation during normal differentiation and development as well as in oncogenesis.
It has been proposed that the core demethylase in cells is kept in an inactive form by interaction with a protein inhibitor. Demethylation could then come about by the removal or modification of the inhibitor. Such an inhibitor could either affect the entire genome resulting in a pattern similar to that seen in the early preimplantation embryo or be confined to the specific sequence influenced by localcis-acting elements and trans-acting factors, as in the case of the κ Ig gene example discussed above.82
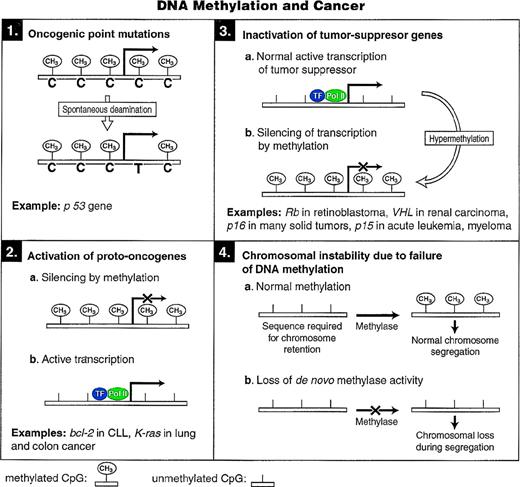
DNA METHYLATION AND CANCER
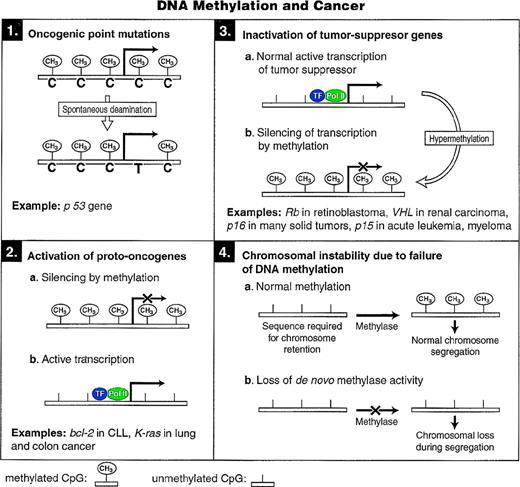
A role for DNA methylation in oncogenesis has been hypothesized for many years. Numerous studies have suggested aberrations in DNA methyltransferase activity in tumor cells.83-85 Transformed cells often have increased total DNA-MTase activity, widespread loss of methylation from normally methylated sites, and more regional areas of hypermethylated DNA.86 The potential contribution of DNA methylation to oncogenesis appears to be mediated by one or more of the following mechanisms that are shown in Fig4.
Models for the different mechanisms through which cytosine methylation can promote oncogenesis.
Models for the different mechanisms through which cytosine methylation can promote oncogenesis.
Signature C→T mutation in cancer cells.
The high mutation rate of cytosine residues within the dinucleotide CpG, the target site of mammalian DNA-MTase, can be accounted for by an increased rate of cytosine to thymine transitions, which are, in turn, a consequence of hydrolytic deamination of 5-mC87 (Fig 1). This mechanism of mutation was first recognized in prokaryotic systems.88
Unmethylated cytosine can also undergo deamination to yield uracil, but the well-characterized Uracil-DNA glycosylase efficiently repairs G:U but not G:T mismatch.89 However, DNA-MTase may block this repair, contributing to C→U→T transition. DNA-MTase may also mediate 5-mC→T transition at CpG dinucleotides under conditions that lead to increased DNA-MTase expression or decreased cellular S-adenosylmethionine levels.90
A striking example of how this process may lead to oncogenesis is shown by the tumor-suppressor gene p53. Mutations in the p53 tumor-suppressor gene occur in more than 50% of human solid tumors.91 An estimated 24% of these mutations are C→T transitions at CpG dinucleotides, suggesting that DNA methylation may contribute to these mutations.92
DNA hypomethylation in cancer.
Decreased levels of overall genomic methylation are common findings in tumorigenesis.93 This decrease in global methylation appears to begin early and before the development of frank tumor formation.94,95 Apart from the overall genomic hypomethylation, specific oncogenes have been observed to be hypomethylated in human tumors. A good inverse correlation between methylation and gene expression was observed in the antiapoptotic bcl-2 gene in B-cell chronic lymphocytic leukemia96 and for the k-ras proto-oncogene in lung and colon carcinomas.97
Hypermethylation of tumor-suppressor genes.
In addition to point mutations or gene deletions, transcriptional repression by hypermethylation of promoter sequences suggests an alternative means for the inactivation of tumor-suppressor genes in cancer. This may result from the increased DNA-MTase levels that have been demonstrated in various cancers83 or it could occur as a result of some other transient event that silences tumor-suppressor gene transcription. The retinoblastoma gene (Rb) was the first classic tumor-suppressor gene in which CpG island hypermethylation was detected. Three of 21 sporadic cases in one study98 and 5 of 32 sporadic cases in another study99 had hypermethylated CpG islands at the 5′ end of the Rb gene. Subsequently, it was also shown that in vitro methylation of the promoter region of Rb directly blocked promoter activation.100
Inactivation of the von Hippel-Lindau (VHL) gene by somatically acquired mutations in one copy of the VHL gene along with loss of the second allele has been implicated as the initiating event in spontaneous cases of clear-cell renal carcinoma.101 Herman et al102 found hypermethylation of the VHL gene CpG island and concomitant lack of expression in 5 of 26 cases of the sporadic form of clear cell renal carcinoma that had no VHL intragenic mutations. Treatment of a clear cell renal carcinoma cell line with 5-deoxyazacytidine, a potent inhibitor of DNA-MTase, resulted in demethylation and expression of a previously silent VHL gene.
One of the most important cell cycle regulatory proteins is p16 (also known as MTS-1 for major tumor suppressor 1, INK4a forINhibitor of cyclin-dependent Kinase 4a, and CDKN2a for Cyclin-Dependent Kinase iNhibitor 2a). The major biochemical effect of p16 is to halt cell-cycle progression at the G1/S boundary, and the loss of p16 function may lead to cancer progression by allowing unregulated cellular proliferation.103 A common genetic alteration in tumor cell lines and to a lesser extent in primary tumors is the loss of heterozygosity at chromosome 9p21, which contains both the related p16 and p15 genes. Among most solid tumors studied, a CpG island in the 5′ region of p16 gene has been found to be frequently methylated, and treatment of cell lines carrying a hypermethylated p16 allele with 5-azacytidine results in transcriptional activation of the gene.104,105 The presence of either coding region deletions or promoter hypermethylation of p16 correlate inversely with the presence of Rb gene mutations in multiple tumor types (reviewed in Baylin et al86). Interestingly, in some instances, p16 promoter hypermethylation may be the sole inactivating event for both alleles of the gene and may be the only lesion associated with loss of the cyclin D-Rb pathway.86 In colon cancer, despite the lack of allelic loss of the 9p region, homozygous deletions of the p16 gene, or Rb gene mutations,106 30% to 40% demonstrate hypermethylated p16 alleles.105 Hypermethylation-mediated inactivation of the p16 gene has been demonstrated in brain, breast, colon, head and neck, and non–small-cell lung cancer and in high grade non-Hodgkin’s lymphoma (reviewed in Baylin et al86).
P15 (INK4b) gene is an inhibitor of the cyclin-dependent kinases CDK4 and CDK6 and appears to play a role in transforming growth factor-β (TGF-β)–mediated growth inhibition responses.107Methylation-mediated inactivation of the p15 gene occurs predominantly in hematopoietic neoplasms such as acute myelogenous leukemia, acute lymphoblastic leukemia, and Burkitt’s lymphoma.86Recently, hypermethylation of the p15 gene has been demonstrated in myelodysplastic syndromes108 and hypermethylation of both p15 and p16 has been found in multiple myeloma.109
Induction of chromosomal instability.
Lengauer et al110 introduced exogenous CpG-rich sequences in the form of a retrovirally contained β-galactosidase gene into 10 colon cancer cell lines. Five of 10 cell lines failed to express the β-gal gene, and these lines were deficient in mismatch activity repair (MMR−), whereas other cell lines competent for mismatch repair (MMR+) expressed the gene. MMR− cell lines were found to be methylation proficient (MET+), and MMR+ cell lines were methylation deficient (MET−) based on Southern blot analysis and 5-azacytidine–induced reactivation of the β-gal gene. It was proposed that in mismatch repair proficient colon cells a methylation defect (MET-) directly facilitates the gain and loss of whole chromosomes, leading to the genomic instability necessary for the development and progression of cancer. In contrast, MMR− cells have normal methylation proficiency and develop the required genomic instability by the alternative pathway of mismatch repair deficiency.111 Consistent with a role for hypomethylation in chromosomal instability were the findings by Chen et al112 in analyzing embryonic stem (ES) cells nullizygous for the DNA-MTase gene, Dnmt I. Gene deletions of selectable marker genes due to mitotic recombination or chromosomal loss were detected at a much higher frequency in the Dnmt 1-deficient ES cells compared with wild-type control cells.
Additional experimental support for the hypothesis linking DNA methylation to chromosomal integrity comes from earlier studies by Feinberg et al,113 who demonstrated an average of 8% and 10% reduction in genomic 5-mC content in colon adenomas and adenocarcinomas, respectively, with no significant difference between benign and malignant tumors. Interestingly, three patients with the highest 5-mC content in their normal colon appeared to have Lynch syndrome (HNPCC), which was subsequently shown to have the MMR− phenotype.113 In a recent study, the occurrence of genome-wide undermethylation, retroviral element amplification, and chromosome remodeling in an interspecific mammalian hybrid (Macropus eugenii × Wallabia bicolor) was demonstrated. Atypically extended centromeres of Macropus eugenii-derived autosomes in the hybrid were composed primarily of unmethylated, amplified retroviral elements not detectable in either parental species, indicating that the failure of DNA methylation to occur and resultant mobile-element activity in the cell hybrids could facilitate rapid karyotypic evolution.114
ALTERED DNA-METHYLTRANSFERASE ACTIVITY IN CANCER
Several studies in the last few years have demonstrated an increase in DNA-MTase activity in neoplastic cells. Kautiainen and Jones83 examined the levels of DNA methyltransferase in nuclei from 9 tumorigenic and 9 nontumorigenic cell lines. In all but 2 cases, the extractable methyltransferase activity was fourfold to 3,000-fold higher in tumorigenic than in nontumorigenic cells.83 Increased DNA-MTase activity has been reported in colon cancers versus normal mucosa from the same patient.84Laird et al85 bred normal mice heterozygous for deletion of the DNA-MTase gene (and having 50% of the DNA-MTase activity compared with wild-type) with mice having a mutant allele of the APC gene.85 When combined with loss of the other APC allele, this mutation results in progressive formation of adenomas throughout the gastrointestinal tract of newborn animals. Intestinal adenomas were reduced by 50% in the offspring mice with both the APC and DNA-MTase gene mutations and were essentially eliminated when treatment with 5-deoxyazacytidine was combined with DNA-MTase allelic deletion.
The mechanism underlying the increased DNA-MTase activity in neoplastic cells has not yet been elucidated. MacLeod et al115proposed that the activation of the ras signal transduction pathway causes increased activity by interacting with AP-1 sites in the presumed DNA-MTase promoter region in a murine adrenocortical tumor cell line. However, this apparent promoter region of the DNA-MTase gene was subsequently found to lie far downstream of the transcription start site.24,116 It has also been demonstrated that overexpression of the c-fos gene results in cellular transformation through increased DMTase activity.117 Further studies are required to understand the regulation of the DNA-MTase genes in normal and neoplastic cells.
The mechanisms responsible for maintaining normal and abnormal methylation patterns in normal and neoplastic cells remain unclear. A recent study demonstrated the binding of DNA-MTase to proliferating cell nuclear antigen (PCNA), an auxiliary factor for DNA replication and repair. This binding occurred in intact cells at foci of newly replicated DNA and did not alter DNA-MTase activity. A peptide derived from the cell cycle regulator p21(WAF1) was shown to disrupt the DNA-MTase-PCNA interaction, suggesting that p21(WAF1) may regulate methylation by blocking access of DNA-MTase to PCNA. The extent of expression of DNA-MTase and p21(WAF1) were found to be inversely related in both SV40-transformed and nontransformed cells.118 Based on these findings, it has been proposed that, in normal cells, the p21 protein negatively regulates DNA-MTase-PCNA interaction in early S phase and protects CpG islands from methylation, whereas diminished effects of p21 in late S phase result in the targeting of DNA-MTase to methylated DNA. In cancer cells, loss of p21 function allows DNA-MTase-PCNA interaction in early S phase, possibly facilitating aberrant increased methylation of CpG islands, whereas no change in decreased relative targeting of DNA-MTase to late S phase foci results in loss of normal methylation.15 This type of mechanism could explain the apparent paradox posed by the observation of both excessive and deficient DNA methylation during tumorigenesis.
CLINICAL AND THERAPEUTIC IMPLICATIONS OF DNA METHYLATION
Just as the vertebrate globin genes were among the first examples of an association between DNA methylation and transcriptional silencing,31,43,44,119 so too were they the first target for clinical intervention based on drugs that affect methylation.120-122 Treatment with 5-azacytidine, an irreversible inhibitor of DNA-MTase, was shown to increase expression of the fetal γ-globin gene in nonhuman primates and subsequently in patients with β-thalassemia and sickle cell anemia. Because of its mutagenicity and the observation that other S-phase active cytotoxic agents that do not inhibit DNA methylation could induce similar increases in γ-globin gene expression,123-1255-azacytidine has not been widely used for this application. These experiences point to the limitations of attempting to alter gene expression through the use of global DNA methylation inhibitors that also possess other potent cellular effects and emphasize the need for a more complete understanding of the specificity of DNA methylation and demethylation control.
The recent advances in understanding of altered DNA methylation in cancer discussed above also have potential clinical implications. Because methylation of many involved genes may represent a process specific to neoplastic cells, this change may be a sensitive index of micrometastases.86 It may also be of some prognostic value in certain situations. For instance, methylation of the ablpromoter in chronic myeloid leukemia has been associated with disease of long-standing duration, most likely associated with a higher probability of imminent blast transformation.126
The increased DNA-MTase activity seen in multiple cancers has prompted targeting of the inhibition of this enzyme as an anticancer strategy. The DNA-MTase inhibitors, 5-azacytidine and 5-azadeoxycytidine, have been used clinically for the treatment of patients with myelodysplastic syndromes or leukemia.127,128 Because, as noted, these agents have effects other than inducing demethylation, have many side effects, and need to be administered by continuous infusion, efforts to develop novel DNA-MTase inhibitors are clearly warranted. In one such approach, Ramchandani et al129 demonstrated that intraperitoneal injection of DNA-MTase antisense oligonucleotides reduced the level of DNA-MTase and inhibited the growth of Y1 adrenocortical carcinoma in syngeneic LAF mice.
THE ROLE OF DNA METHYLATION IN EVOLUTION
Two hypotheses have been proposed for the evolutionary role of DNA methylation. The first hypothesis is derived from the perspective of gene numbers and biologic complexity.12 The size of the genome in free-living organisms has increased from a few thousand genes in prokaryotes (eg, Escherichia coli with 4,000 genes) to 7,000 to 25,000 genes in nonvertebrate eukaryotes and 50,000 to 100,000 genes in vertebrates. Bird12 has proposed that, if the number of tissue-specific genes is to increase during evolution, the efficiency of gene repression must be high. This repression, or transcriptional noise reduction, in eukaryotes has been attributed to two features (the nuclear envelope and histones) that are present in eukaryotes but not in prokaryotes. Vertebrates, in turn, have several fold more genes than the nonvertebrate eukaryotes, and one possible additional repression mechanism for vertebrates could be DNA methylation. A comparison of methylation patterns in invertebrates versus vertebrates shows some important differences. In the invertebrates, methylation of cytosine occurs at only a minor fraction of the CpG dinucleotides in the genome and in some cases, such asDrosophila, cannot be detected at all. It is likely that, in the vast majority of eukaryotes, DNA methylation functions as part of a system that silences potentially damaging DNA elements such as transposons, viral genomes, etc. However, vertebrates have the bulk of their DNA methylated, except for CpG islands. Bird12proposes that DNA methylation in vertebrates provides a novel layer of global repression, further reducing the transcriptional noise and thereby allowing vertebrates to accumulate and selectively use the extra genes that are crucial to their development.
The alternative theory proposed is that cytosine methylation in mammals is a nuclear host-defense system that evolved primarily to counter the threats posed by endogenous parasitic mobile genetic elements.5 Cytosine methylation inactivates the promoter of most viruses and transposons, including retroviruses and Alu elements, and such sequences are methylated in the DNA of differentiated cells. In fact, the large majority of 5-mCs in the genome lie within these elements. Demethylating drugs have been shown to activate transcription of endogenous transposons.130 Also, cloned, unmethylated human L1 elements transpose at a higher rate in transfected human cells,131 and this transposition rate is far in excess of the rate of the identical but methylated endogenous elements. In an interspecific mammalian hybrid involving Macropus eugenii andWallabia bicolor, the occurrence of genome-wide undermethylation, retroviral element amplification, and chromosome remodeling has been demonstrated.114 Yoder et al5 also argue that there is little compelling evidence for a role for reversible promoter methylation in developmental gene control. However, our laboratory has shown that methylation does appear to play a role in specific developmental gene control.52-54
METHYLATION AND FOREIGN GENE SILENCING
In both cultured cells transfected with foreign DNA and transgenic organisms, the newly integrated foreign DNA frequently becomes de novo methylated.132 It has been proposed that de novo methylation constitutes a cellular defense mechanism to silence integrated foreign DNA or genes.132
Evidence over the past several years suggests that DNA methylation is involved in the inactivation of virally introduced genes in vivo. The MoMuLV has been shown to be completely inactive in embryonic stem cell and embryonic carcinoma cell lines, and the inactivity is accompanied by de novo methylation of the proviral sequences.133,134Orend et al135 have shown that, upon integration, de novo methylation spreads from the center of the integrated collinear viral DNA. Methylation of specific sites in the adenoviral promoter results in promoter inactivation.136 Herpes virus also undergoes de novo methylation in mammalian cells.137 Epstein-Barr virus DNA has been found to be methylated in the normal lymphocytes of healthy volunteers.138
In transduced murine hematopoietic cells, transcriptional inactivation of the proviral MoMuLV-LTR was shown to be associated with methylation of the proviral-LTR.139 We have recently demonstrated that an in vitro-methylated retroviral LTR fragment containing 13 CpG dinucleotides was able to compete in binding assays for an MeCP complex that has a mobility similar to MeCP-1,140 although the role of methylation in LTR silencing remains unresolved.
Methylation-mediated inactivation of foreign gene expression in specific cell types has important therapeutic and pharmacological implications in that inhibition of the methylation of therapeutically introduced genes might enhance gene therapy significantly by preventing transcriptional silencing.
CONCLUSION
DNA methylation clearly plays an important regulatory role in vertebrates, as evidenced by its vital role in embryonic development. Whether the primary evolutionary role of DNA methylation is through transcriptional silencing or as a host defense system against endogenous and exogenous parasitic sequence elements remains to be fully determined. However, there is substantial evidence that DNA methylation plays a critical role in silencing specific genes during development and cell differentiation. The intrinsic mutagenicity of 5-mC, activation of proto-oncogenes through hypomethylation, transcriptional inactivation of tumor-suppressor genes through hypermethylation, and defects in chromosomal segregation due to failure of de novo methylation may all contribute to neoplasia. Selective modulation of DNA methylation may therefore have important clinical implications for the prevention and treatment of cancer. To develop safe and effective strategies for therapeutic alteration of DNA methylation, the factors that regulate the specificity of both the methylation and demethylation processes must be more fully understood. Likewise, understanding the factors involved in DNA methylation-induced gene silencing will facilitate attempts to selectively affect gene expression. Recent studies have linked two global mechanisms of gene regulation, DNA methylation, and histone deacetylation.61 62 Further investigations are necessary to understand the complex links between the methyltransferases, demethylases, methyl cytosine binding proteins, histone acetylation, and the transcriptional activity of genes.
ACKNOWLEDGMENT
The authors acknowledge the helpful assistance of Catharine W. Tucker in preparing their manuscript and thank Dr Steven Snyder for comments.
Supported by National Institutes of Health Grant No. DK29902 and by the Massey Cancer Center (G.D.G.) and the Feist-Weiller Cancer Center (R.S.).
REFERENCES
Author notes
Address reprint requests to Gordon D. Ginder, MD, Professor of Internal Medicine and Human Genetics, Director, Massey Cancer Center, Virginia Commonwealth University, 401 College St, PO Box 980037, Richmond, VA 23298-0037; e-mail: gginder@mcc1.mcc.vcu.edu.

![Fig. 2. (A) Illustration of the steps involved in determining the methylation status of cytosines in a known DNA sequence by the bisulfite conversion method. (B) In vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are only seen in adult erythroid cells. Positions indicated are relative to the transcription start site. Cytosines that are not associated with CpG dinucleotides (sequence shown in [C]) have all been converted to thymidines in both 5-day and adult erythroid cells. (C) Graphic representation of the in vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are clearly seen in (A) (data not shown for CpG dinucleotide at position −15). Primers R and F indicate the sequence of rho globin promoter used for designating internal primers and for sequencing.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/12/10.1182_blood.v93.12.4059/4/m_blod41240002ax.jpeg?Expires=1769407355&Signature=ZoZy~iinYjHw0oqM8QJ6HFBLXiZCK330bjSyZUcBD2LgJ-WgxkanRoPkwrH9VjFia~Ntv1EzDZslVb49aTiEQnf9En~3BMUWAfvDdTuKX2IHUJ-RxmDFKNYostxHjPPmYBJ7jms72I1Ebh5D5U1lXMHnsA5-QmirwdeHsi4ux-Q6M5AldAiqvIMXY77TIK-wN99Wx6D1brgJaBxp3ZkKT7DQDeqS3kpVBGFuh7HEkaTOcudTPbKl8La0d9xkuPEDpquYmmu1lwmv1w4M-mt4C3KXNEB~foGpwlj4xfCWSD2lfFpdqJIOwU6YNqInUue46v~mYqTOWDzXq3AM1zL-IQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. (A) Illustration of the steps involved in determining the methylation status of cytosines in a known DNA sequence by the bisulfite conversion method. (B) In vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are only seen in adult erythroid cells. Positions indicated are relative to the transcription start site. Cytosines that are not associated with CpG dinucleotides (sequence shown in [C]) have all been converted to thymidines in both 5-day and adult erythroid cells. (C) Graphic representation of the in vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are clearly seen in (A) (data not shown for CpG dinucleotide at position −15). Primers R and F indicate the sequence of rho globin promoter used for designating internal primers and for sequencing.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/12/10.1182_blood.v93.12.4059/4/m_blod41240002bw.jpeg?Expires=1769407355&Signature=EwZ3KtTscUqW~jqfvgnsZDz7Dw03d5Rx~hP9W7vNdrPv-C74xDZV8hU4irIT0lB~qIwLdj4lTHcLZ0tfce0GALkcYyS2tjQeRrMmRIv~h7Wf-iN~HJGWVlHpLffrWw8tMQ9bxyUHvAX3n08cGp-jsfpTfmoxjMoBX4A0-Di1~Jnn~~~1RtCCXs~aRXQmKEOymueAoi0i2MWj-ojWTN41jWtjRy1wuFG7G73mFytSbB4qLW4sKLFl2YiM2iI9InEJ6RXEDlc9yPgt3V0MyoIuXCqIYlggEXST4RXdsytA1mTsbzEhALYiUYqRGnuNLn72Bjg~CQFaCdb2niy6WtdmbQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 2. (A) Illustration of the steps involved in determining the methylation status of cytosines in a known DNA sequence by the bisulfite conversion method. (B) In vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are only seen in adult erythroid cells. Positions indicated are relative to the transcription start site. Cytosines that are not associated with CpG dinucleotides (sequence shown in [C]) have all been converted to thymidines in both 5-day and adult erythroid cells. (C) Graphic representation of the in vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are clearly seen in (A) (data not shown for CpG dinucleotide at position −15). Primers R and F indicate the sequence of rho globin promoter used for designating internal primers and for sequencing.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/12/10.1182_blood.v93.12.4059/4/m_blod41240002ax.jpeg?Expires=1769407356&Signature=x-TVF~Un49kLHpj10TRkh0e-g9PYk2X3wUHmsO7PzhO0J2G~V6YEUoeHCs8ikL68ipLMsrMiL-1wmxsQyHNy4ivbpLVVRXTcxY~bHijnIMdRuXrX-JVbK5sCxiDIc2opvTWogoq9Q-clDoaKFZYIpQwm-EHRAxGgIu2QxP4aK8RUrNVgTVd7s4LsRl8c9f-QSWvC0WT-Vy7fO0OYakiNub3MzYqF~pWr0KR8Rk5CPIVTiQ1vRvs5jb-oHXQ3BQtClT4ASh~Bdk~tw1pa12LrP8QifFBgQHYThfvEMOLj7m6kjMnGXJRwqeXGVw5eYU8holNdafnJYovNCPxUQslGaA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. (A) Illustration of the steps involved in determining the methylation status of cytosines in a known DNA sequence by the bisulfite conversion method. (B) In vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are only seen in adult erythroid cells. Positions indicated are relative to the transcription start site. Cytosines that are not associated with CpG dinucleotides (sequence shown in [C]) have all been converted to thymidines in both 5-day and adult erythroid cells. (C) Graphic representation of the in vivo methylation of CpG dinucleotides of rho promoter in 5-day and adult chicken erythroid cells using bisulfite conversion technique. Arrows indicate methylated cytosines and are clearly seen in (A) (data not shown for CpG dinucleotide at position −15). Primers R and F indicate the sequence of rho globin promoter used for designating internal primers and for sequencing.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/12/10.1182_blood.v93.12.4059/4/m_blod41240002bw.jpeg?Expires=1769407356&Signature=gBG4zJjDaujuxFPTdKdFfCjUY3ro0FrSx6oAUv-G2FlW23FJd47xgdr6Fwdm42sHhOq1l38heedcSGwPqTZ1vWC48jFKc5HdszudchujZqvGxsjhucayu5vyAE2xuiGyNbK-a6n6KPWhkWKRRp8X~2xIsGzPtWOQSb-RVN2xUrn6FM2yHUX94CDcSPglIw692TxOVazMK5RUwJOloQxX-wjxWIMrXK0yGu8tHDg76BedWfN0O7YvI~eCQqeO7-Z11PLXRCNnpTGg4ev0hV6lIbtfGrAZq-8Pva9AC-OXV5xFaoDwO9ZuAVObSmPYAG6kWOTa9-qUG6RerpawzjnUIQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)