Abstract
The surrogate light chain (ΨL) associates with μ and Ig-Igβ chains to form the preB-cell receptor that plays a critical role in early B-cell differentiation. Discrepancies exist in human concerning the existence of ΨL+μ− proB cells and the biochemical structure of such a proB-cell complex remains elusive. Among new antihuman VpreB monoclonal antibodies (MoAbs), 5 of the γκ isotype bound to recombinant and native VpreB protein with high affinity. They recognized 4 discrete epitopes, upon which 2 were in the extra-loop fragment. Such MoAbs detected the ΨL at the cell surface of either preB or on both proB and preB cells. The previously reported SLC1/SLC2 MoAbs recognize a conformational epitope specific for the μ/ΨL association in accordance with their preB-cell reactivity. Using the proB/preB 4G7 MoAb, ΨL cell surface expression was detected on normal bone marrow, not only on CD34−CD19+ preB but also on CD34+CD19+ proB cells. Futhermore, this MoAb identified ΨL+μ− fresh proB leukemic cells of the TEL/AML1 type. Biochemical studies showed that, at the proB stage, the ΨL is associated noncovalently with two proteins of 105 and 130 kD. Triggering of this complex induces intracellular Ca2+ flux, suggesting that the ΨL may be involved in a new receptor at this early step of the B-cell differentiation.
THE FIRST STEPS OF B-cell differentiation take place in the bone marrow, where precursor hematopoietic stem cells become proB, preB, and then immature B lymphocytes. This period of differentiation is antigen-independent and is essentially devoted to the generation of the basic Ig repertoire. This results from a complex sequence of events that involves multiple gene rearrangements. These processes are strictly coordinated, with at least two quality control checkpoints, one dependent on the preB receptor at the transition from large to small preB cells and the second due to the B-cell receptor at the immature B-cell stage.
All of these steps are defined by Ig gene rearrangements (H→κ→λ) and the expression of a well-defined set of surface molecules, among which, in humans, are CD34, CD10, CD19, and the surrogate light chain (ΨL) markers. The ΨL is composed of two polypeptides encoded by λ-like1,2 (or λ5 in mice3) and VpreB4,5 genes, both related to Cλ and Vλ Ig light chain domains, respectively. Theoretical models of the μ/ΨL structure have been proposed,6,7 based on the fact that λ5/λ-like and VpreB interact with each other and with the heavy chain in a way somewhat similar to a regular Fab fragment. In these models, the λ5 polypeptide contributes the equivalent of the CL domain, whereas the VpreB, together with a short segment of λ-like, may be considered a VL equivalent domain. In addition, it was also proposed6 7 that the COOH-terminal region of VpreB (25 aa) and the NH2-terminal portion of λ-like (50 aa) loop out from the 2 main Ig domains.
In humans, CD34−CD10+CD19+preB cells express at the cell surface a small amount of the preB receptor composed of the μ/ΨL complex associated with the Igα-Igβ transducing module.8-11 Although this receptor is expressed at a very low level at this stage, several reports showed its implication in signal transduction.8,12,13 It is now well established that this receptor is required to drive (1) the transition from large to small preB, (2) the repertoire selection of preB cells,14-18 (3) the amplification of preB cells, and (4) the control of allelic exclusion at the H chain locus.19
In addition to being expressed at the cell surface of preB cells, the ΨL has been detected on μ− murine proB-cell lines in association with proteins of 200, 130, 105, and 65-35 kD.20 In humans, expression of ΨL on CD34+CD10+CD19+μ−proB cells remains controversial. Using anti-ΨL monoclonal antibodies (MoAbs; SLC1, SLC2), Lassoued et al10,21 concluded that ΨL cell surface expression was restricted to the preB stage of B-cell differentiation. In contrast, using an anti-VpreB MoAb (688), Sanz and De La Hera22 detected ΨL on the surface of both μ− proB and μ+ preB-cell lines. Moreover, 75% of normal cells labeled with this MoAb were CD34+, most of them being CD10brightCD19dull, pointing to their proB status.22,23 In a previous publication, we also reported ΨL cell surface expression on a human proB-cell line and on normal CD34+CD38+ proB cells using anti-VpreB MoAbs.24
Identification of cells expressing ΨL not associated to μ chains obviously raises the question of a possible function for the proB complex. In λ5−/− knockout mice, the preB compartment is affected, but a normal number of proB cells are present,25 suggesting a ΨL-independent development of proB cells. However, the λ5−/− phenotype may not be identical to that obtained upon inactivation of the entire ΨL (ie, λ5 and VpreB), and the possible function of a proB-cell complex remains unanswered.
In addition to the above-mentioned discrepancies regarding ΨL cell surface expression at the proB-cell stage that are presumably due to subtle differencies in MoAb specificity, it was difficult to clearly identify the components associated with ΨL in this proB complex, partially due to the fact that most MoAbs were of the μ isotype.
By using a new series of antihuman VpreB MoAbs of the γκ isotype with a high affinity for the VpreB protein, the cell surface expression, biochemical structure, and transduction capacity of the proB-cell complex was investigated. The fine specificities of these MoAbs were compared with those of SLC1/SLC2 MoAbs, definitively establishing the existence of surface ΨL chain-expressing compartments in both human proB and preB cells.
MATERIALS AND METHODS
Cells
RS4.11,26 JEA2,24 and REH (kindly provided by Dr H.G. Drexler, German Collection of Microorganisms and Cell Cultures, Department of Cell Cultures, Braunschweig, Germany) are μ− proB-cell lines. NALM627 and LAZ 22128 are μ+ preB-cell lines and Namalwa (NWA) is a μ+/λ+ mature B-cell line. Cells were maintained at 37°C in 7% CO2 in RPMI medium supplemented with penicillin, streptomycin, 10% fetal calf serum (FCS), 2 mmol/L L-glutamine, and 1 mmol/L sodium pyruvate. JEA2 proB-cell line was cultivated for 4 days in RPMI medium containing 20 ng/mL of recombinant human interleukin-7 (IL-7; Immugenex Corp, Los Angeles, CA).
After we received informed parental consent, normal human bone marrow was obtained by iliac crest aspiration from 3 young donors (6 months to 3 years of age) and from femurs of a 24-week-old fetus. Bone marrow from 12 patients suffering from acute lymphocytic leukemia (ALL) with a TEL-AML1 translocation were obtained from the Timone Hospital (Marseille, France). These leukemias are characterized by a B-cell precursor phenotype CD34+CD10+CD19+cTdT+cIgM−. For normal and ALL bone marrows, the mononuclear cells were isolated by centrifugation over Ficoll-Hypaque gradients.
Preparation of Antihuman VpreB MoAbs
MoAbs were prepared against the human VpreB protein. Balb/c mice were first immunized with 25 μg of soluble recombinant VpreB protein in CFA. The mice were boosted intraperitonealy with the same dose in IFA (at weeks 2 and 4). At 6 weeks, the last injection was performed in phosphate-buffered saline (PBS). Four days later, spleen cells were fused to the mouse myeloma X63-Ag8.653 in the presence of PEG 1500. Supernatants of hybridomas were tested on flat-bottom microtiter plates coated with rabbit anti-VpreB antibodies24 onto which was adsorbed the human recombinant VpreB. As negative controls, plates coated with rabbit anti–λ-like antibodies and with recombinant human λ-like protein were used. All antibodies selected by enzyme-linked immunosorbent assay (ELISA) were further characterized by surface plasmon resonance using the BIAcore apparatus (Pharmacia Biosensor, Saint Quentin-Yvelines, France) and by immunoprecipitation, cell surface staining, and Western blotting.
Antibodies
Surface staining of cell lines, bone marrow, and ALL cells was performed using the following antibodies: fluorescein isothiocyanate (FITC)-labeled anti-CD10 or anti-CD19 and phycoerythrin (PE)-labeled anti-VpreB (4G7) or irrelevant PE-labeled IgG1 (Immunotech, Marseille, France); PE-labeled polyclonal goat antimouse IgG+IgM (H+L; Jackson Immunoresearch, West Grove, PA); peridinin chlorophyll (PerCP)-labeled anti-CD34 and Streptavidin-PerCP (Becton Dickinson & Co, Moutain View, CA); FITC-labeled anti-CD24 (TEBU, CLB, Amsterdam, The Netherlands); FITC-labeled anti-CD40 (SBA Southern Biotechnology Associates, Birmingham, AL); and polyclonal FITC-conjugated rabbit antihuman IgM F(ab′)2, polyclonal FITC-conjugated rabbit F(ab′)2 negative control, and FITC-conjugated rabbit antihuman κ F(ab′)2 and anti-λ F(ab′)2(Dakopatts, Glostrup, Denmark). The anti-VpreB and irrelevant anti-HEL MoAbs (kindly provided by P. Machy, CIML, Marseille, France) were purified from ascitic fluid on a protein A column (Pharmacia, Uppsala, Sweden). Antihuman surrogate light chain MoAbs (SLC1 and SLC2) were kindly provided by K. Lassoued and M. Cooper.10 21
Immunoprecipitations were performed using mouse anti-IgM MoAb (Immunotech), anti-VpreB (4G7), and anti-HEL γ1 irrelevant control. In Western blot experiments, the VpreB was detected by the anti-VpreB 4G7 MoAb followed by horseradish peroxidase (HRPO)-conjugated goat antimouse IgG (Sigma, Saint Quentin Fallavier, France), and the μ chain was detected using the HRPO-conjugated anti-μ MoAb (SBA Southern Biotechnology Associates).
Calcium flux stimulations were performed with mouse anti-CD19 and anti-IgM MoAbs (Immunotech) followed by cross-linking with a PE-labeled goat F(ab′)2 fragment antimouse IgG (H+L; Jackson Immunoresearch).
Flow Cytometric Analysis
For direct staining of cell lines, 106 cells were incubated at 4°C for 20 minutes in 70 μL PBS, 0.2% bovine serum albumin (BSA), 0.1% sodium azide, and 0.1% normal mouse serum containing 0.25 μg of PE-labeled anti-VpreB 4G7. For indirect staining, the same incubation conditions were used for the anti-VpreB 10G5, 14G3, and 15D3. Staining was shown with the PE-labeled goat F(ab′)2 fragment antimouse IgG (H+L). PE-labeled IgG1 and anti-HEL IgG1 served as isotype-matched control MoAbs. Staining using SLC1 or SLC2 was performed using 50 μg/mL of MoAbs and the revelation was performed as described above. Direct staining of cell surface μ chain was achieved using FITC-labeled polyclonal rabbit antihuman IgM F(ab′)2.
Normal bone marrow and ALL cells triple-staining was performed using PerCP-labeled anti-CD34, FITC-labeled anti-CD19, and PE-labeled anti-VpreB or PerCP-labeled anti-CD34, PE-labeled anti-CD19, and either FITC-labeled anti-CD10, anti-CD40, or anti-CD24 MoAbs.
For the detection of cytoplasmic μ and κ/λ chains in ALL, cells were first stained with PE-labeled anti-CD19 and PerCP-labeled anti-CD34 and then fixed and permeabilized using the reagent and protocol supplied with IntraPrep Permeabilization Reagent kit from Immunotech. The μ and κ/λ chain detection was performed using FITC-labeled polyclonal rabbit antihuman IgM F(ab′)2or anti-κ and λ antibodies. A polyclonal FITC-conjugated rabbit F(ab′)2 served as negative control. Stained cells were analyzed using a Becton Dickinson FACScan device.
Recombinant Proteins
Human VpreB, scΨL (λ-like and VpreB covalently joined by a linker), two Fdμ (VH-CH1) from NAML6 and 1E8 preB-cell lines, and the two corresponding Fab-like (Fdμ-scΨL) recombinant proteins were produced in baculovirus as described previously.18
BIAcore Analysis
Surface plasmon resonance measurements were performed on a BIAcore apparatus (Pharmacia Biosensor, Saint Quentin-Yvelines, France). Recombinant VpreB, scΨL, Fab-like NALM6, and Fab-like 1E8 were immobilized covalently to carboxyl groups on the dextran layer of a Sensor Chip CM5.18 MoAbs (20 μL at 20 μg/mL) were injected on sensor chips, and the amount of bound MoAb to immobilized antigen was monitored. All measurements were performed with a continuous flow of 5 μL/min of HEPES-buffered saline (HBS) buffer. At the end of each cycle, the surface of the sensor chip was regenerated by 10-μL injections of 10 mmol/L NaOH for 1 minute.
Cell-Surface Biotinylation and Immunoprecipitation
Viable cells (50 × 106) in 2 mL ice-cold PBS were incubated with 1 mg of Sulfo-NHS-LC-Biotin (Pierce Chemical Co, Rockford, IL) for 30 minutes at 4°C. After 5 washes with ice-cold PBS, labeled cells were lysed with 1 mL of NP-40 lysis buffer (1% NP-40, 150 mmol/L NaCl, 20 mmol/L Tris, pH 8, 1 mmol/L phenylmethyl sulfonyl fluoride [PMSF], pepstatin, leupeptin, antipain, and iodoacetamide). After 3 successive incubations with 100 μL of protein A-Sepharose saturated with 4% nonfat milk, cell lysates were incubated for 6 hours at 4°C with the indicated antibodies (10 μg) preadsorbed overnight on protein G sepharose (50 μL). Immunoprecipitates were washed, suspended in 70 μL of reducing Laemmli sample buffer, boiled for 10 minutes, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 5% to 15%) in reducing conditions. The proteins were transferred to Immobilon-P membranes (Millipore Corp, Bedford, MA). After blocking with 5% BSA in PBST (PBS, 0.1% Tween 20) for 1 hour, membranes were shown by incubation with a 1:5,000 dilution of streptavidin-HRPO (Amersham, Les Velis, France). On the same membranes, the presence of VpreB and μ proteins were detected, with the 4G7 anti-VpreB MoAb (1 μg/mL) shown by incubation with a 1:3,000 dilution of HRPO-conjugated goat antimouse IgG MoAb (Sigma) and with a 1:2,500 dilution of HRPO-conjugated antihuman μ MoAb (SBA Southern Biotechnology Associates), respectively. For two-dimensional analysis, precipitates were run in the first dimension under nonreducing conditions using a 5% to 15% gradient SDS-PAGE. The relevant strips were then cut out, incubated in Laemmli sample buffer with 20 mmol/L dithiothreitol for 30 minutes, and run in the second dimension on a 5% to 15% gradient SDS-PAGE. Proteins were transferred to immobilon P membranes (Millipore) and treated as described above.
Pronase treatment.
After cell surface biotinylation, 50 × 106 cells were washed 2 times in ice-cold PBS, resuspended in 1 mL of PBS containing 50 μg of pronase (Boehringer Mannheim GmbH, Mannheim, Germany), and incubated at 37°C for 5 minutes. Enzymatic activity was stopped by the addition of 1 mL ice-cold PBS containing 5% BSA and 100 μg/mL of DNase. After 3 washes in PBS 5% BSA, any remaining pronase activity was inactived by 0.5 mmol/L PMSF for 10 minutes on ice. Cells were then lysed and subjected to immunoprecipitation procedures, as described above.
PNGase F treatment.
Biotinylated immunoprecipitated proteins (from 50 × 106 cells) were resuspended in 30 μL of nonreducing sample buffer and boiled for 10 minutes. Released proteins were diluted 4 times in PBS, 2% Triton X100 buffer, followed by the addition of 0.4 U PNGase F (N-glycosidase F; Boehringer Mannheim) and overnight incubation at 37°C. Proteins were precipitated by 1 mL of acetone at −20°C, suspended in 70 μL of Laemmli sample buffer, and analyzed by SDS-PAGE under reducing conditions.
Calcium Flux
JEA2, LAZ 221, and Namalwa cells (5 × 106) in 1 mL of IMDM medium, pH 7.0, 10 mmol/L HEPES were incubated at 37°C for 30 minutes with 2.5 μmol/L of acetoxymethyl ester indo-1 (Molecular Probes, Eugene, OR). An equal volume of IMDM medium, pH 7.4, supplemented with 5% FCS was then added and cells were incubated for 30 minutes at 37°C. After centrifugation, cells were transferred in IMDM medium containing 5% normal goat serum (1 mL per 106cells). The fluorescence ratio of 405:485 nm emitted from Indo-1/AM was measured under basal conditions or after stimulation either with anti-VpreB, anti-CD19, anti-IgM, or irrelevant IgG1 MoAb, followed by cross-linking with PE-labeled polyclonal goat antimouse IgG on a FACStar apparatus.
RESULTS
Fine Characterization of Antihuman VpreB MoAbs
MoAbs against the human VpreB recombinant protein produced inEscherichia coli have been described previously.18In brief, hybridoma supernatants were first tested for their reactivity in ELISA using rabbit anti-VpreB antibodies onto which was adsorbed the human recombinant VpreB protein and were subsequently tested for detection of recombinant VpreB protein by Western blotting. Ten MoAbs were selected, from which 5 (4G7, 4E7, 10G5, 14G3, and 15D3) were characterized for their fine specificities. All 5 MoAbs had binding constant values between 10−7 and 10−10 mol/L and detected the intracellular VpreB protein as a single band at 16 kD (or as a doublet, depending on the cell lines) from proB-and preB-cell line lysates by Western blotting. All MoAbs were of the γ1κ isotype, except 15D3, which was γ3κ.18
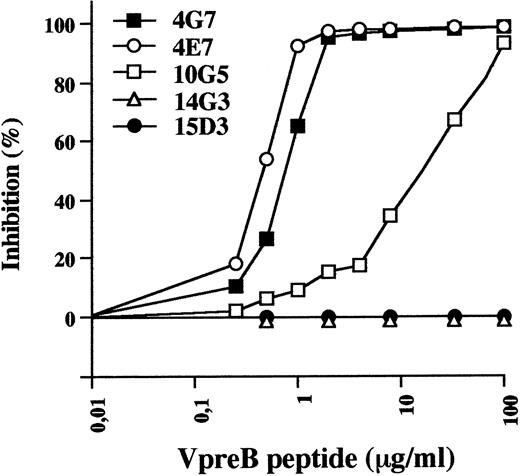
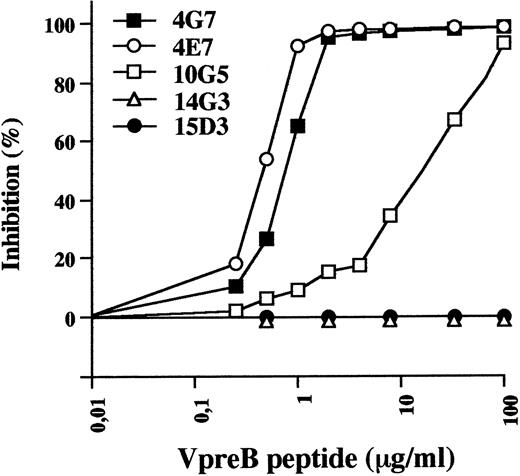
Localization of the discrete VpreB epitopes was first aproached by competitive binding of the various pairs of MoAbs to recombinant VpreB using the Biacore system. Four discrete epitopes were identified by 4G7/4E7 (2 MoAbs that completely inhibit each other), 10G5, 14G3, and 15D3 MoAbs (data not shown). To localize more precisely the epitopes, we tested the ability of the COOH-terminal 25-residue–long VpreB peptide to inhibit the binding of the various MoAbs to the immobilized VpreB protein. This peptide corresponds to the extra loop of the VpreB molecule.7 As shown in Fig 1, fixation of 4G7 and 4E7 was similarly inhibited in a dose-dependent manner by the peptide, indicating that they were specific of an extra-loop epitope. Binding of 10G5 was inhibited on a less efficient basis, suggesting that it defines a closely overlapping epitope. Absence of inhibition with 14G3 and 15D3 indicate that these MoAbs recognize epitopes on the Ig-like domain of the VpreB protein.
Binding inhibition of anti-VpreB MoAbs to immobilized VpreB by the COOH-terminal V-preB peptide, using the BIAcore apparatus. Anti-VpreB MoAbs (20 μg/mL) were preincubated for 30 minutes in HBS buffer with serial concentrations of the COOH terminal VpreB peptide (0 to 100 μg/mL) and were injected at a flow rate of 5 μL/min in HBS buffer on a sensor chip surface containing 1,200 RU of recombinant human VpreB.
Binding inhibition of anti-VpreB MoAbs to immobilized VpreB by the COOH-terminal V-preB peptide, using the BIAcore apparatus. Anti-VpreB MoAbs (20 μg/mL) were preincubated for 30 minutes in HBS buffer with serial concentrations of the COOH terminal VpreB peptide (0 to 100 μg/mL) and were injected at a flow rate of 5 μL/min in HBS buffer on a sensor chip surface containing 1,200 RU of recombinant human VpreB.
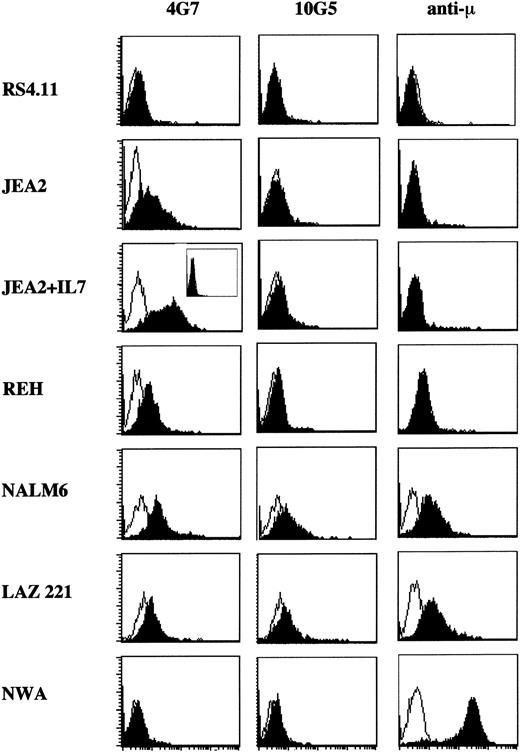
We further tested the capacity of the 4 different MoAbs to recognize the VpreB protein at the cell surface of proB-cell (RS4.11, JEA2 and REH), preB-cell (NALM6, LAZ 221), and B-cell (Namalwa) lines by flow cytometry (Fig 2). The Namalwa B-cell line that did not express the VpreB protein served as a negative control for the different MoAbs. The 14G3 and 15D3 MoAbs that recognize the VpreB Ig-like domain failed to label any cell line. The lack of 15D3 recognition of preB cells is in agreement with our previous BIAcore analysis that indicates that this epitope is lost when ΨL is associated with μ.18 The lack of positive signal on proB cells suggests that this epitope is also lost when ΨL is bound to components of the proB-cell complex. Regarding 14G3, because it was able to interact with the recombinant Fab-like molecule when tested with the BIAcore device, its failure to label precursor cells may result from its lower affinity, as compared with that of other MoAbs.18 By contrast, MoAbs that were specific for the extra-loop epitopes (ie, 10G5 and 4G7/4E7) gave positive signals in FACS analysis, but had discrete behaviors. The 10G5 MoAb detected VpreB only when associated with μ chains on the surface of the two preB-cell lines, whereas 4G7/4E7 recognized both the proB-cell (JEA2 and REH) and the preB-cell (NALM6 and LAZ 221) lines (Fig 2). The large increase of VpreB surface expression on JEA2 cell line induced by IL-724 is exclusively detected by the 4G7/4E7 MoAbs. The labeling specificity is confirmed by the fact that we obtained a complete FACS inhibition using as inhibitor the COOH-terminal 25-residue–long VpreB peptide (boxed inset in Fig 2). As previously observed and despite intracellular expression of the VpreB protein, the RS4.11 proB-cell line was negative for cell surface labeling with all anti-VpreB MoAbs.
Surface FACS analysis of VpreB and μ chains on human proB-cell (RS4.11, JEA2, REH), preB-cell (NALM6, LAZ 221), and B-cell (NAMALWA) lines, using the anti-VpreB (4G7 and 10G5) and anti-μ MoAbs. The 4G7 MoAb is PE-labeled, whereas the 10G5 is indirectly shown by a PE-conjugated goat antimouse IgG+IgM (H+L). Isotype-matched MoAbs served as negative controls. When indicated (line 3), JEA2 cells were cultured with 20 ng/mL of IL-7 for 4 days. In that case, the 4G7 anti-VpreB labeling is completely inhibitable by 2 μg/mL of the COOH terminal VpreB peptide (boxed inset).
Surface FACS analysis of VpreB and μ chains on human proB-cell (RS4.11, JEA2, REH), preB-cell (NALM6, LAZ 221), and B-cell (NAMALWA) lines, using the anti-VpreB (4G7 and 10G5) and anti-μ MoAbs. The 4G7 MoAb is PE-labeled, whereas the 10G5 is indirectly shown by a PE-conjugated goat antimouse IgG+IgM (H+L). Isotype-matched MoAbs served as negative controls. When indicated (line 3), JEA2 cells were cultured with 20 ng/mL of IL-7 for 4 days. In that case, the 4G7 anti-VpreB labeling is completely inhibitable by 2 μg/mL of the COOH terminal VpreB peptide (boxed inset).
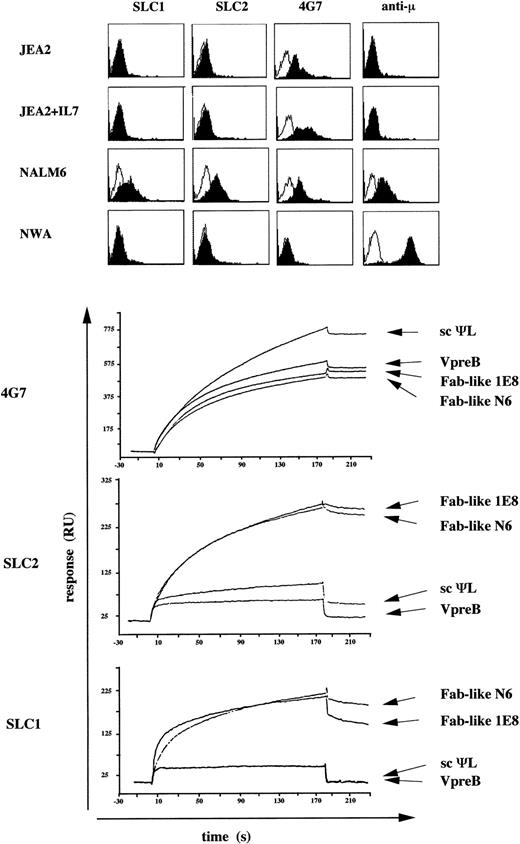
Because 4G7 MoAb recognized the VpreB protein both in the presence or absence of μ chains, its reactivity was compared with the previously reported SLC1 (γ1κ) and SLC2 (μκ) antisurrogate light chain MoAbs10,21 using FACS and BIAcore analysis (Fig 3). Immunofluorescence experiments confirm that both SLC1 and SLC2 MoAbs are preB-specific reagents, because they recognized only the NALM6 preB-cell line and do not stain the JEA2 proB-cell line cultivated in absence or presence of IL-7 (Fig3, top). Using the BIAcore technology, we determined the fine specificity of these MoAbs on different VpreB-containing recombinant proteins (Fig 3, bottom). SLC1 and SLC2 MoAbs interact with the two Fab-like proteins, whereas the free scΨL or VpreB proteins are not or poorly recognized by these MoAbs. In the same conditions, the 4G7 bound all the recombinant proteins18 and had a higher affinity for the two Fab-like MoAbs than the SLC1/SLC2 MoAbs. It thus appears that SLC1/SLC2 MoAbs recognized conformational epitopes depending on the association of the scΨL with the Fdμ fragments in Fab-like complexes, whereas 4G7 detected the VpreB protein regardless of whether it was associated or not with the Fdμ chains. These data are in agreement with the immunofluorescence analysis and confirm the preB-cell specificity of the SLC1/SLC2 reagents. By contrast, the 4G7 MoAb detects both cell surface expression of ΨL+μ+ preB-cell and ΨL+μ− proB-cell complexes.
Comparison of the fine specificities of anti-VpreB MoAbs by FACS and BIAcore analysis. (Top) Surface FACS analysis of VpreB and μ chains on human proB-cell (JEA2), preB-cell (NALM6), and B-cell (NAMALWA) lines, using the 4G7 anti-VpreB, SLC1, SLC2,10 21and anti-μ MoAbs. The 4G7 MoAb is PE-labeled and unconjugated SLC1 and SLC2 are shown by a PE-conjugated goat antimouse IgG+IgM (H+L). Wherever indicated (line 2), JEA2 cells were cultured with 20 ng/mL of IL-7 for 4 days. (Bottom) Binding of anti-VpreB MoAbs to immobilized VpreB-containing recombinant proteins using the BIAcore apparatus. In two separate experiments, 20 μL (20 μg/mL) of 4G7, SLC1, and SLC2 was injected at a flow rate of 5 μL/min in HBS buffer on four surfaces containing 700, 410, 1,400, and 900 RU of VpreB, scΨL, Fab-like NALM6, and Fab-like 1E8, respectively. The resulting sensorgrams are superimposed.
Comparison of the fine specificities of anti-VpreB MoAbs by FACS and BIAcore analysis. (Top) Surface FACS analysis of VpreB and μ chains on human proB-cell (JEA2), preB-cell (NALM6), and B-cell (NAMALWA) lines, using the 4G7 anti-VpreB, SLC1, SLC2,10 21and anti-μ MoAbs. The 4G7 MoAb is PE-labeled and unconjugated SLC1 and SLC2 are shown by a PE-conjugated goat antimouse IgG+IgM (H+L). Wherever indicated (line 2), JEA2 cells were cultured with 20 ng/mL of IL-7 for 4 days. (Bottom) Binding of anti-VpreB MoAbs to immobilized VpreB-containing recombinant proteins using the BIAcore apparatus. In two separate experiments, 20 μL (20 μg/mL) of 4G7, SLC1, and SLC2 was injected at a flow rate of 5 μL/min in HBS buffer on four surfaces containing 700, 410, 1,400, and 900 RU of VpreB, scΨL, Fab-like NALM6, and Fab-like 1E8, respectively. The resulting sensorgrams are superimposed.
Characterization of proB- and preB-Cell Compartments in Normal Bone Marrows
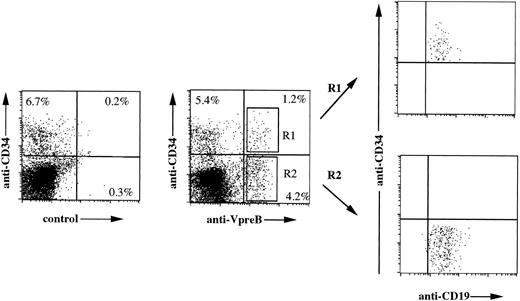
Because the 4G7 MoAb labels proB- and preB-cell lines, it was used to define their normal counterpart in fetal or young donor bone marrows. We performed three-color immunofluorescence analysis using anti-CD34, anti-CD19, and anti-VpreB MoAbs on 4 bone marrow samples. As shown in Fig 4, the 4G7 anti-VpreB, in comparison with the irrelevant γ1κ MoAb, labeled 5.4% of lymphoid fetal bone marrow cells. All cells, whether CD34+VpreB+(R1) or CD34−VpreB+ (R2), expressed the CD19 marker (Fig 4, right). This points to the identification of two distinct populations, ie, proB (CD34+CD19+VpreB+) and preB (CD34−CD19+VpreB+) cells. Depending on samples, proB cells ranged from 0.3% to 1.2% (0.84% ± 0.45%) and preB cells ranged from 0.8% to 4.2% (2.64% ± 1.62%) of total bone marrow cells.
Three-color immunofluorescence analysis of normal bone marrow cells shows cell surface expression of ΨL on both the proB and preB compartments. Fetal bone marrow lymphoid cells were incubated with PerCP-labeled anti-CD34, FITC-labeled anti-CD19, and PE-labeled anti-VpreB 4G7 or PE-labeled irrelevant γ1 control MoAbs. Both the CD34+VpreB+ (R1) and the CD34−VpreB+ (R2) cells were CD19+, identifying proB- and preB-cell compartments, respectively.
Three-color immunofluorescence analysis of normal bone marrow cells shows cell surface expression of ΨL on both the proB and preB compartments. Fetal bone marrow lymphoid cells were incubated with PerCP-labeled anti-CD34, FITC-labeled anti-CD19, and PE-labeled anti-VpreB 4G7 or PE-labeled irrelevant γ1 control MoAbs. Both the CD34+VpreB+ (R1) and the CD34−VpreB+ (R2) cells were CD19+, identifying proB- and preB-cell compartments, respectively.
Characterization of Surface ΨL+μ−proB-Cell Leukemias
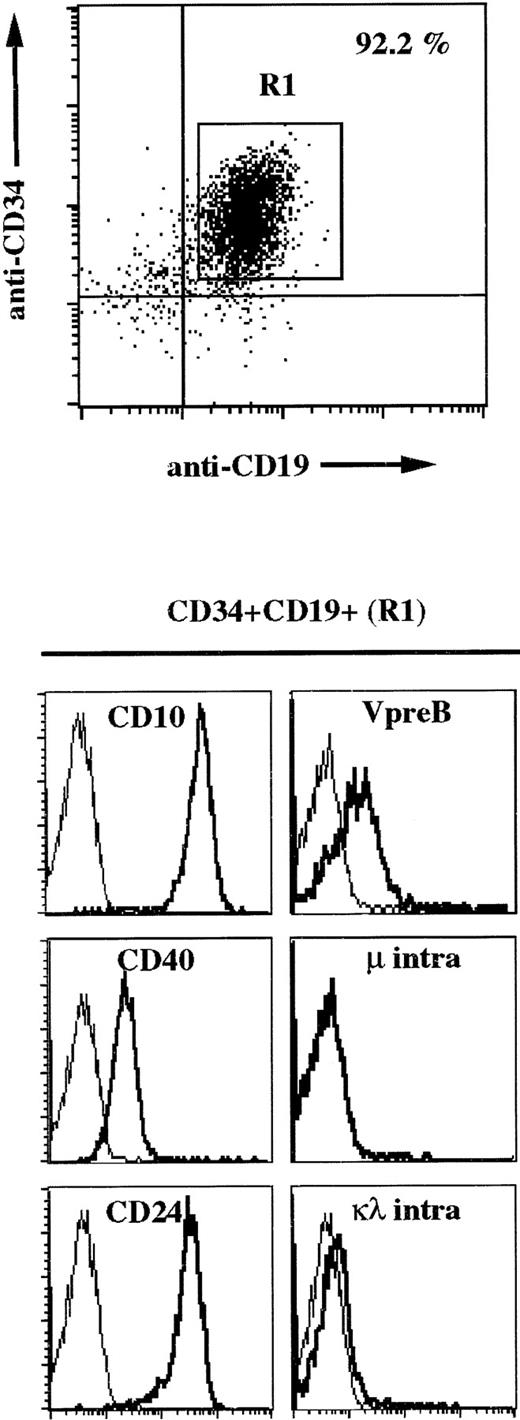
Using the 4G7 MoAb, we characterized two proB ΨL+μ− leukemic cell lines from which one, REH, expressed the TEL/AML1 fusion transcripts resulting from the t(12;21)(p13;q22) translocation. This observation prompted us to analyze fresh TEL/AML1-positive leukemias. This was performed on 12 leukemic cell samples that had been shown previously to express the fusion transcript by a PCR approach.29 These cells were considered to be proB, because they were positive for the CD34, CD19, and CD10 markers. We performed three-color immunofluorescence analysis using anti-CD34, anti-CD19, and either anti-CD10, anti-CD40, anti-CD24, and anti-VpreB on intact cells or anti-μ and anti-κ/λ antibodies on permeabilized cells. One representative analysis is presented for one patient’s bone marrow cells in Fig 5and indicates that all leukemic blasts (92.2% on gate R1) that coexpress CD34 and CD19 are surface VpreB+. Moreover, we confirm that these cells are negative for intracytoplasmic μ or κ/λ chains (Fig 5, bottom) and are positive for the expression of the TdT protein (data not shown), thus confirming the proB status of these leukemic cells. From 12 analyzed cases, 8 had the same phenotype and 4 expressed the VpreB protein only intracellularly (data not shown).
Three-color immunofluorescence analysis of TEL/AML1 leukemia bone marrow cells shows cell surface expression of ΨL in the absence of μ chain. Leukemic blasts coexpressing CD34 and CD19 were gated (R1) and analyzed for the other markers. Cells were stained with PerCP-labeled anti-CD34, FITC-labeled anti-CD19, and PE-labeled anti-VpreB or PerCP-labeled anti-CD34, PE-labeled anti-CD19, and either FITC-labeled anti-CD10, anti-CD40, or anti-CD24 MoAbs. For μ and κ/λ intracellular staining, cells were first labeled with PerCP-labeled anti-CD34 and PE-labeled anti-CD19 and then permeabilized and treated with FITC-conjugated rabbit antihuman IgM F(ab′)2 or FITC-conjugated rabbit antihuman κ F(ab′)2 plus antihuman λ F(ab′)2.
Three-color immunofluorescence analysis of TEL/AML1 leukemia bone marrow cells shows cell surface expression of ΨL in the absence of μ chain. Leukemic blasts coexpressing CD34 and CD19 were gated (R1) and analyzed for the other markers. Cells were stained with PerCP-labeled anti-CD34, FITC-labeled anti-CD19, and PE-labeled anti-VpreB or PerCP-labeled anti-CD34, PE-labeled anti-CD19, and either FITC-labeled anti-CD10, anti-CD40, or anti-CD24 MoAbs. For μ and κ/λ intracellular staining, cells were first labeled with PerCP-labeled anti-CD34 and PE-labeled anti-CD19 and then permeabilized and treated with FITC-conjugated rabbit antihuman IgM F(ab′)2 or FITC-conjugated rabbit antihuman κ F(ab′)2 plus antihuman λ F(ab′)2.
Biochemical Analysis of the proB- and preB-Cell Surface Complexes
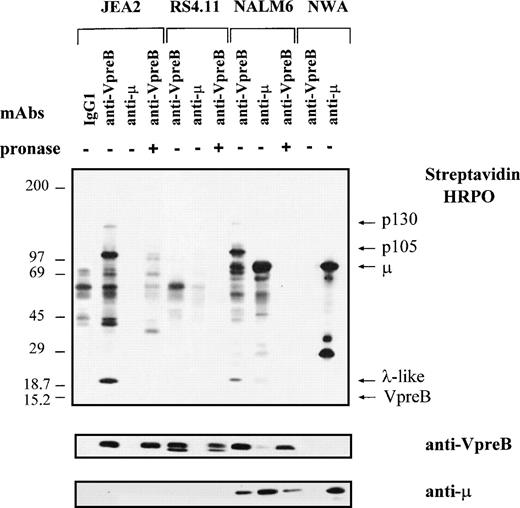
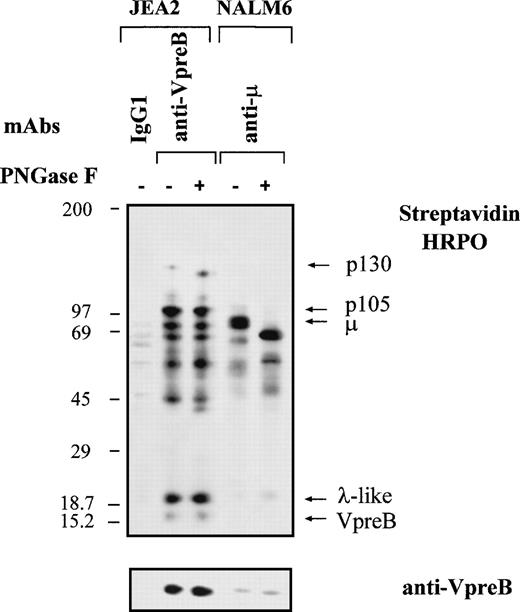
Besides the identification of the early steps of B-cell differentiation in normal bone marrow, the 4G7 MoAb was used to identify molecular components associated to ΨL at the cell surface of proB-cell lines. It also allowed us to compare the biochemical structure of the proB- and preB-cell complexes. Biotinylated cell surface proteins from a series of representative cell lines corresponding to proB, preB, and mature B stages were immunoprecipitated using anti-VpreB (4G7), anti-μ, and irrelevant γ1 MoAbs; were subjected to SDS-PAGE; were transfered to membrane filter; and were then shown by streptavidin-HRPO (Fig 6). In agreement with FACS analysis, we did not observe specific VpreB- or μ-associated complexes using the RS4.11 cell line. By contrast, several proteins were immunoprecipitated from the JEA2 cell line, using the anti-VpreB (4G7) MoAb: (1) the VpreB protein, faintly labeled but clearly visible after immunoblotting; (2) the λ-like component at 20 kD; (3) a p105 that was strongly biotinylated; and (4) a p130 that was always faintly labeled. Other weaker bands could be visualized in the range of 42 to 70 kD in some experiments. As expected with this cell line, no μ protein was detected either at the cell surface or intracellularly. Similar results were observed whenever iodination was used instead of biotinylation, except that the labeling of the VpreB protein was more intense (data not shown). Using the anti-μ MoAb on the NALM6 (Fig 6) or on the LAZ 221 (data not shown) preB-cell lines, preB-cell receptor components were immunoprecipitated, ie, μ heavy chains and weakly labeled ΨL. Surprisingly, using the 4G7 MoAb, besides the ΨL components and the μ chain (that is clearly identified in the μ Western blot), the p105 and the p130 proteins were also immunoprecipitated. Thus, the two proB- and preB-cell complexes were present at the cell surface of the two preB-cell lines. In some experiments, the 14G3 anti-VpreB MoAb that does not stain proB- and preB-cell lines is used as a control and gave the same pattern of background bands as the γ1 irrelevant control (data not shown). Pronase treatment of biotinylated proteins from all three precursor cell lines led to the disappearance of labeled proteins upon anti-VpreB immunoprecipitation without affecting the size of the intracellular VpreB pool, as demonstrated by the anti-VpreB Western blot (Fig 6). This confirms that biotinylation had been targeted mainly to cell surface molecules. Finally, as expected, the Namalwa mature B-cell line expressed only the conventional μ and λ chains.
Biochemical characterization of the proB-cell surface complex on proB- and preB-cell lines. The proB-cell (JEA2, RS4.11), preB-cell (NALM6), and B-cell (Namalwa) lines were surface labeled with biotin and lysed with 1% NP-40 lysis buffer. Lysates (5 × 107 cells) were incubated with either IgG1 control, anti-VpreB (4G7), or anti-μ MoAbs. Immunoprecipitates were submitted to a gradient SDS-PAGE (5% to 15%) under reducing conditions and transferred onto immobilon P membrane. Cell surface biotinylated proteins were detected by streptavidin-peroxidase. The membrane was then successively incubated with the anti-VpreB 4G7 MoAb, which was shown by a peroxidase-conjugated goat antimouse IgG and finally with a peroxidase-conjugated mouse antihuman μ MoAb.
Biochemical characterization of the proB-cell surface complex on proB- and preB-cell lines. The proB-cell (JEA2, RS4.11), preB-cell (NALM6), and B-cell (Namalwa) lines were surface labeled with biotin and lysed with 1% NP-40 lysis buffer. Lysates (5 × 107 cells) were incubated with either IgG1 control, anti-VpreB (4G7), or anti-μ MoAbs. Immunoprecipitates were submitted to a gradient SDS-PAGE (5% to 15%) under reducing conditions and transferred onto immobilon P membrane. Cell surface biotinylated proteins were detected by streptavidin-peroxidase. The membrane was then successively incubated with the anti-VpreB 4G7 MoAb, which was shown by a peroxidase-conjugated goat antimouse IgG and finally with a peroxidase-conjugated mouse antihuman μ MoAb.
Further characterization of the surface proB-cell complex was achieved by testing the N-linked oligosaccharides status of the different components. The treatment of the JEA2 biotinylated anti-VpreB immunoprecipitated with PNGase F changed the apparent molecular weight of the p130 protein to p115 and led to the appearance of a p40 derived from an unknown protein (Fig 7). Digestion seemed to be complete, as demonstrated by the complete shift of the μ chains from 76 to 62 kD in the similarly treated NALM6 biotinylated anti-μ immunoprecipitate.
Effect of deglycosylation on components of the proB-cell complex. Cell surface labeling and immunoprecipitation conditions are as described in Fig 6. The anti-VpreB 4G7 immunoprecipitates from the JEA2 proB and the anti-μ immunoprecipitates from the NALM6 preB-cell lines were incubated with or without PNGase F, submitted to a gradient SDS-PAGE (5% to 15%) under reducing conditions, and transferred onto immobilon P membrane. Cell surface biotinylated proteins were detected by streptavidin-peroxidase and the membrane was incubated with the anti-VpreB 4G7 MoAb and shown by a peroxidase-conjugated goat antimouse IgG.
Effect of deglycosylation on components of the proB-cell complex. Cell surface labeling and immunoprecipitation conditions are as described in Fig 6. The anti-VpreB 4G7 immunoprecipitates from the JEA2 proB and the anti-μ immunoprecipitates from the NALM6 preB-cell lines were incubated with or without PNGase F, submitted to a gradient SDS-PAGE (5% to 15%) under reducing conditions, and transferred onto immobilon P membrane. Cell surface biotinylated proteins were detected by streptavidin-peroxidase and the membrane was incubated with the anti-VpreB 4G7 MoAb and shown by a peroxidase-conjugated goat antimouse IgG.
The presence of disulfide bridges between the different components of the proB cell complex was investigated by two-dimensional SDS-PAGE analysis. JEA2 cell surface biotinylated proteins were immunoprecipitated with either 4G7 or irrelevant γ1κ MoAbs and subjected to nonreducing/reducing two-dimensional SDS-PAGE (Fig 8). All of the proteins that coimmunoprecipitated with the VpreB molecule were detectable on the diagonal, indicating that they were noncovalently associated to ΨL complexes.
Two-dimensional SDS-PAGE analysis of proteins associated with ΨL at the cell surface of the JEA2 proB-cell line. JEA2 cell surface labeling and anti-VpreB (4G7) or IgG1 control immunoprecipitation conditions are as described in Fig 6. Immunoprecipitates were run in the first and second dimensions under nonreducing and reducing conditions, respectively, using for both dimensions a 5% to 15% gradient SDS-PAGE. After transfer onto immobilon P membrane, biotinylated proteins were shown by streptavidin-peroxidase.
Two-dimensional SDS-PAGE analysis of proteins associated with ΨL at the cell surface of the JEA2 proB-cell line. JEA2 cell surface labeling and anti-VpreB (4G7) or IgG1 control immunoprecipitation conditions are as described in Fig 6. Immunoprecipitates were run in the first and second dimensions under nonreducing and reducing conditions, respectively, using for both dimensions a 5% to 15% gradient SDS-PAGE. After transfer onto immobilon P membrane, biotinylated proteins were shown by streptavidin-peroxidase.
Functional Analysis of the proB-Cell Surface Complex
ProB complex signal transduction ability was investigated by measuring the release of Ca2+ after anti-VpreB stimulation of the JEA2 proB-cell line. ProB-cell triggering was compared with anti-VpreB– and anti-μ–induced Ca2+ flux in the LAZ 221 preB-cell line that was chosen rather than NALM6, which did not express CD45, or in the Namalwa B-cell line (Fig 9). For the different cell lines, the Ca2+ response induced by the anti-CD19 stimulation served as positive control, whereas background level was determined by an irrelevant isotype-matched γ1κ MoAb for JEA2 and LAZ 221 or by the anti-VpreB (4G7) MoAb for Namalwa. An additional negative control using the 14G3 anti-VpreB MoAb was also introduced for JEA2 cell line (data not shown). The effect of anti-VpreB stimulation was significantly different in preB or proB cells. In preB cells, Ca2+release started quickly after anti-VpreB stimulation, peaked, and decreased within a few minutes. Anti-μ stimulation generated the same profile, although the signal intensity was stronger than with an anti-VpreB triggering. In proB cells, a lag was observed before any Ca2+ release that was of lower intensity and reached a plateau. Thus, anti-VpreB triggering results in different and specific responses in B-cell precursors depending on whether ΨL is involved in a proB- or preB-cell complex. These differences may be due to the nature of the transducing components, because the Igα/Igβ complex that is part of the μ+/ΨL+ preB receptor is present in proB-cell lines but is not associated with the μ−/ΨL+ proB-cell complex.24
Transduction ability of the proB-cell surface complex as compared with that of the preB- and B-cell receptor. The proB (JEA2), preB (LAZ 221), and B (Namalwa) cells were loaded with Indo-1 as described in Materials and Methods. Anti-VpreB 4G7 (30 μg/mL), anti-CD19 (30 μg/mL), anti-μ (30 μg/mL), or irrelevant IgG1 (30 μg/mL) MoAbs were added and cross-linked by a goat antimouse IgG+IgM (H+L) at the times indicated by arrows. Fluorescence variation of indo-1 was measured by FACstar and the percentage of activated cells was determined by the MultiTime software (Phoenix Flow Systems, San Diego, CA).
Transduction ability of the proB-cell surface complex as compared with that of the preB- and B-cell receptor. The proB (JEA2), preB (LAZ 221), and B (Namalwa) cells were loaded with Indo-1 as described in Materials and Methods. Anti-VpreB 4G7 (30 μg/mL), anti-CD19 (30 μg/mL), anti-μ (30 μg/mL), or irrelevant IgG1 (30 μg/mL) MoAbs were added and cross-linked by a goat antimouse IgG+IgM (H+L) at the times indicated by arrows. Fluorescence variation of indo-1 was measured by FACstar and the percentage of activated cells was determined by the MultiTime software (Phoenix Flow Systems, San Diego, CA).
By analogy to the preB-cell receptor, these data suggest that ΨLs in proB-cell complexes may function as proB-cell receptors.
DISCUSSION
Among newly generated antihuman VpreB MoAbs of the γκ isotype, five have been selected for their ability to recognize recombinant and native intracellular VpreB proteins.18 They identify 4 VpreB epitopes, two on the extra-loop and two on the Ig-like domain of the VpreB molecule, of which only the former gave signals by FACS analysis. Furthermore, these MoAbs defined two cell surface reactivities (Fig 2). The 10G5 labeled only the preB-cell lines, suggesting that the corresponding epitope remains accessible within the preB-cell receptor but is lost in the proB-cell complex. By contrast, 4G7 and 4E7 that define a common epitope on the COOH-terminal VpreB peptide and have a high affinity (kd 10−10 mol/L) for VpreB detect the ΨL at the surface of both proB and preB cells.
The distinctive behavior of the different anti-VpreB shown by FACS analysis prompted us to compare these MoAbs with the previously reported SLC1/SLC2 using the same techniques (Fig 3). Biacore analysis showed that SLC1 and SLC2 fixation to free VpreB or ΨL chain was very weak, whereas they bound to Fab-like molecules strongly, suggesting that they recognized predominantly a conformational epitope generated upon association of ΨL to the μ chain. FACS analysis showed that this epitope was only detected at the surface of preB cells, confirming the preB-cell specificity of these reagents.10,21 However, the SLC1 (γ1κ) was reported to immunoprecipitate the ΨL in the absence of the μ chain after internal labeling of the RS4.11 precursor cell line.10 One way to account for this result is to assume that p98, p60, and p40 coimmunoprecipitated proteins confer to ΨL a conformation similar to that induced upon its interaction with μ chain, resulting in the expression of the SLC1 epitope.
Taking advantage of the 4G7 antibody, a ΨL+ proB compartment was detected in normal CD34+CD19+bone marrow cells (Fig 4). These cells did not contain intracytoplasmic μ chains (data not shown), in agreement with previous data in humans.22-24 Because ΨL cell surface expression was identified on the REH proB-cell line that is a typical prototype of TEL/AML1 leukemias, we analyzed 12 fresh TEL/AML1 cell populations. These leukemias that represent the most common genetic translocation in the B-lineage childhood ALL exhibited a proB phenotype, because they expressed the CD34, CD19, CD10, and the TdT markers and were negative for the expression of intracytoplasmic μ chain. It was indeed found that all of the 12 tested cases were positive for the 4G7 epitope, with 8 having the proB complex expressed at the cell surface. This observation, in addition to stressing the common expression of the proB complex at the cell surface of B precursors, also extends the previous observation from the cell lines to fresh cells that may be considered closer to the physiological state in vivo. Altogether, these data suggest that the ΨL may be a good marker for both normal and pathological proB cells.
In humans, characterization of proteins that allow ΨL to be expressed at the cell surface (ie, surrogate H chain) has been limited. Sanz and De La Hera22 identified a p125 protein that coimmunoprecipitated with other minority components at 200, 100, and 70-40 kD using the 688 anti-VpreB MoAb of the μ isotype and the REH proB-cell line. Using the same approach with the 4G7 MoAb, we found that the major protein associated with the ΨL at the cell surface of JEA2 was a p105 (Fig 6). In addition, we consistently detected a weakly labeled p130 and, depending on the experiments, components at p60-40 Mw. A p125 protein was also identified by Sanz and De La Hera22 at the surface of the 697 preB-cell line, suggesting that the proB-cell complex was present on preB cells. We confirm these data, because we observed that the 4G7 MoAb was capable of immunoprecipitating the ΨL/p105p130 proB-cell complex on the NALM6 and LAZ 221 preB-cell lines. However, this MoAb, in contrast to 688, immunoprecipitates the ΨL in both proB and preB complexes at the cell surface of preB cells.
The pattern of surrogate H chain proteins that we obtained is close but not identical to that detected in a μ− precursor line in mice.20 In this latter case, a complex containing glycoproteins at 200, 130, 105, and occasional bands at 65-35 kD and λ5 dimers and VpreB was identified. The potential homologies between human and mouse proteins include p130 and p105 proteins that are noncovalently linked to the ΨL. The mouse p130 is a N-linked glycoprotein with a protein core at approximately 100 kD, but it is the strongest labeled protein after cell surface iodination of the proB complex, whereas p105 was the major band in humans after cell surface biotinylation. These labeling intensity differences are not simply due to the use of different reagents for cell surface labeling of mouse and human cell lines, because we obtained similar relative band intensity for the human p105 and the p130 proteins when iodine was used instead of biotin. Human and mouse p105 differ also in that the latter exists as homodimers and is sensitive to PNGase F. The identity of this protein remains unresolved, but a search for a possible chaperone30 in the vicinity of this molecular weight failed to identify the grp 94 chaperone among the associated proteins (data not shown).
The transducing ability of the proB-cell complex has been demonstrated for the JEA2 proB-cell line by the capacity of the 4G7 anti-VpreB MoAb to induce a Ca2+ flux. In mice, a Ca2+ influx induced by anti-λ5 triggering were also reported with a μ−ΨL+ progenitor B-cell line.12 By comparison with the preB receptor, proB complex triggering is characterized by a slow and sustained Ca2+response, suggesting that the inactivation mechanisms that operate after the initial stimulus are less efficient in proB than in preB-cell lines. Furthermore, in contrast to the preB-cell receptor8,12 and in accordance with previous results in the mouse,30 we did not detect any tyrosine phosphorylation after stimulation of the proB-cell complex (data not shown). This may be explained by the difference in sensitivity between calcium and tyrosine phosphorylation tests. The nature of the transducing module associated with the proB complex remains to be determined, because Igα and Ιgβ, although present in the JEA2 proB-cell line, were shown not to be associated with the ΨL chain.24 31
Besides the biochemical and functional characterization of a proB-cell complex, its possible functions have to be raised. In λ5−/− mice, a proB to preB differentiation block exists and normal amounts of proB cells are present.25 However, we do not know if the λ5−/− phenotype may be extrapolated to the entire ΨL (λ5, VpreB1, and VpreB2) inactivation. We favor the hypothesis that this complex might be implicated in the delivery of positive signals (differentiation and/or proliferation) to the proB cells by interacting with bone marrow stromal cells. The recent demonstration that mutation within the λ-like gene in humans32 leads to a severe immunodeficiency affecting the proB/preB-cell transition appears compatible with this proposal.
ACKNOWLEDGMENT
The technical expertise of N. Brun-Roubereau and M. Barad (cytofluorometry) and of C. Forneli (PE-MoAb conjugation) is gratefully acknowledged. We thank Drs H. Chambost and J. Gabert for providing us with the TEL/AML1 leukemic cells and thank C. Fossat, D. Sainty, and C. Arnoulet for their help in cell surface analysis studies. Antihuman surrogate light chain MoAbs (SLC1 and SLC2) were kindly provided by K. Lassoued and M. Cooper. We also thank A. Trautmann for helping us with measuring the release of Ca2+ and thank K. Lassoued, E. Meffre, and J. Ewbank for their comments on the manuscript.
L.G. and V.G.-F. contributed equally to this work.
Supported by Centre National de la Recherche Scientifique (CNRS), Institut National de la Santé et de la Recherche Médicale (INSERM), Institut Universitaire de France (IUF), Association de Recherche contre le Cancer (ARC; Grant No. 6345). V.G.-F. was supported by the Vaucluse Lyons Club.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Claudine Schiff, PhD, Centre d’Immunologie de Marseille-Luminy, Case 906, 13288 Marseille Cedex 9, France; e-mail: schiff@ciml.univ-mrs.fr.