Abstract
Cyclin D3 plays a major role in the development of polyploidy in megakaryocytes. The expression of cyclin D3 gene and the level of cyclin D3 protein are increased by the Mpl ligand in the Y10/L8057 megakaryocytic cell line, as indicated by Northern and Western blot analyses, and by nuclear run-on assays and transfection experiments with cyclin D3 promoter constructs. DNase I footprinting of the promoter region showed protected segments, at −75 to −60 bp and at −134 to −92 bp, which display binding sites for the Sp family of transcription factors. Gel mobility shift assay and supershifts with specific antibodies indicate that Sp1 binds to these regions in the cyclin D3 promoter and that Sp1 binding activity is significantly increased by Mpl ligand. Mutation of either Sp1 site both decreases the basal promoter activity and eliminates the induction by Mpl ligand. We find that the nonphosphorylated form of SP1 has greater affinity for the cyclin D3 promoter and that the majority of Sp1 in the cells is nonphosphorylated. Mpl ligand treatment results in increased levels of Sp1 protein, which also appears as nonphosphorylated. Okadaic acid, which inhibits protein phosphatase 1 (PP1) and shifts Sp1 to a phosphorylated form, decreases cyclin D3 gene expression and suppresses Mpl ligand induction. Our data point to the potential of Mpl ligand to activate at once several Sp1-dependent genes during megakaryopoiesis.
PROGRESSION THROUGH the proliferating eukaryotic cell cycle is controlled by cyclin-dependent kinases (Cdks). The activities of Cdks rely on the association with specific regulatory subunits, cyclins, to trigger different downstream processes of the cell cycle. D-type cyclins (D1, D2, and D3), in conjunction with their major catalytic partners Cdk2, Cdk4, Cdk5, and Cdk6, appear to govern progression through G1 phase in response to various extracellular signals, including growth factors.1,2 D-type cyclins are induced as part of the delayed early response to mitogenic stimulation by growth factors, such as colony-stimulating factor-1 (CSF-1), and are required for S-phase entry.3,4Overexpression of D-type cyclins accelerates progression through the G1 phase and reduces dependence on exogenous growth factors.5 The primary target of the cyclin D-Cdk complexes is thought to be the retinoblastoma protein (pRB) and/or related proteins p107 and p130.6 The phosphorylation state of pRB, in turn, has been shown to regulate transcription mediated by specific transcription factors such as E2F,7,8 ATF-2,9Myc,10,11 and Elf-1.12 These transcription factors have been shown to induce the expression of several genes that are required for DNA replication in S phase. However, the regulation of cyclin gene transcription remains poorly understood, with most attention to date focused on the posttranscriptional control of cyclin gene products.
Mature megakaryocytes, the precursors of circulating platelets, display nuclear polyploidization, increased cell size, and increased cytoplasmic mass.13 Megakaryocytopoiesis is regulated by a number of cytokines with either stimulatory or inhibitory effects.13 The physiological c-Mpl ligand, referred to as thrombopoietin (TPO), is a hematopoietic growth factor considered to be the primary regulator of megakaryocytopoiesis and thrombopoiesis.14 A truncated, PEGylated c-Mpl ligand that is related to TPO and has its biological activities is referred to as PEG-rHuMGDF.15,16 Mpl ligand has been reported to activate both JAK-STAT and Shc-MAPK signal transduction pathways.17-19 However, the involved signal transduction mechanisms are not fully understood, and these pathways are currently not well connected with gene regulation in megakaryocytes.
We previously reported that cyclin D3 expression is important for the production of large megakaryocytes in primary cultures of mouse bone marrow cells.20 We also found that the level of cyclin D3 mRNA in primary megakaryocytes is induced by the Mpl ligand.21 In the current study, we attempted to elucidate the mechanism through which cyclin D3 levels are regulated by the Mpl ligand in megakaryocytes. We found that the elevation of cyclin D3 mRNA is due to an increased rate of transcription and that the nonphosphorylated form of Sp1 transcription factor, which plays an important role in cyclin D3 gene expression, is detected at higher levels in cells treated with Mpl ligand.
MATERIALS AND METHODS
Cell cultures, acetylcholinesterase assay, and ploidy analysis.
L8057 is a murine megakaryocytic cell line22 obtained from Dr H. Sasaki (Yokohama City Medical College, Yokohama, Japan). Cells were maintained in F12 medium (GIBCO BRL, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal calf serum (GIBCO BRL). To select for cells with high differentiation potential by the Mpl ligand, L8057 cells were cultured in Iscove’s modification of Dulbecco’s medium (IMDM; GIBCO BRL) supplemented with 10% heat-inactivated fetal calf serum and subcloned by limiting dilution. Cloned cells were screened for an ability to express acetylcholinesterase (a unique marker for rodent megakaryocytes)23 in the presence of 25 ng/mL PEG-rHuMGDF (MGDF; generous gift of Amgen Inc, Thousand Oaks, CA). This compound was reported to have the biological features of native Mpl ligand, such as effects on megakaryopoiesis, ploidy, and platelet level.15,16 One of the subclones, Y10, showed strong induction of acetylcholinesterase activity and increased polyploidization in the presence of 25 ng/mL of MGDF. Acetylcholinesterase activity was assayed as described elsewhere.20 L8057 or Y10 cells were stained with propidium iodide and subjected to ploidy analysis as we described before.24 Time course and titration experiments with MGDF indicated that the Y10 cells reached the highest ploidy level only after 3 days of incubation with 25 ng/mL MGDF.25
Nuclei isolation and nuclear run-on assay.
Nuclei were prepared from Y10 cells cultured in the absence or presence of 25 ng/mL MGDF for 3 days as described elsewhere.26Isolated nuclei were frozen in liquid nitrogen in 50 mmol/L Tris, pH 8.3, 5 mmol/L MgCl2, 0.1 mmol/L EDTA, and 40% glycerol in aliquots of approximately 1 × 107 nuclei/100 μL. Nuclear run-on assay was performed essentially as described before,26 with some modifications. Briefly, 100 μL of 2× reaction buffer (10 mmol/L Tris, pH 8.0, 5 mmol/L MgCl2, 0.3 mol/L KCl, 1 mmol/L ATP, 1 mmol/L CTP, 1 mmol/L GTP, and 50 mmol/L dithiothreitol [DTT]) and 12.5 μL [α-32P]UTP (10 mCi/mL) were incubated with 100 μL nuclei for 30 minutes at 30°C. One hundred micrograms of yeast tRNA (GIBCO BRL) and 25 μg of DNase I (Sigma, St Louis, MO) were added into the reaction mixture and incubated for 10 minutes at 30°C. The nuclear run-on reaction was extracted with 1 mL Tri-Zol (GIBCO BRL) according to the manufacturer’s instructions. The resultant transcribed RNA was resuspended in 250 μL of a solution containing: 20 mmol/L HEPES, pH 7.5, 5 mmol/L EDTA, followed by treatment with 200 mmol/L NaOH for 10 minutes on ice. HEPES (free acid) at a concentration of 250 mmol/L was added to stop hydrolysis. pUC19 DNA and plasmids containing cDNA for cyclin D3, 18 S rRNA, each at 10 μg were applied to nitrocellulose with a slot-blot apparatus. The resultant transcription reactions (1 × 106 cpm/mL) were then added to prehybridized filters and hybridization was performed in 50% formamide, 5× Denhardt’s, 0.5% sodium dodecyl sulfate (SDS), 5× SSPE, and 200 μg/mL salmon sperm DNA at 42°C for 24 hours. The filters were washed twice with 1× SSC, 0.1% SDS for 20 minutes at 65°C and once with 0.1× SSC, 0.1% SDS for 20 minutes at 65°C and exposed on an X-ray film for up to 24 hours.
Plasmids.
pD3GH, the human growth hormone (HGH) reporter gene plasmid containing 1680 bp of mouse cyclin D3 promoter sequences, was generated as described before.27 The deletion constructs pD3-957 (with +1 being the transcription start site), pD3-446, pD3-190, pD3-99, and pD3-37 were generated by excising sequences between HindIII at −1691 and BamHI (−957), Xho I (−446),Xcm I (−190), Bsm I (−99), andBssHII (−37) from pD3GH plasmid, respectively. The Sp1 mutation constructs in the full promoter construct pD3GH were pD3-m1, pD3-m2, and pD3-m1/m2, in which Sp1 sites at positions −64 to −72, −121 to −129, or both were mutated, respectively. These constructs were made by polymerase chain reaction (PCR)-directed site-specific mutagenesis, using a procedure described elsewhere.28 The following are the primers used for each construct, with the mutated site underlined: pD3-m1 (TCGTCGCGAGGAATGGGGCGCCTGTCAGGG); pD3-m2 (TTCCGACTGCGGCCCCATTCCTTAGAACGTT); and pD3-m1/m2 (both primers used). All mutations were verified by DNA sequencing. The oligonucleotides that we used throughout this study were synthezied by GIBCO BRL. The expression vector pCMVSp1, which contains the Sp1 cDNA driven by the cytomegalovirus (CMV) promoter, is a generous gift of Robert Tjian (University of California at Berkeley, Berkeley, CA).
Transfection and reporter gene assays.
Y10 cells were stably transfected by electroporation as follows: the cells were washed with phosphate-buffered saline (PBS; 136 mmol/L NaCl, 8 mmol/L Na2HPO4, 2.6 mmol/L KCl, and 1.4 mmol/L KH2PO4, pH 7.4) and resuspended in electroporation buffer (30.8 mmol/L NaCl, 120.7 mmol/L KCl, 8.1 mmol/L Na2HPO4, 1.46 mmol/L KH2PO4, 5 mmol/L MgCl2). Each of the HGH gene reporter plasmids (100 μg) along with CMVβ-galactosidase control plasmid (pCMVβ-gal; 20 μg) and the pcDNA3 plasmid (10 μg) containing neomycin resistant gene (Invitrogen, San Diego, CA) were added to 1 × 107cells in 0.8 mL electroporation buffer. Electroporation was performed at a setting of 400 V and 500 μF, using the Bio-Rad Electroporator (Bio-Rad, Hercules, CA). Forty-eight hours after electroporation, the cells were cultured for 3 weeks in medium containing 400 μg/mL G418 (GIBCO BRL). The HGH expression assay was performed in medium of cells29 cultured in the absence or presence of 25 ng/mL MGDF for 3 days, unless otherwise indicated. Populations of cells transfected with two different deletion constructs were subcloned by limiting dilution. During preliminary experiments, we found that, for each clone derived, the level of expression of HGH was higher as compared with the expression in noncloned cells, but the relative levels of expression of the two constructs were similar in the cloned and noncloned cells. We thus continued to assay all constructs in noncloned cells. We resorted to stable transfection, because the efficiency of transient transfection of Y10 cells by any available method was very poor. In most experiments, we used electroporation for transfection, but in a few experiments we used the nonliposomal FuGENE transfection reagent (Boehringer Mannheim, Indianapolis, IN) as a method of transfection. It should be pointed out that MGDF increases the activity of CMV or SVTK promoters by about 1.2- to 1.6-fold (observed in at least 10 determinations). Our data on cyclin D3 promoter activity are normalizing for efficiency of transfection in control and in MGDF-treated cells, based on the values obtained for β-galactosidase activity driven by the CMV promoter. Thus, our presented values (see Figs 2 and 6A) may be an underestimation of the extent of induction of cyclin D3 by MGDF. We have elected doing so, rather than to multiply values by a constant factor, because the extent of induction of β-galactosidase varies somewhat between different sets of experiments. This way we are also certain that cyclin D3 promoter induction is real and not a result of a numerical manipulation.
Preparation of nuclear extracts and electrophoretic mobility shift assay (EMSA).
Nuclear proteins were extracted according to a method described before.30 Y10 cells were cultured in the absence or in the presence of MGDF (25 ng/mL) for 3 days, unless otherwise indicated, washed twice with PBS, and lysed in lysis buffer (10 mmol/L Tris, pH 7.4, 10 mmol/L NaCl, 3 mmol/L MgCl2 0.5% NP-40) for 5 minutes on ice. Cell extracts were then spun at 800g for 10 minutes to pellet the nuclei. An equal volume of nuclear binding protein extraction buffer (20 mmol/L HEPES, pH 7.9, 0.4 mol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 25% glycerol, 0.5 mmol/L DTT, 1.0 mmol/L phenylmethylsulfonyl fluoride [PMSF], 0.5 μg/mL of leupeptin, 2.0 μg/mL of aprotinin, and 0.7 μg/mL of pepstatin; all from Sigma) was added to the packed nuclei. The nuclear extraction mixtures were incubated on ice for 20 minutes, followed by centrifugation at 14,000 rpm in a microcentrifuge at 4°C. The extracts were aliquoted and stored at −80°C. Two to 10 μg of nuclear protein was mixed with 2 μg of poly(dI-dC)-poly(dI-dC) (Pharmacia, Piscataway, NJ) in 30 μL of binding buffer (20% glycerol, 20 mmol/L HEPES, pH 7.9, 4 mmol/L Tris, pH 7.9, 60 mmol/L KCl, 1 mmol/L EDTA, and 1 mmol/L DTT), along with approximately 40,000 to 100,000 cpm of end-labeled probe on ice for 30 minutes. The probe was labeled with γ-32P-ATP, using T4 polynucleotide kinase (New England Biolabs, Inc, Beverly, MA). In reactions using competitor DNA or oligonucleotides, a 50 molar excess of the indicated unlabeled competitor DNA was used. In antibody supershift experiments, 1 μg of anti-Sp1 or anti-Sp3 (Santa Cruz Biotechnology, Inc, San Diego, CA) was preincubated with the nuclear extract for 30 minutes on ice before adding the 32P-labeled probe. The reactions were then analyzed in a 4.5% nondenaturing polyacrylamide gel (Acrylamide:Bisacrylamide = 39:1). In the assays with purified Sp1, 10 μg bovine serum albumin was incubated in the reaction mixture. The sequences of all the oligonucleotides used for gel electromobility shift assays are as follows: oligomer −199/−170, GCCCAGCGGACCAATGGGGCTGCGGGGAGG; oligomer −183/−152, GGGCTGCGGGGAGGGAGGGAGGGTATAGCGCC; oligomer −139/−90, CCGACTGCGGCCCCGCCCCTTAGAACGTTGTGACGTAGGAGCATTCCACG; oligomer −139/−90mATF, CCGACTGCGGCCCCGCCCCTTAGAACGTTGAATACCAGGAGCATTCCACG; oligomer −139/−90mSp1, CCGACTGCGGCCCCATTCCTTAGAACGTTGTGACGTAGGAGCATTCCACG. When relevant, the mutations in putative binding sites are underlined. The oligonucleotides that we used throughout this study were synthezied by GIBCO BRL.
DNase I footprint analysis.
The DNA fragment containing the cyclin D3 promoter sequence from −446 to −37 was single end-labeled as follows: pD3GH was digested with either Xho I (−446) or BssHII (−37); end-labeled with α-32P-dATP or with dGTP and Klenow (New England Biolabs, Beverly, MA); digested with BssHII or Xho I, respectively; run on a polyacrylamide gel; and purified from the gel. The labeled fragments were incubated with nuclear extracts derived from Y10 cells cultured in the presence or in the absence of 25 ng/mL MGDF for 3 days. The nuclear extracts were dialyzed at 4°C against a buffer containing 20 mmol/L HEPES, pH 7.9, 20% glycerol, 100 mmol/L KCl, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, and 0.5 mmol/L PMSF. The DNase I footprinting reaction contained 50 μg of nuclear extract, 2 μg of poly(dI-dC)-poly(dI-dC), 10 mmol/L HEPES, pH 7.9, 10% glycerol, 50 mmol/L KCl, 1 mmol/L EDTA, 0.25 mmol/L PMSF, 0.25 mmol/L DTT, 1 μg calf thymus DNA (Boehringer Mannheim), and 10 to 25 fmol of one single end-labeled DNA probe (2 × 104 cpm) in a total volume of 50 μL. The reaction mixture was incubated for 60 minutes on ice, and an equal volume of solution containing 5 mmol/L CaCl2, 10 mmol/L MgCl2 was added. The mixture was left for 1 minute at room temperature before 0.1 U of DNase I (Boehringer Mannheim) was added. The mixture was incubated at room temperature for 1 minute and the reaction was immediately terminated by the addition of 90 μL of a stop solution (200 mmol/L NaCl, 30 mmol/L EDTA, 1% SDS, and 100 μg/mL yeast tRNA). After phenol/CHCl3 extraction and ethanol precipitation, the DNA samples were dissolved in 8 μL of the sequencing gel loading buffer, followed by analysis on a 6% DNA sequencing gel.
Western blot analysis.
Western blot analysis was performed as we previously described.24 Y10 cells were cultured in the absence or presence of 25 ng/mL MGDF, as indicated. Cells were lysed in lysis buffer (0.5% NP-40, 50 mmol/L Tris, pH 7.4, 250 mmol/L NaCl, 5 mmol/L EDTA, 1 mmol/L PMSF, 10 μg/mL n-tosyl-l-phenylalanine chloromethyl ketone [TPCK], and 10 μg/mL soybean trypsin inhibitor; all from Sigma). Ten micrograms of lysed proteins was separated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE; for cyclin D3) or by 7% SDS-PAGE (for Sp1) and electrophoretically transferred from the gel onto a nitrocellulose membrane (Schleicher & Schuell, Keene, NH) in a buffer containing 25 mmol/L Tris, 192 mmol/L glycine, and 20% methanol. For Sp1 Western blot analysis, 10 μg nuclear extracts derived from cells, which were cultured in the absence or presence of MGDF for 2 days before the addition of 100 nmol/L okadiac acid (OA; Sigma) for an additional 24 hours, were loaded per each lane. Because OA was found to be toxic to the cells during prolonged incubations (>24 hours), we resorted to the above-noted mode of addition of OA that resulted in no significant decrease in cell number.31,32 Cell number was determined with a hemocytometer and trypan blue exclusion. When indicated, the Western blot was incubated for 1 hour at room temperature with a cyclin D3 antibody (Pharmingen, San Diego, CA) or Sp1 antibody (Santa Cruz Biotechnology) followed by detection with the ECL system (Amersham Life Science Inc, Arlington, IL), as we previously described.24
Phosphorylation and dephosphorylation of Sp1.
A total of 0.25 footprint (fp) units of human recombinant Sp1 (hrSp1; Promega, Madison, WI) or 10 μg nuclear extract from Y10 cells was incubated with 1 U of casein kinase II (CKII; New England Biolabs) in a buffer containing 20 mmol/L Tris, pH 7.5, 50 mmol/L KCL, 10 mmol/L MgCl2, and 200 mmol/L ATP at 30°C for 30 minutes to allow Sp1 phosphorylation. For dephosphorylation, hrSp1 or nuclear extract was incubated with 1 U of protein phosphatase 1 (PP1; New England Biolabs) in the presence or absence of 1 μg PP1 inhibitor 2 (I-2; New England Biolabs) in a binding buffer (20% glycerol, 20 mmol/L HEPES, pH 7.9, 4 mmol/L Tris, pH 7.9, 60 mmol/L KCl, 1 mmol/L EDTA, and 1 mmol/L DTT) at 30°C for 30 minutes. CKII phosphorylated or PP1 dephosphorylated hrSp1 or nuclear extract was used for electromobility shift DNA binding assays, as described above.
RESULTS
Effect of the Mpl ligand on cyclin D3 gene expression in Y10 cells.
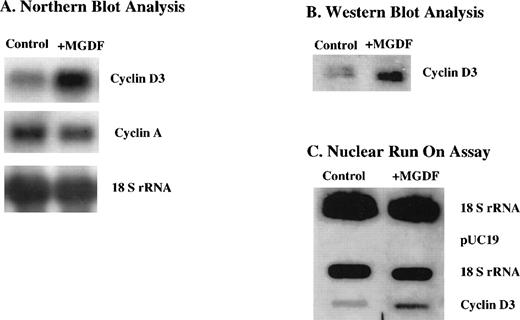
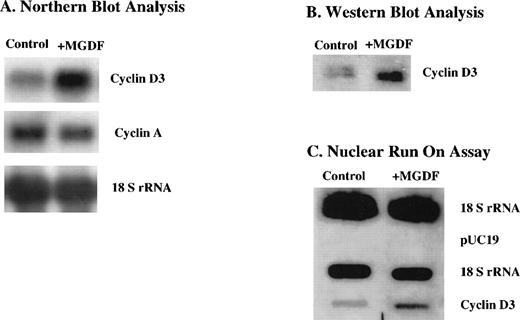
We have shown in a previous study that cyclin D3 mRNA expression is upregulated in megakaryocytes of mice injected with Mpl ligand.21 Because of the rarity of megakaryocytes in the marrow, these data were obtained by visualizing individual cells via in situ hybridization. To explore the mechanism by which cyclin D3 mRNA is induced by Mpl ligand, we resorted to a megakaryocytic cell line. In a recent study, we described the subcloning of the megakaryocytic cell line L8057, yielding Y10/L8057 cells that respond to Mpl ligand through increases in ploidy and other markers, such as acetylcholinesterase activity.25 Our current studies showed that cyclin D3 mRNA levels in Y10 cells were also induced by MGDF (∼3-fold induction derived from 3 experiments) as indicated by Northern blot analysis (Fig 1A). Similarly, cyclin D3 protein was also elevated (∼3-fold) as determined by Western blot analysis (Fig1B). It is interesting to note that, in contrast to cyclin D3, cyclin A mRNA levels were not induced by MGDF treatment of these cells (Fig 1A).
Cyclin D3 gene expression is upregulated by Mpl ligand in Y10 cells, a subclone of the mouse megakaryocytic cell line L8057, at the (A) mRNA level, (B) protein level, and (C) transcriptional level. Y10 cells were cultured in the presence or absence of MGDF, as detailed in Materials and Methods. (A) Northern blot analysis of 15 μg total RNA, using cDNAs of cyclin D3 and cyclin A as probes. Equal loading of RNA per lane was confirmed by probing the blot with cDNA encoding 18S rRNA. (B) Western blot analysis of 10 μg protein from whole cell lysates of Y10 cells, using a cyclin D3 antibody. (C) Nuclear run-on assays, performed as described in Materials and Methods. Ten micrograms of plasmid containing cDNA for cyclin D3, 2 μg of plasmid containing cDNA for 18 S rRNA, or 10 μg pUC 19 was applied to nitrocellulose. The intensities of the bands were quantitated using the Electrophoresis Documentation and Analysis System. The data presented are of a representative experiment out of three performed.
Cyclin D3 gene expression is upregulated by Mpl ligand in Y10 cells, a subclone of the mouse megakaryocytic cell line L8057, at the (A) mRNA level, (B) protein level, and (C) transcriptional level. Y10 cells were cultured in the presence or absence of MGDF, as detailed in Materials and Methods. (A) Northern blot analysis of 15 μg total RNA, using cDNAs of cyclin D3 and cyclin A as probes. Equal loading of RNA per lane was confirmed by probing the blot with cDNA encoding 18S rRNA. (B) Western blot analysis of 10 μg protein from whole cell lysates of Y10 cells, using a cyclin D3 antibody. (C) Nuclear run-on assays, performed as described in Materials and Methods. Ten micrograms of plasmid containing cDNA for cyclin D3, 2 μg of plasmid containing cDNA for 18 S rRNA, or 10 μg pUC 19 was applied to nitrocellulose. The intensities of the bands were quantitated using the Electrophoresis Documentation and Analysis System. The data presented are of a representative experiment out of three performed.
To determine whether the increase in the steady-state levels of cyclin D3 mRNA resulted from increased transcription of the cyclin D3 gene, nuclear run-on assays were performed on nuclei isolated from cells cultured in the absence or presence of MGDF. Figure 1C indicates that cyclin D3 gene expression is induced (∼3-fold) transcriptionally by MGDF in Y10 cells. Thus, transcriptional activation is at least partially responsible for the observed increase in cyclin D3 mRNA and protein upon MGDF treatment.
Effect of Mpl ligand on cyclin D3 promoter activity.
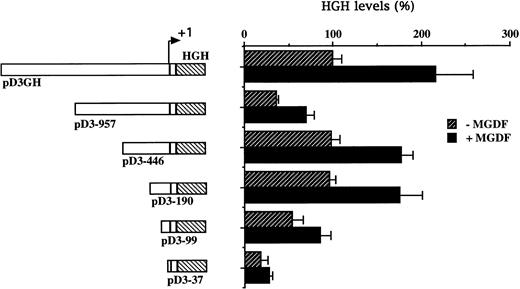
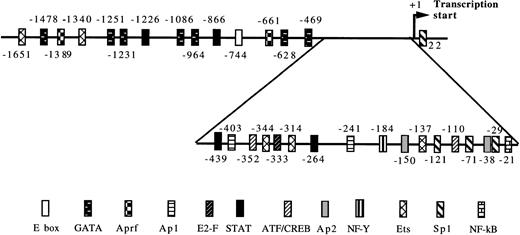
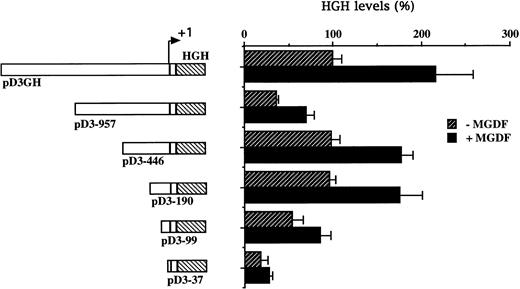
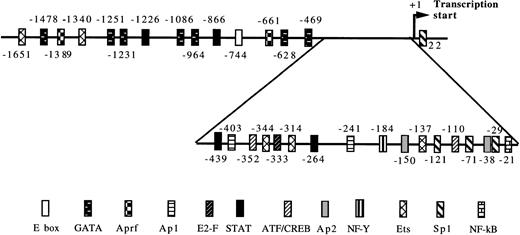
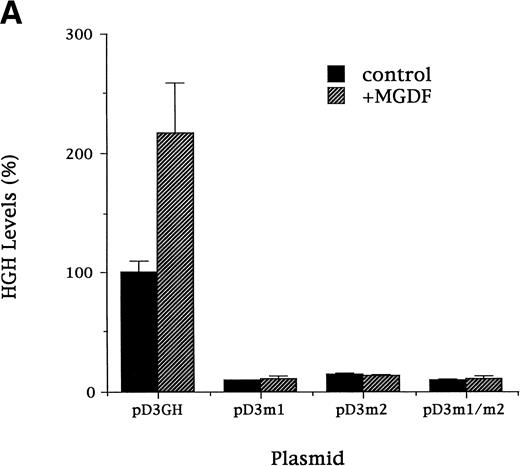
To explore the inducibility of the cyclin D3 promoter by Mpl ligand and to delineate its important regulatory sequences, we created a series of 5′-deletions of the promoter linked to the HGH reporter gene (Fig 2). When pD3GH, which includes the 5′ noncoding sequences from +85 to −1680, was stable transfected into Y10 cells, a twofold induction of HGH activity was observed upon MGDF treatment. This indicated the presence of an Mpl ligand-inducible element within this segment of the cyclin D3 promoter. When promoter deletion constructs were transfected into Y10 cells, the segment spanning the region from −190 bp to the transcription initiation site gave nearly full promoter activity (95%), as compared with the activity of the 1.68-kb promoter construct, whereas this activity was reduced to 18% when the promoter was truncated to −37. Deleting sequences from −190 to −99 reduced Mpl ligand induction significantly, indicating that an Mpl ligand-responsive element may be located in this region. The data shown in Fig 2 also indicate the presence of a negative regulatory element in the cyclin D3 promoter between −446 and −957 bp. The nature of this element will require further exploration in the future. The signaling pathways of c-mpl, the Mpl ligand receptor, in megakaryocytes are thought to involve the Janus family of nonreceptor tyrosine kinases (JAKs), as well as the signal transducers and activators of transcription (STATs).33 Phosphorylation of STATs by Janus kinases results in their translocation to the nucleus and subsequent binding to specific promoters.34 There are several putative STAT binding sites in the cyclin D3 promoter, including the putative STAT1 or STAT 3 sequence TTATGGGAA at −264 and the putative STAT 1 or STAT 5 sequence TTCCTAGAA at −439 (Fig 3). However, none of these sites is located in the region between −190 and the transcription start site.
Cyclin D3 promoter activity is induced by Mpl ligand. A series of 5′-deletions of the cyclin D3 promoter linked to the HGH reporter gene was stably transfected into Y10 cells, which were subsequently cultured in the absence or presence of MGDF as detailed in Materials and Methods. Schematic representation of respective reporter gene constructs is shown adjacent to the presentation of the corresponding HGH levels. The constructs are designated by numbers corresponding to the length of cyclin D3 sequences upstream of the transcriptional start site. The HGH level produced by cells transfected with pD3GH and cultured in the absence of MGDF was designated as 100%. All experiments were normalized for transfection efficiency by cotransfection with pCMVβ-galactosidase, as described in Materials and Methods. The data are the means of four determinations, with standard deviations indicated by the error bars.
Cyclin D3 promoter activity is induced by Mpl ligand. A series of 5′-deletions of the cyclin D3 promoter linked to the HGH reporter gene was stably transfected into Y10 cells, which were subsequently cultured in the absence or presence of MGDF as detailed in Materials and Methods. Schematic representation of respective reporter gene constructs is shown adjacent to the presentation of the corresponding HGH levels. The constructs are designated by numbers corresponding to the length of cyclin D3 sequences upstream of the transcriptional start site. The HGH level produced by cells transfected with pD3GH and cultured in the absence of MGDF was designated as 100%. All experiments were normalized for transfection efficiency by cotransfection with pCMVβ-galactosidase, as described in Materials and Methods. The data are the means of four determinations, with standard deviations indicated by the error bars.
Putative transcription factor binding sites in the cyclin D3 promoter. The numbers indicate the 5′ end of the indicated transcription factor recognition site in relation to the transcriptional start (+1).27
Putative transcription factor binding sites in the cyclin D3 promoter. The numbers indicate the 5′ end of the indicated transcription factor recognition site in relation to the transcriptional start (+1).27
Identification of nuclear factor binding sites by DNase I footprinting in the cyclin D3 promoter.
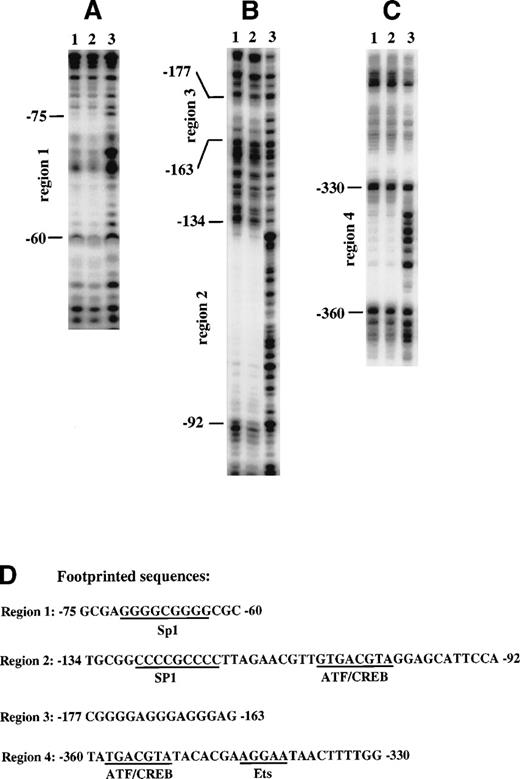
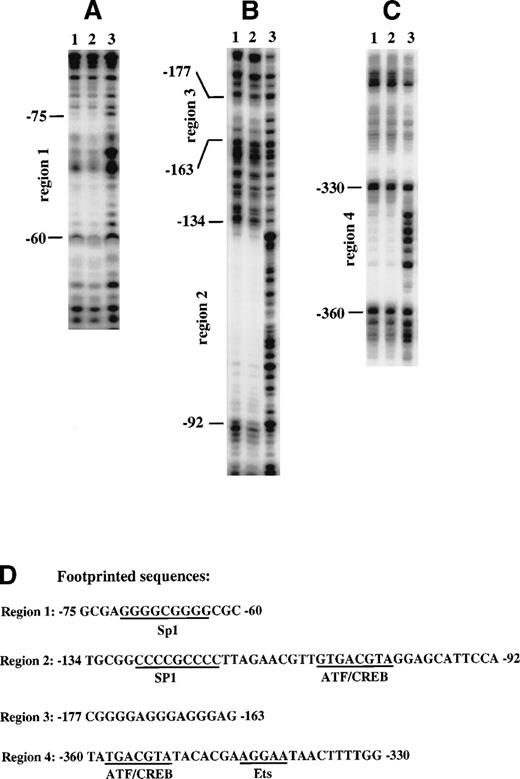
To identify nuclear factor binding sites within the region of the cyclin D3 promoter that responds to Mpl ligand, a DNA fragment containing sequences from −446 to −37 was analyzed by DNase I footprinting assays with nuclear extract from Y10 cells. Four footprinted areas were detected. Two of them, from −75 to −60 (Fig 4A) and from −134 to −92 (Fig 4B), were found to contain potential binding sites for Sp1 (Fig 4D), and both were within the region −190 to −37, which was shown to contain positive regulatory elements (Fig 2). The area from −134 to −92 also contains a putative ATF/CREB binding site. The third protected area, extending from −177 to −163, is a GA-rich region (Fig 4B). The fourth protected area, extending from −360 to −330 (Fig 4C), contains a putative ATF/CREB binding site and a putative Ets binding site (Fig 4D).
Identification of nuclear factor binding sites by DNase I footprinting of the cyclin D3 promoter. A DNA fragment from the cyclin D3 gene promoter spanning sequences from −446 to −43 was end-labeled either at −43 (A and B) or at −446 (C). The fragment was incubated with 50 μg nuclear extract from Y10 cells cultured in the absence (lane 1) or presence (lane 2) of MGDF or with no nuclear extract (lane 3) and then digested with 1 U/mL (lane 1 and 2) or 0.1 U/mL (lane 3) of DNase. The protected sequences were identified by a sequencing reaction using primers end-labeled either at −37 or at −446. The nucleotide positions are indicated on the left. The sequences of the protected regions and nuclear factor putative binding sites are shown in (D). The data presented are of a representative experiment out of three performed.
Identification of nuclear factor binding sites by DNase I footprinting of the cyclin D3 promoter. A DNA fragment from the cyclin D3 gene promoter spanning sequences from −446 to −43 was end-labeled either at −43 (A and B) or at −446 (C). The fragment was incubated with 50 μg nuclear extract from Y10 cells cultured in the absence (lane 1) or presence (lane 2) of MGDF or with no nuclear extract (lane 3) and then digested with 1 U/mL (lane 1 and 2) or 0.1 U/mL (lane 3) of DNase. The protected sequences were identified by a sequencing reaction using primers end-labeled either at −37 or at −446. The nucleotide positions are indicated on the left. The sequences of the protected regions and nuclear factor putative binding sites are shown in (D). The data presented are of a representative experiment out of three performed.
Induction of Sp1 binding to the cyclin D3 promoter by Mpl ligand.
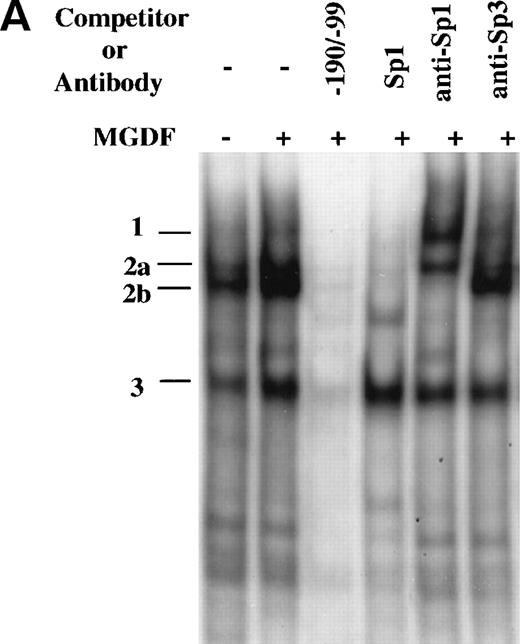
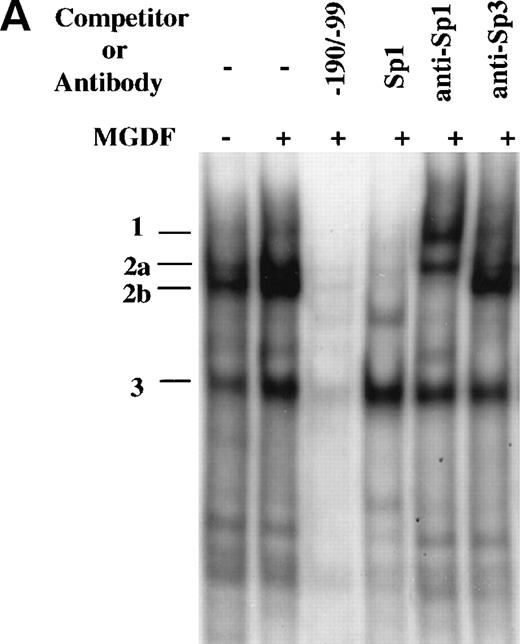
As indicated above, only the regions corresponding to SP1 and ATF/CREB consensus binding sites were found to be protected by footprint analysis of the region −190 to −99. This assay, which is not quantitative, does not allow a detection of clear differences in binding to the same DNA region by different samples. To further explore the interaction of proteins with this promoter region, we used a DNA fragment that extends from −190 to −99 (−190/−99) in a gel mobility shift assay. Several distinctive DNA-protein complexes were observed when this probe was incubated with nuclear extracts prepared from Y10 cells cultured in the absence or presence of MGDF, of which the major ones were designated as complex number 1, 2, and 3 (Fig 5A). These complexes were specific for the probe, because they were competed away by an excess of identical unlabeled competitor (Fig 5A), but not by several unlabeled random synthetic oligonucleotides (data not shown). Complexes 1, 2a, and 2b, but not complex 3, were competed by an unlabeled consensus Sp1 oligonucleotides. Because the Sp1 oligomers can bind to Sp1 as well as to other members of the Sp family of transcription factors, we identified protein components of these complexes through the use of specific antibodies in supershift assays. As shown in Fig5A, complex 2 was clearly supershifted by anti-Sp1, whereas the mobility of complex 2a was marginally affected by anti-Sp3. This indicated that Sp1 is a major component of complex 2. Of great interest is the observation that MGDF significantly enhanced the appearance of complex 2. It should be pointed out that, whereas there was a consistent enhancement by MGDF of complex 2 formation (an average of 2.5-fold derived from 4 experiments), the induction of complex 3 was variable, ranging from 1.5- to 2.5-fold over four experiments. All data were quantitated using the Electrophoresis Documentation and Analysis System (Eastman Kodak, Rochester, NY).
Induction of Sp1 binding activity to cyclin D3 promoter by Mpl ligand. (A) A DNA fragment that extends from −190 to −99 bp was used in a gel mobility shift assay with nuclear extracts derived from Y10 cells cultured in the absence or presence of MGDF, as detailed in Materials and Methods. The resulting major distinctive protein complexes were indicated as 1, 2a, 2b, and 3. A 50-fold molar excess of unlabeled DNA fragment (−190/−99) or Sp1 consensus oligonucleotides were added as indicated. The sequence of the oligomer containing the Sp1 consensus site (underlined) was 5′ATTCGATCGGGGCGGGGCGAGC3′. Supershift analysis was conducted by incubating the reaction mix with Sp1 or Sp3 antibodies. Similar competition and supershift assays were also performed with nuclear extracts derived from cells not treated with MGDF, yielding data similar to those described in this figure (not shown). The intensities of the bands were quantitated using the Electrophoresis Documentation and Analysis System. (B) The larger DNA fragment spanning the region −190 to −37 (−190/−37wt) was used as a probe for gel mobility shift assay along with nuclear extract derived from Y10 cells treated with MGDF. A 50-fold molar excess of unlabeled −190/−37(Wt) DNA fragment was added as a competitor and a Sp1 antibody was added when indicated. Similar shifts were conducted with probes containing mutations in one (−190/37m1 or −190/37m2) or both (−190/−37m1/m2) of the Sp1 sites at −64 to −72 and −121 to −129, respectively. The nucleotides in each mutated Sp1 site are GGAATGGGG (as compared with GGGGCGGGG in wild-type). The intensities of the bands were quantitated using the Electrophoresis Documentation and Analysis System, indicating a 2.7-fold increase in the extent of Sp1 binding in extracts derived from MGDF-treated cells, as compared with control nontreated cells (not shown). (C) Oligonucleotides spanning the region from −139 to −90, in which the ATF/CREB site at position −103 to −110 was mutated (−139/−90mATF), or oligonucleotides spanning the region from −139 to −90, in which the Sp1 site at −121 to −129 was mutated (−139/−90mSp1), and oligonucleotides extending from −139 to −90 (−139/−90) were used as probes in gel mobility shift assays. The nature of the mutations is described in Materials and Methods. A 50-fold molar excess of unlabeled Sp1 consensus oligonucleotides or 1 μg anti-Sp1 was added as indicated. The intensities of the bands were quantitated using the Electrophoresis Documentation and Analysis System, indicating no significant change in the extent of ETF/CREB binding in extracts derived from MGDF-treated cells, as compared with control nontreated cells (not shown). The data shown are taken from a representative experiment of four performed.
Induction of Sp1 binding activity to cyclin D3 promoter by Mpl ligand. (A) A DNA fragment that extends from −190 to −99 bp was used in a gel mobility shift assay with nuclear extracts derived from Y10 cells cultured in the absence or presence of MGDF, as detailed in Materials and Methods. The resulting major distinctive protein complexes were indicated as 1, 2a, 2b, and 3. A 50-fold molar excess of unlabeled DNA fragment (−190/−99) or Sp1 consensus oligonucleotides were added as indicated. The sequence of the oligomer containing the Sp1 consensus site (underlined) was 5′ATTCGATCGGGGCGGGGCGAGC3′. Supershift analysis was conducted by incubating the reaction mix with Sp1 or Sp3 antibodies. Similar competition and supershift assays were also performed with nuclear extracts derived from cells not treated with MGDF, yielding data similar to those described in this figure (not shown). The intensities of the bands were quantitated using the Electrophoresis Documentation and Analysis System. (B) The larger DNA fragment spanning the region −190 to −37 (−190/−37wt) was used as a probe for gel mobility shift assay along with nuclear extract derived from Y10 cells treated with MGDF. A 50-fold molar excess of unlabeled −190/−37(Wt) DNA fragment was added as a competitor and a Sp1 antibody was added when indicated. Similar shifts were conducted with probes containing mutations in one (−190/37m1 or −190/37m2) or both (−190/−37m1/m2) of the Sp1 sites at −64 to −72 and −121 to −129, respectively. The nucleotides in each mutated Sp1 site are GGAATGGGG (as compared with GGGGCGGGG in wild-type). The intensities of the bands were quantitated using the Electrophoresis Documentation and Analysis System, indicating a 2.7-fold increase in the extent of Sp1 binding in extracts derived from MGDF-treated cells, as compared with control nontreated cells (not shown). (C) Oligonucleotides spanning the region from −139 to −90, in which the ATF/CREB site at position −103 to −110 was mutated (−139/−90mATF), or oligonucleotides spanning the region from −139 to −90, in which the Sp1 site at −121 to −129 was mutated (−139/−90mSp1), and oligonucleotides extending from −139 to −90 (−139/−90) were used as probes in gel mobility shift assays. The nature of the mutations is described in Materials and Methods. A 50-fold molar excess of unlabeled Sp1 consensus oligonucleotides or 1 μg anti-Sp1 was added as indicated. The intensities of the bands were quantitated using the Electrophoresis Documentation and Analysis System, indicating no significant change in the extent of ETF/CREB binding in extracts derived from MGDF-treated cells, as compared with control nontreated cells (not shown). The data shown are taken from a representative experiment of four performed.
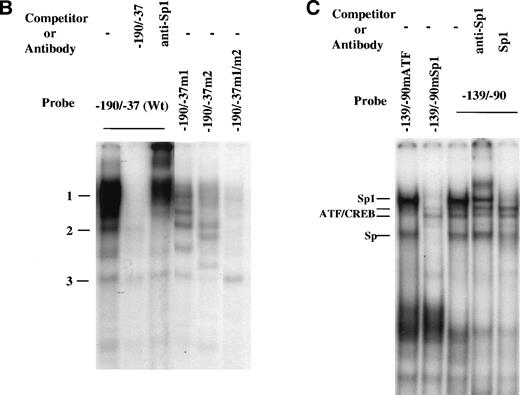
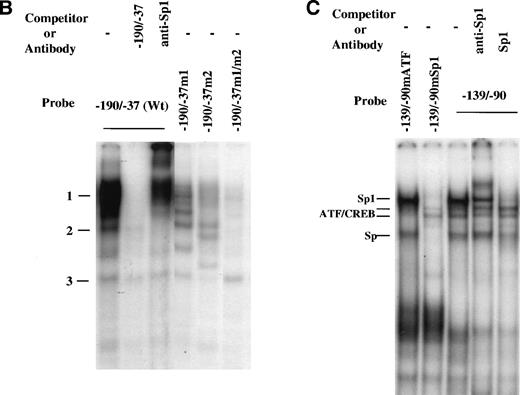
Whereas specific nuclear factors may bind to short segments in DNA, thus forming low molecular weight complexes, larger complexes may form in the context of longer promoter regions, potentially due to the interaction of proteins once bound to looped regions. Thus, we also used the entire region spanning −190 to −37, which contains two Sp1 binding sites (Fig 4), for gel mobility shift assay (Fig 5B). Similar induction of DNA binding by MGDF was found using this probe. However, under these conditions, the intensity of the bands supershifted by anti-Sp1 was significantly increased (by ∼4-fold; compare Fig 5B with 5A). Similar shifts were conducted with probes having the Sp1 binding site either at −64 to −72 (−190/−37m1) or at −121 to −129 (−190/−37m2) mutated to verify if all the shifts observed resulted from binding to Sp1 sites. Mutation of either Sp1 site significantly reduced (by ∼70% to 80%) the nuclear proteins binding to the region supershifted by anti-Sp1. When both Sp1 sites were mutated (−190/−37m1/m2), almost all binding of DNA-protein complexes disappeared, except for complex 3. This is consistent with the footprinting results (Fig 4A and B), suggesting that Sp1 binds to both Sp1 binding sites.
Binding of proteins to ATF/CREB consensus sites in the cyclin D3 promoter.
Footprinting results also suggested that a putative ATF/CREB binding site (starting at position –103) may be important for Mpl ligand induction of cyclin D3 promoter activity, because this site was within the region found by transfection studies to be important for this ligand effect. To confirm binding of proteins to the putative ATF/CREB site, an oligonucleotide spanning a sequence that covers footprinted region 2 (see Fig 4D), which contains a Sp1 binding site and a putative ATF/CREB site, was used in gel mobility shift analysis (probe −139/−90; Fig 5C). Four major bands were detected in this assay. Sp1 consensus oligomers competed complexes designated in Fig 5C as Sp1 and Sp, and anti-Sp1 supershifted the major complex, designated as Sp1. Oligonucleotides −139/−90, in which the ATF/CREB site at −110 was mutated but the Sp1 site was kept intact (−139/−90mATF), and −139/−90, in which the Sp1 site at −121 was mutated but the ATF/CREB site was kept intact (−139/−90mSp1), were also used as probes for gel mobility shift assay. When −139/−90mSp1 was used as a probe, none of the complexes labeled as Sp was formed, but a very weak complex designated as ATF/CREB became apparent. This latter complex was competed away by an excess (50-fold) of unlabeled −139/−90mSp1 or of unlabeled ATF/CREB oligomer (TGTGACGTAG), but not by an excess of unlabeled −139/−90mATF (not shown). With −139/−90mATF as a probe, the Sp complexes were formed, but the one designated as ATF/CREB was absent (Fig 5C). These data indicate that a very weak binding occurs at this consensus ATF/CREB binding site or that a binding occurs at a different site within the region –139/−90 with full dependency on an intact ATF/CREB consensus site. Our data also indicate that this association is not essential for the binding of Sp1 transcription factor to its consensus site.
Mutation of Sp1 sites, but not of ATF/CREB site, decreases activation of the cyclin D3 promoter and Mpl ligand effects on this promoter.
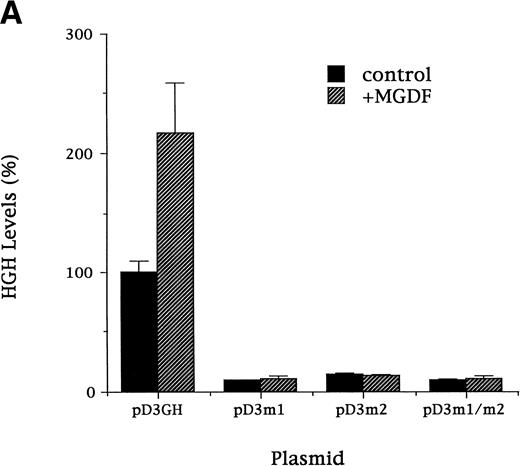
To confirm the importance of the Sp1 consensus sites for high activity of the mouse cyclin D3 promoter and for Mpl ligand effect, the following mutation constructs were made in the context of the full promoter (pD3GH): pD3-m1, pD3-m2, and pD3-m1/m2, which contain mutations in the Sp1 site at −64 to −72, at −121 to −129, or in both Sp1 sites, respectively. In each of four experiments, the mutation constructs were transfected into Y10 cells and assayed for HGH production in cells cultured for 3 days in the absence or presence of MGDF (Fig 6A). Each of these mutations reduced (by ∼80%) the basal transcriptional activity and significantly lessened MGDF induction. We are aware of the argument that inducibility may not be relevant when the basal promoter activity is eliminated, but we wish to emphasize that this was not the case in the Sp1 mutations. Approximately 20% of promoter activity was still detected in cells transfected with pD3m2. These results confirm the importance of each of the Sp1 sites at −64 to −72 or at −121 to −129 for basal promoter activity and for Mpl ligand effect. It should be pointed out that two experiments involving the use of a promoter in which the ATF/CREB site at −110 was mutated (to AATACC) indicated a reduced cyclin D3 promoter activity (25% reduction) in the absence of Mpl ligand, but the stimulatory effect of Mpl ligand was not altered (not shown).
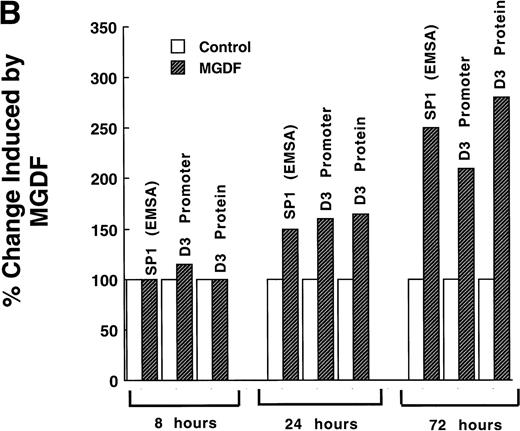
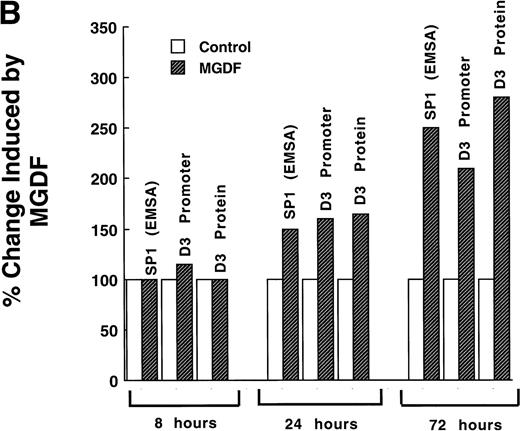
(A). Mutation of Sp1 sites decreases activation of the cyclin D3 promoter. Three mutation constructs were made in the context of the full promoter (pD3GH) as follows: pD3-m1, pD3-m2, and pD3-m1/m2, which contain mutations in the Sp1 site at −64 to −72, at −121 to −129, or in both Sp1 sites, respectively. The nucleotides mutated in each Sp1 site are as described in the legend to Fig 5. The mutation constructs were transfected into Y10 cells, which were then cultured in the presence or absence of MGDF and assayed for HGH production, as detailed in Materials and Methods. All experiments were normalized for transfection efficiency by cotransfection with pCMVβ-galactosidase, as described in Materials and Methods. The data are the means of four determinations with standard deviations indicated by the error bars. (B) Sp1 binding-ability, cylin D3 promoter activity, and cyclin D3 levels in cells cultured with MGDF for different times. Sp1 binding ability to the fragment –190/−99 of the cyclin D3 promoter was determined by EMSA, as in Fig 5. Cyclin D3 promoter activity (pD3GH) and cyclin D3 protein level were determined as in Figs 2 and 1, respectively. The data shown are of a representative experiment out of two performed. The intensities of the bands (in EMSA and Western blots) were quantitated using the Electrophoresis Documentation and Analysis System.
(A). Mutation of Sp1 sites decreases activation of the cyclin D3 promoter. Three mutation constructs were made in the context of the full promoter (pD3GH) as follows: pD3-m1, pD3-m2, and pD3-m1/m2, which contain mutations in the Sp1 site at −64 to −72, at −121 to −129, or in both Sp1 sites, respectively. The nucleotides mutated in each Sp1 site are as described in the legend to Fig 5. The mutation constructs were transfected into Y10 cells, which were then cultured in the presence or absence of MGDF and assayed for HGH production, as detailed in Materials and Methods. All experiments were normalized for transfection efficiency by cotransfection with pCMVβ-galactosidase, as described in Materials and Methods. The data are the means of four determinations with standard deviations indicated by the error bars. (B) Sp1 binding-ability, cylin D3 promoter activity, and cyclin D3 levels in cells cultured with MGDF for different times. Sp1 binding ability to the fragment –190/−99 of the cyclin D3 promoter was determined by EMSA, as in Fig 5. Cyclin D3 promoter activity (pD3GH) and cyclin D3 protein level were determined as in Figs 2 and 1, respectively. The data shown are of a representative experiment out of two performed. The intensities of the bands (in EMSA and Western blots) were quantitated using the Electrophoresis Documentation and Analysis System.
The data shown in Fig 6B indicate that the effect of MGDF on Sp1 binding ability to cyclin D3 promoter, on cyclin D3 promoter activity, and on the level of cyclin D3 protein are not immediate. A significant effect on all these parameters is observed only after 24 hours of exposure to MGDF, suggesting that in this case Sp1 is not activated due to an immediate postranslational modulation.
Mechanisms of effect of Mpl ligand on Sp1 binding to the cyclin D3 promoter.
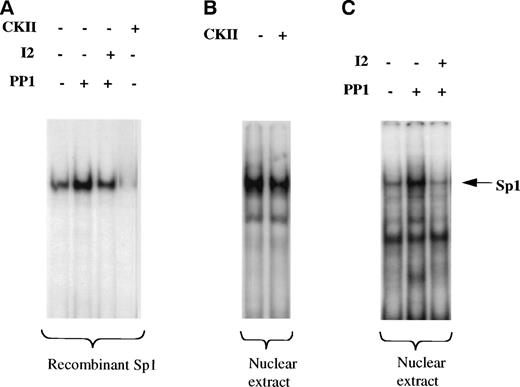
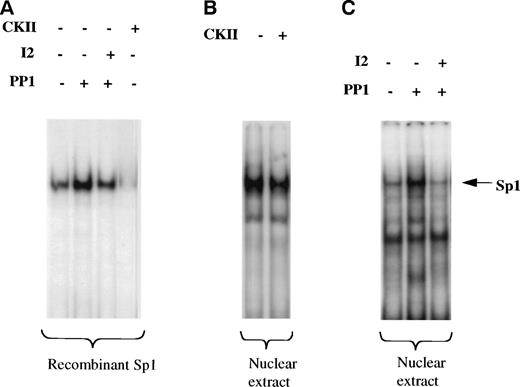
Sp1-mediated activation of several genes has been attributed to an increase in Sp1 level, to a change in glycosylation or phosphorylation state of SP1, or to cooperation with other factors.35 We first wished to establish if the phosphorylated or nonphosphorylated form of Sp1 binds to cyclin D3 promoter. As shown in Fig 7A, treatment of human recombinant Sp1, purified from HeLa cells (hrSp1), with casein kinase II (CKII), one of the enzymes known to phosphorylate Sp1,36 abolished the binding of Sp1 to the cyclin D3 promoter fragment −190/−99. Treatment of hrSp1 with protein phosphatase 1 (PP1) increased Sp1 binding activity, whereas the PP1 specific inhibitor 2 (I2), a 204 amino acid peptide, reduced the binding. Similarly, CKII reduced the binding of nuclear extract derived from Y10 cells to the cyclin D3 promoter region −190/−99 (Fig 7B). Also, PP1 and I2 had similar effects on Y10 nuclear extract (Fig 7C) as on hrSp1. This indicated that the nonphosphorylated form of Sp1 has higher binding activity towards the cyclin D3 promoter.
Nonphosphorylated Sp1 has higher DNA binding activity to the cyclin D3 promoter. A cyclin D3 DNA fragment that extends from −190 to −99 bp was used in a gel mobility shift assay. (A) Before being used in the gel mobility shift assay, human recombinant Sp1 (hrSp1) was subjected to dephosphorylation by protein phosphatase 1 (PP1) in the absence or presence of PP1 inhibitor 2 (I2) or to a phosphorylation reaction with casein kinase II (CKII), under conditions shown before to indeed affect the phosphorylation state of SP1 (as in Fig 8). (B) Nuclear extracts from Y10 cells were subjected to a kinase reaction with CKII, before being used in the gel mobility shift assay. (C) Nuclear extracts from Y10 cells were subjected to dephosphorylation with PP1 in the absence or presence of I2, before being used in the gel mobility shift assay. The buffer required for PP1 activity is different from that for CKII activity (see Materials and Methods), thus resulting in a somewhat different intensity of Sp1 binding under the two buffer conditions. The arrow points to the Sp1 complex, initially confirmed by supershift experiments with anti-Sp1 (as in Fig 5).
Nonphosphorylated Sp1 has higher DNA binding activity to the cyclin D3 promoter. A cyclin D3 DNA fragment that extends from −190 to −99 bp was used in a gel mobility shift assay. (A) Before being used in the gel mobility shift assay, human recombinant Sp1 (hrSp1) was subjected to dephosphorylation by protein phosphatase 1 (PP1) in the absence or presence of PP1 inhibitor 2 (I2) or to a phosphorylation reaction with casein kinase II (CKII), under conditions shown before to indeed affect the phosphorylation state of SP1 (as in Fig 8). (B) Nuclear extracts from Y10 cells were subjected to a kinase reaction with CKII, before being used in the gel mobility shift assay. (C) Nuclear extracts from Y10 cells were subjected to dephosphorylation with PP1 in the absence or presence of I2, before being used in the gel mobility shift assay. The buffer required for PP1 activity is different from that for CKII activity (see Materials and Methods), thus resulting in a somewhat different intensity of Sp1 binding under the two buffer conditions. The arrow points to the Sp1 complex, initially confirmed by supershift experiments with anti-Sp1 (as in Fig 5).
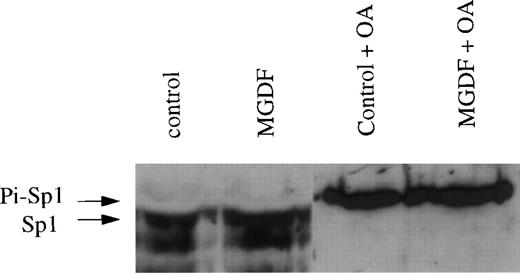
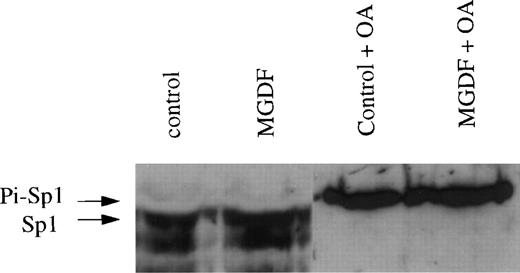
We next determined the levels of phosphorylated and nonphosphorylated Sp1 in nuclear extracts obtained from Y10 cells treated or not treated with Mpl ligand, using Western blot analysis. In other studies, it has been shown that phosphorylated Sp1 migrates on SDS-PAGE slightly slower (∼115 kD) than nonphosphorylated Sp1 (appearing as 105- and 95-kD proteins). In these studies, it has also been shown that okadaic acid (OA), which inhibits PP1, shifts Sp1 to the slower migrating, phosphorylated form.31,32 The OA-based determination of the relative migration of the phosphorylated and nonphosphorylated forms of Sp1 correlates well with data derived from experiments involving the use of labeled ATP to identify phosphorylated Sp1 on SDS-PAGE.37 We found that the vast majority of Sp1 in nuclear extracts from megakaryocytes is nonphosphorylated (Fig 8), as also reported for other cell types.31 32 The dephosphorylation of Sp1 in Y10 cells is likely to be dependent on PP1 activity, because OA shifted all Sp1 to a phosphorylated form. The level of nonphosphorylated Sp1 increased by 2.7-fold in the presence of Mpl-ligand, as quantitated by an Electrophoresis Documentation and Analysis System (Fig 8). The mechanism by which Mpl ligand increases the total level of Sp1 protein is yet to be explored. We conclude that PP1-mediated changes in the phosphorylation state of Sp1 affect cyclin D3 gene expression, because treatment of Y10 cells with 100 nmol/L OA reduces the levels of cyclin D3 mRNA and HGH production in pD3GH-transfected cells by 75% and abolishes Mpl ligand induction (experiments performed as in Figs 1 and6; data not shown). Residual cyclin D3 promoter activity due to transcription factors, other than Sp1, could have been anticipated (also deduced from Fig 6). Because of the potential pluripotent effect of OA on cellular functions, we are only able to conclude that, under conditions in which phosphorylation of Sp1 is increased (perhaps among other changes), cyclin D3 expression is dramatically reduced.
The levels of nonphosphorylated Sp1 are increased by Mpl-ligand in Y10 megakaryocytes. In a Western blot of a 7% SDS-PAGE, lanes were loaded each with 10 μg nuclear extract prepared from Y10 cells treated for 3 days with or without 25 ng/mL MGDF in the absence or presence 100 nmol/L OA, as indicated. The major bands detected were 105 and 95 kD, as also reported by others.31,32 One arrow points to the major phosphorylated form of Sp1 (designated as Pi-Sp1 of ∼115 kD), and a second arrow points to the nonphosphorylated form of Sp1 (105 kD). The blot was treated with the ECL system as indicated in Materials and Methods. Equal loading of protein per lane was confirmed by staining with Ponceau S (not shown),24 followed by destaining and antibody reaction. The results shown are of a representative experiment out of three performed.
The levels of nonphosphorylated Sp1 are increased by Mpl-ligand in Y10 megakaryocytes. In a Western blot of a 7% SDS-PAGE, lanes were loaded each with 10 μg nuclear extract prepared from Y10 cells treated for 3 days with or without 25 ng/mL MGDF in the absence or presence 100 nmol/L OA, as indicated. The major bands detected were 105 and 95 kD, as also reported by others.31,32 One arrow points to the major phosphorylated form of Sp1 (designated as Pi-Sp1 of ∼115 kD), and a second arrow points to the nonphosphorylated form of Sp1 (105 kD). The blot was treated with the ECL system as indicated in Materials and Methods. Equal loading of protein per lane was confirmed by staining with Ponceau S (not shown),24 followed by destaining and antibody reaction. The results shown are of a representative experiment out of three performed.
DISCUSSION
Platelets, which play a central role in thrombosis and hemostasis, fragment from polyploid megakaryocytes. Cyclins have been shown to be key regulators of the cell cycle. Cyclins D1, D2, and D3 have unique cell- and tissue-specific expression patterns. Of the three D-type cyclins, cyclin D3 has been found to be present at significant levels in megakaryocytes, whereas cyclin D1 is found in relatively lower amounts and cyclin D2 is not measurable.20,24 It was reported that CDK2/cyclin D3 protein kinase activity was increased in polyploid megakaryocytic HEL cells.38 We recently reported that cyclin D3 gene expression in primary megakaryocytes is induced by the Mpl ligand, and overexpression of cyclin D3 in megakaryocytes of transgenic mice results in an increase in megakaryocyte number and ploidy.21 The human cyclin D3 promoter activity has been shown not to be induced when starved rat VSMCs were serum stimulated,39 whereas the DNA binding activities of rat cyclin D3 promoter were induced by mitogen (prolactin) treatment in the early G1-arrested Nb2 cells.40 In the current report, we have shown that levels of cyclin D3 mRNA increased in response to the Mpl ligand in the mouse megakaryocytic cell line, Y10. The increase of transcription rate correlated with increased levels of cyclin D3 mRNA and protein upon Mpl ligand-treatment (Fig 1). The goal of the current study has been to examine the mechanism of activation of the mouse cyclin D3 gene by Mpl ligand.
As to the regulation of cyclin D3 gene transcription in other cell types, DNase I footprinting of the human cyclin D3 gene, using HeLa nuclear extracts, showed two protected regions. Both of these lie within a positive regulatory element identified by deletion analysis and contain potential binding sites for Sp1.39 DNase I footprinting analysis of the rat cyclin D3 gene, using nuclear extracts from Nb2 cells stimulated by prolactin, also showed two protected regions. Region I contains putative binding sites for ATF/CREB, Sp1 and Ap2, whereas region II contains a binding site for MCBF/MGF.40 In those studies, none of the putative binding sites was further examined. To explore the inducibility of the cyclin D3 promoter in megakaryocytes by Mpl ligand and to identify its important regulatory sequences, we created a series of 5′-deletions of this promoter linked to the HGH reporter gene. Our reporter gene transfection experiments identified positive regulatory elements important for cyclin D3 promoter activity in Y10 megakaryocytes. The response to Mpl ligand was dependent on the region from −190 to −37, which contains putative binding sites for Sp1, CREB/ATF, Ets, Ap2, and NF-Y transcription factors. Our DNase I footprinting analysis indicated binding to Sp1, CREB/ATF, and Ets sites within the region −446 to −43 bp in the mouse cyclin D3 gene. Our EMSA and expression studies with mutated constructs indicated that the levels of Sp1 bound to mouse cyclin D3 promoter are enhanced by Mpl-ligand and that Sp1 is a crucial regulator of the mouse cyclin D3 gene and is likely to be important for the induction of this gene by Mpl ligand in megakaryocytes. EMSA experiments and mutation studies (Figs 5 and 6) indicated that putative CREB/ATF binding sites are not essential for cyclin D3 gene expression or for Mpl induction. The putative Ets binding site within the region −360 to −330 (Fig 4C) is also not essential for cyclin D3 promoter activity, as shown by 5′ deletion experiments (Fig 2). As in megakaryocytes, in other systems as well Sp1 has been shown to mediate cytokine stimulation of different genes. For example, Sp1 mediates epidermal growth factor (EGF) stimulation of the gastrin promoter,41 insulin-like growth factor-I (IGF-I) regulation of the elastin gene,42transforming growth factor-β (TGF-β) activation of both the a2(I) collagen43 and p15INK4Bgenes,44 platelet-derived growth factor (PDGF) activation of the vascular endothelial growth factor gene,45 and nerve growth factor (NGF) induction of p21WAF1/CIP.
The Sp1 family members, including Sp1, Sp3, and Sp4, contain three zinc finger DNA binding motifs that recognize the classical GC box (GGGCGGG) and related motifs.46,47 Sp1 and Sp3 are ubiquitous proteins, whereas Sp4 is expressed predominantly in the brain.48,49 Sp3 recognizes the same DNA sequences as Sp1 and may act as a repressor of Sp1-mediated action.50 Sp1 has traditionally been considered to be a constitutively active transcription factor.51 However, it is becoming increasingly clear that modulation of Sp1 activity plays a critical role in cellular growth and differentiation. Sp1 has been implicated in the expression of many genes involved in DNA synthesis and cell cycle control, most of them without TATA boxes in their promoters, such as cdk2, cdc2, p27Kip1, p15INK4B, cyclin A, thymidine kinase (TK), and dihydrofolate reductase (DHFR). Cooperation between Sp1 and other transcription factors is required for activation of some cell-type specific promoters and cell cycle-regulated promoters. These include GATA-1 for the γ-globin gene in erythroid cells and for the tal-1 gene in hematopoietic lineages,35,52 Ets for the aIIb gene in megakaryocytes,53 Ap2 for the acetylcholine receptor a3 gene,54 and E2F1 for the DHFR gene and TK gene.55,56 It has been shown that transcription from a TATA-less promoter requires a multisubunit TFIID complex, including TATA-binding protein (TBP), TBP-associated factors (TAFs), coactivators, and Sp1.57 This is noted with interest, because the cyclin D3 gene has a TATA-less promoter.27 Sp1 has been shown to interact directly with TAFII 110.58 It can also interact with other Sp1 molecules to loop out the intervening DNA and establish interactions between promoters and distant regulatory elements.59 60 This is certainly a possibility in the case of the cyclin D3 gene, in which each of the proximal Sp1 binding sites plays an important role in gene expression. Indeed, our mobility shift assays, using single or double mutations of proximal Sp1 sites, indicate that these two sites may interact and/or affect each other’s binding activity; it was found that, whereas mutations of both Sp1 sites almost completely abolished the formation of the Sp1 complex, single mutations of either Sp1 site (at −71 or −121) resulted in approximately a 70% to 80% reduction in the complex formed (Fig 5C). If these sites did not depend on each other for binding to Sp1, we would have expected to only find a 50% reduction in complex formation for each individual mutation.
Sp1 was initially thought to regulate ubiquitously expressed housekeeping genes, but the role of Sp1 in regulation of cell-type–specific and differentiation-specific genes has now been established. The level of Sp1 expression changes during development and varies in different cell types, with highest levels seen in developing hematopoietic cells, fetal cells, and spermatids.61 Sp1 is involved in transcription of many hematopoietic cell-specific genes, such as the myeloid-specific integrin gene CD 11b,62 the monocyte specific gene CD14,63 the erythroid specific gene γ-globin,35 and megakaryocytic- and platelet-specific genes such as a2 integrin, aIIb integrin, and platelet thromboxane receptor.53,64,65 We note this with interest, because, of the D-type cyclins, cyclin D3 is highly expressed in megakaryocytes. Regulation of transcription from promoters containing Sp1-binding sites is thought to involve several poorly characterized mechanisms, including the following: posttranslational modification of Sp1, including glycosylation and phosphorylation31,38,66-69; control of Sp1 abundance70; cooperative interaction with tissue-specific transcription factors71,72; or control of Sp1 affinity for DNA and transcription factors within the TFIID complex.73Western blot analysis indicated that Mpl ligand increases the level of nuclear Sp1 in megakaryocytes as a consequence of ongoing differentiation. It should be pointed out that, because of the low efficiency of transient transfection of Y10 megakaryocytes by any method used (<5% of the cells), we could not achieve an appreciable increase in Sp1 levels in cells cotransfected with pD3GH and a plasmid containing Sp1 cDNA driven by the CMV promoter (at various molar ratios). Indeed, with such a low number of cotransfected cells, one may calculate that an increase in cyclin D3 promoter activity will be detected only if cyclin D3 induction is in the order of greater than 30-fold. Because we found that Sp1 levels as well as cyclin D3 promoter activation are induced by Mpl ligand by up to threefold, a coactivation experiment in Y10 megakaryocytes is not feasible. However, this level of induction is sufficient to promote cyclin D3 expression and megakaryocyte growth and ploidy in a transgenic mouse model.21
The Mpl ligand induces activation of at least two distinct signaling pathways: a specific Tyk2-JAK2/STAT1-STAT3-STAT5 signaling cascade and a common Shc/Vav/Ras/Raf-1/MAP kinase-kinase/MAP kinase signaling cascade.17-19 Our EMSA experiments suggested that dephosphorylation of Sp1 by PP1 significantly improved its binding to the cyclin D3 promoter. Our findings are in accordance with other studies in which it was reported that dephosphorylation of Sp1 by PP1 is essential for Sp1-mediated activation of several genes such as the acetyl-CoA carboxylase gene and aldolase A and pyruvate kinase M2 genes.31,32 Experiments involving the use of a PP1 inhibitor suggested that PP1 activity is needed for maintenance of high levels of nonphosphorylated Sp1 in Y10 megakaryocytes and for high expression of cyclin D3 gene. Interestingly, Western blot analysis of total rat bone marrow cells indicated that, in this system also, Sp1 appears mostly as nonphosphorylated (data not shown). It should be also pointed out that we and others have found that Mpl ligand activates MAP kinase in Y10 cells (not shown) and in other cell types,17-19 whereas MAP kinase pathway has been implicated in phosphorylation and activation of PP1.74-76 Future detailed signaling experiments are required to examine if PP1 is downstream of MAP kinase signaling in megakaryocytes and the potential importance of this latter pathway for cyclin D3 gene expression.
In summary, we have identified two critical Sp1 sites regulating the transcription of the cyclin D3 gene. Mutations at these sites prevent Sp1 binding, decrease cyclin D3 promoter activity, and abolish the induction by Mpl ligand. Our data also indicate that Mpl ligand increases the level of Sp1 protein. Sp1 appears as nonphosphorylated, which has greater affinity for the cyclin D3 promoter. It should be pointed out that our findings do not preclude other factors as potentially being activated by Mpl ligand and consequently affecting other clusters of genes. For example, Doubeikovski et al77recently reported an effect of Mpl ligand on the GPIIb gene via an increase in PU.1 levels. Because the expression of several genes depends on Sp1 activation, Mpl ligand is likely to activate many genes at once during megakaryopoiesis. As detailed above, we know of several genes expressed in megakaryocytes and that are activated by Sp1, such as the GPIIb and thromboxane receptor genes. One may also predict that other cytokines that are able to affect the levels and/or phosphorylation state of Sp1 should be able to influence genes, cyclin D3 among them, that are activated by Sp1 binding. Future studies with a variety of cytokines and genes in different cell types would address these possibilities.
ACKNOWLEDGMENT
The authors thank Dr Hideki Sasaki for providing us with the original L8057 cells.
K.R. is an Established Investigator with the American Heart Association. Supported by NIHBI Grants No. HL53080 and HL58547 to K.R. Z.W. was supported by a National Institutes of Health Training Grant No. AG00115.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Katya Ravid, Dsc/PhD, Department of Biochemistry, K225, Boston University School of Medicine, 80 E Concord St, Boston, MA 02118.