Abstract
The t(12;13)(p13;q12) is a rare, recurrent translocation reported in a range of hematological malignancies. We have analyzed the molecular basis of this lesion in three patients with acute myeloid leukemia (AML), two of whom were known to have chromosome 12 breakpoints within the ETV6 gene. Fluorescence in situ hybridization (FISH) with ETV6 cosmids indicated that this gene was also disrupted in the third patient, while the normal ETV6 allele was retained. 3′ rapid amplification of cDNA ends (RACE) polymerase chain reaction (PCR) from bone marrow mRNA of this individual identified a novel sequence fused to ETV6 that was homologous to a region just upstream of the mouse CDX2 homeobox gene, the human homologue of which has previously been mapped to chromosome 13q12. PCR primers designed to amplify an ETV6-CDX2 fusion identified two major transcripts from this patient. First, a direct in-frame fusion between exon 2 of ETV6 and exon 2 of CDX2, and second, a transcript that had an additional sequence of unknown origin spliced between these same exons. Surprisingly, apparently normal CDX2 transcripts, usually expressed only in intestinal epithelium, were also detectable in cDNA from this patient. Neither normal nor fusion CDX2 mRNA was detectable in the two other patients with a t(12;13), indicating that this translocation is heterogeneous at the molecular level. Reverse transcription-PCR analysis showed that CDX2 mRNA, but not ETV6-CDX2 mRNA, was strongly expressed in 1 of 10 patients with chronic myeloid leukemia in transformation, suggesting that deregulation of this gene may be more widespread in leukemia. CDX2 is known to regulate class I homeobox genes and its expression in hematopoietic cells may critically alter the balance between differentiation and proliferation.
TRANSLOCATIONS BETWEEN the long arm of chromosome 13 and the short arm of chromosome 12 are rare nonrandom abnormalities in acute lymphoblastic leukemia (ALL),1-3acute myeloid leukemia (AML),4 secondary AML,5myelodysplastic syndrome,6-8 and blast crisis of chronic myeloid leukemia (CML-BC).9 The breakpoints are reported to vary from 12p11 to p13 and from 13q11 to q14 but it is uncertain whether this represents a true heterogeneity at the molecular level. The region 13q12-14 is also the site of other abnormalities associated with hematological malignancies, such as the t(8;13)(p11;q12)10 or the interstitial deletions that are often seen in patients with chronic lymphocytic leukemia or primary myelofibrosis. Rearrangements of 12p (deletions and balanced or unbalanced translocations) are common findings in diverse hematological malignancies. In many cases these rearrangements target ETV6 (initially called TEL), a gene at 12p13 that has been reported to form fusions with multiple partners: PDGFRB,11 MN1,12ABL,13 CBFA2 (AML1),14 MDS1/EVI1,15STL,16 and JAK2.17 In many cases the normal ETV6 allele is deleted in patients with these abnormalities.
Here we describe a novel fusion between ETV6 and CDX2 at 13q12 in a patient with AML and a t(12;13). In addition, aberrant expression of CDX2 was found in a case of CML-BC who did not have any cytogenetically visible abnormality of chromosome 13. CDX2 is a homeobox gene related to the Drosophila gene caudal and is not normally expressed in hematopoietic cells. Although mutations of CDX2 may be associated with rare cases of colorectal cancer,18 abnormalities of this gene have not been described previously in leukemia. It is becoming increasingly clear that homeobox genes are involved in the processes of normal hematopoietic cell proliferation and differentiation, and that disruption of their normal patterns of expression can lead to leukemia.
MATERIALS AND METHODS
Case report and patient material.
A 66-year-old man presented with a short history of palpable lymph nodes in both axillae and easy bruising for the previous 2 weeks. Laboratory findings showed a leukocyte count of 144 × 109/L with 98% blasts, platelet count of 23 × 109/L, and a hemoglobin level of 10.2 g/dL. A bone marrow (BM) biopsy showed virtually all cells to be myeloblasts with occasional Auer rods and little differentiation; cytogenetic analysis demonstrated a translocation t(12;13)(p13;q12). A diagnosis of AML M1 was made. The patient achieved complete remission after three courses of chemotherapy. However, 38 months later he relapsed with a leukocyte count of 134 × 109/L, platelet count of 51 × 109/L, and a hemoglobin level of 12.4 g/dL. Cytogenetic analysis showed a deletion of 6q in addition to the t(12;13). Chemotherapy was initiated and BM examination showed less than 5% blasts after completing the second course. Unfortunately, fever occurred during neutropenia and a pneumonia was diagnosed. The patient developed generalized sepsis despite antibiotic treatment and died 2 months after the diagnosis of relapse.
For this patient (patient no. 1), peripheral blood and BM cells that had been cryopreserved at relapse were available for analysis. We also analyzed cDNA from two other t(12;13) patients (patients nos. 2 and 3) with AML M0 who have been described previously.4
As controls, peripheral blood (PB) or BM was obtained from 10 patients with AML, 10 patients with myeloid blast crisis of CML, and 5 normal individuals.
Fluorescence in situ hybridization (FISH).
Cytogenetic cell suspension was not available for patient no. 1 and cryopreserved cells failed to grow in culture. Therefore, for FISH analysis we used cytospin preparations of interphase nuclei prepared from cryopreserved cells which were fixed and stored at −20°C in 3:1 methanol/acetic acid until required. The cosmids 179A6 and 148B6 (kindly provided by Dr P. Marynen, Leuven, Belgium), which contain ETV6 exons 1 and 8, respectively, were labeled with biotin and digoxigenin by nick-translation and detected with rhodamine and FITC, respectively. CDX2 cosmids ICRFc108G2043QD2 and ICRFc108B0629QD2 were labeled with digoxigenin. FISH was performed according to standard procedures.19 Images were captured using a Vanox fluorescence microscope (Olympus Optical Co, Ltd, Tokyo, Japan) equipped with a CCD camera and SmartCapture software (Digital Scientific, Cambridge, UK).
Rapid amplification of cDNA ends (RACE) polymerase chain reaction (PCR).
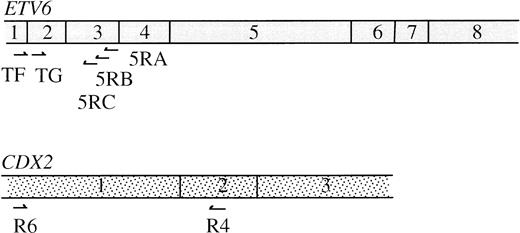
RNA was extracted from cryopreserved cells using an RNeasy Mini Kit (Qiagen, Hilden, Germany). For the 3′ RACE, the primer Ro-Ri-dT17 was used for reverse transcription (5′-AAGGATCCGTCGACATCGACAATCCTA-CGACTCACTATAGGGATTTTTTTTTTTTTTTTT-3′), an oligo-dT sequence joined to a “double adapter” sequence that allows a two-step RACE amplification.20 RACE PCR was performed using the nested ETV6 exon 2 primers TF (5′-CAGGAACGAATTTCATATACACCT-3′) and TG (5′-CCAGTGCCGAGTTACGCTTCCT-3′) and outer and inner adapters Ro(5′-AAGGATCCGTCGACATCGAT-3′) and Ri(5′-GACATCGATCCTACGACTCA-3′). Amplified products were cloned using a TA Cloning Kit (Invitrogen, Leek, The Netherlands). Clones that were positive for an internal ETV6 exon 2 probe were sequenced. For the 5′ RACE, reverse transcription (RT) was performed using the ETV6 exon 3 primer 5RA (5′-TTTGCCATTCATTTCAAA-3′). The 5′ cDNA ends were poly-A tailed, first-step PCR was performed with Ro-Ri-dT17, Ro and 5RB (5′-GTGTTGCTGTCAATTGGCCT-3′), and second step with Ri and 5RC (5′-GAAAACTCATTTTCAGCCCAC-3′). Products were cloned as above and those that were positive for an internal ETV6 exon 3 probe but negative for an ETV6 exon 2 probe were sequenced. The position of all primers used is shown in Fig 4.
RT-PCR.
Patient RNA was extracted and reverse transcribed with random hexamer primers. Quality of cDNA was confirmed in all cases by amplification of the normal ABL gene.21 All amplifications were performed for 32 cycles with an annealing temperature of 60°C to 66°C.
DNA sequencing and analysis.
Clones and PCR products were directly sequenced by thermal cycling with fluorescent dye terminators. Reactions were analyzed on an automated sequencer (model ABI 373A; Applied Biosystems, Foster City, CA).
Physical mapping.
CDX2 cosmids were isolated by screening a gridded chromosome 13 specific library, obtained from the Ressourcen Zentrum Primär Datenbank (RZPD; Berlin, Germany). CDX2 positive yeast artificial chromosomes (YACs) were identified by screening a panel of clones that were known to map to 13q12-14 (obtained from the UK HGMP Resource Centre, Hinxton, UK).
RESULTS
FISH.
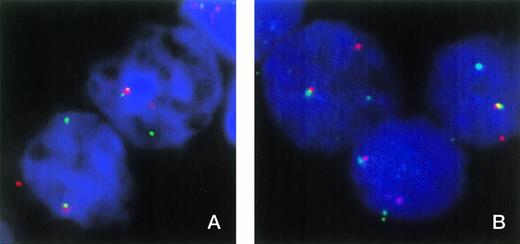
FISH was used to show the involvement of ETV6 in patient no. 1. In normal nuclei, FISH with cosmids for ETV6 exons 1 and 8 shows two fusion signals indicating two intact ETV6 genes. Patient material showed one fusion signal and single, separate red and green signals, indicating a breakpoint in one ETV6 allele and that the normal allele is not grossly deleted (Fig 1A). However, this result does not exclude the possibility of a small intragenic deletion or other mutation that may have inactivated the normal copy of ETV6.
(A) FISH analysis of two nuclei from patient no. 1 with cosmids 179A6 (detected with rhodamine) and 148B6 (detected with FITC), which hybridize to ETV6 exons 1 and 8, respectively. Colocalized signals indicate the normal ETV6 allele, while separate red and green signals indicate a break within ETV6. (B) Three nuclei from patient no. 1 hybridized with cosmids 179A6 (detected with rhodamine) and ICRFc108G2043QD2 (CDX2 exon 1 plus upstream sequence, detected with FITC). Colocalization of these cosmids indicates a chromosome 13 break upstream of CDX2 exon 1.
(A) FISH analysis of two nuclei from patient no. 1 with cosmids 179A6 (detected with rhodamine) and 148B6 (detected with FITC), which hybridize to ETV6 exons 1 and 8, respectively. Colocalized signals indicate the normal ETV6 allele, while separate red and green signals indicate a break within ETV6. (B) Three nuclei from patient no. 1 hybridized with cosmids 179A6 (detected with rhodamine) and ICRFc108G2043QD2 (CDX2 exon 1 plus upstream sequence, detected with FITC). Colocalization of these cosmids indicates a chromosome 13 break upstream of CDX2 exon 1.
RACE PCR.
Because the FISH results had indicated a break between ETV6 exons 1 and 8, 3′ RACE PCR using primers designed to ETV6 exon 1 was performed to capture any novel sequence that may have become fused to ETV6. 3′ RACE PCR resulted in 11 ETV6+ clones. Of these, 10 contained ETV6 sequence only and one, clone 3R30, contained 547 bp of novel sequence immediately after ETV6 exon 2 (Fig2A). A BLAST search showed that part of this sequence was homologous to a region shortly upstream of the mouse Cdx2 gene (Genbank accession no. U00454, bases 475-541). The human homologue, CDX2 (originally referred to as CDX3), has been cloned22 and mapped to 13q12 by FISH.23Therefore, it was likely that CDX2 was the partner gene. 5′ RACE PCR from ETV6 exon 3 failed to find any novel sequences.
(A) DNA sequence of clone 3R30. The end of ETV6 exon 2 shown in capitals and the sequence with partial homology to a region upstream of the mouse CDX2 gene is underlined. (B) Amino acid and mRNA sequences of normal ETV6, normal CDX2 and the smaller ETV6-CDX2 product at the site of fusion. (C) Sequence of larger ETV6-CDX2 fragment. The end of ETV6 exon 2 and the start of CDX2 exon 2 are shown in capitals. The stop (TAA) and potential start (ATG) codons referred to in the text are shown underlined.
(A) DNA sequence of clone 3R30. The end of ETV6 exon 2 shown in capitals and the sequence with partial homology to a region upstream of the mouse CDX2 gene is underlined. (B) Amino acid and mRNA sequences of normal ETV6, normal CDX2 and the smaller ETV6-CDX2 product at the site of fusion. (C) Sequence of larger ETV6-CDX2 fragment. The end of ETV6 exon 2 and the start of CDX2 exon 2 are shown in capitals. The stop (TAA) and potential start (ATG) codons referred to in the text are shown underlined.
RT-PCR and sequence analysis.
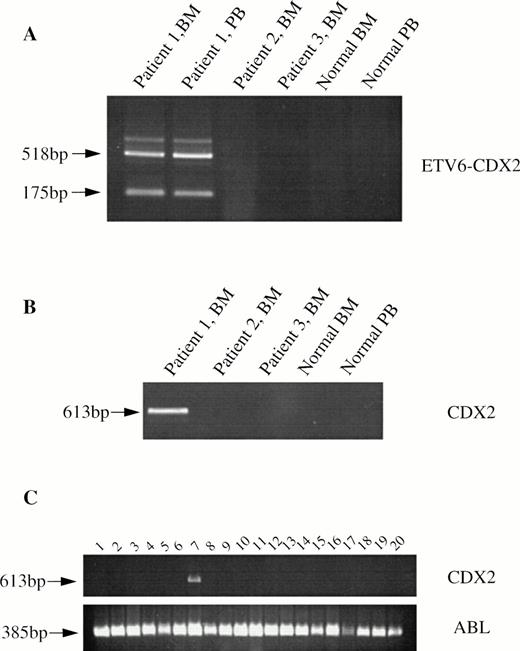
Primers designed to detect an ETV6-CDX2 fusion, TG (ETV6 exon 2) and R4 (CDX2 exon 2, 5′-AGTGAAACTCCTTCTCCAGGTC-3′) resulted in the amplification of two major and several weaker bands from cDNA derived from both PB and BM leukocytes of patient no. 1 (Fig3A). No amplification products were obtained from cDNAs made from patient nos. 2 and 3, from normal BM or normal PB (Fig 3A). Amplification of the normal ABL gene from these samples indicated that the quality of the cDNA was good (not shown). Sequencing showed that the smaller of the two major bands from patient no. 1 was a direct in-frame fusion between exon 2 of ETV6 and exon 2 of CDX2 (Fig 2B). The larger band had an additional sequence inserted between these same exons (Fig 2C), which was related to the novel sequence identified by 3′ RACE PCR. Two of the weakly amplified bands were also sequenced. Both were identical to the large major transcript except for the addition of 97 bp of ETV6 intron 2 sequence in one case and the deletion of the first 83 bp of the inserted sequence in the second. No amplification products were obtained from any individual using primers to CDX2 exon 1 and ETV6 exon 3, indicating that the reciprocal CDX2-ETV6 fusion is not expressed (not shown).
RT-PCR analysis from patient and control cDNAs. (A) ETV6-CDX2 (primers TG and R4) from patients 1, 2, and 3, normal BM and PB leukocytes. (B) CDX2 (primers R6 and R4) from the same patients and (C) CDX2 and ABL from 10 patients with myeloid blast crisis of CML (lanes 1 through 10) or AML (lanes 11 through 20).
RT-PCR analysis from patient and control cDNAs. (A) ETV6-CDX2 (primers TG and R4) from patients 1, 2, and 3, normal BM and PB leukocytes. (B) CDX2 (primers R6 and R4) from the same patients and (C) CDX2 and ABL from 10 patients with myeloid blast crisis of CML (lanes 1 through 10) or AML (lanes 11 through 20).
Using primers (R6, 5′-ATGTACGTGAGCTACCTCCTGG-3′, and R4) to CDX2 exons 1 and 2 (Fig 4), we confirmed that transcripts of this gene are undetectable in normal PB and BM leukocytes. CDX2 mRNA was also undetectable in patients no. 2 and 3. Surprisingly, however, in view of the fusion sequences described above, a strong amplification product was obtained from patient no. 1 (Fig3B). The identity of this product was confirmed by sequencing. This finding shows that, in addition to the ETV6-CDX2 fusion, patient no. 1 expresses CDX2 transcripts that are predicted to be translated into normal, full-length CDX2 protein.
Normal ETV6 and CDX2 mRNAs showing position of primers described in the text.
To determine if CDX2 is expressed in other patients with leukemia, we screened 20 patients who did not have cytogenetic evidence of chromosome 13 abnormalities by RT-PCR with primers R6 and R4. Surprisingly, CDX2 mRNA was detected in one patient with CML-BC (Fig3C). Further RT-PCR with various combinations of CDX2 and ETV6 primers failed to identify an ETV6-CDX2 fusion in this patient (not shown).
Physical mapping.
Screening of a 13q YAC panel by PCR with primers R4 and R6, which amplify the first two exons and a short intron, identified the CEPH YAC 272G3 to be positive for CDX2. The 540-kb YAC 272G3 has been partially mapped and shown to contain the FLT1 and FLT3 genes.24
The fact that the 3′ RACE PCR product and the larger ETV6-CDX2 fusion transcript contained a sequence that matched a region shortly upstream of the mouse Cdx2 gene strongly suggested that the chromosome 13 breakpoint must lie upstream of the human CDX2 gene. This hypothesis is supported by the fact that no reciprocal CDX2-ETV6 transcripts were detected. To confirm the position of the breakpoint, two chromosome 13 cosmids containing the region immediately upstream of human CDX2 were isolated by screening a library with the inserted sequence from the larger fusion transcript. PCR analysis showed that clone ICRFc108G2043QD2 also contained CDX2 exon 1 and that clone ICRFc108B0629QD2 contained upstream sequence only. Both cosmids hybridized to chromosome 13q12 only in normal control metaphases. Two-color FISH analysis on nuclei from patient no. 1 showed colocalization of both these cosmids with the ETV6 exon 1 cosmid (Fig1B), thus confirming that the break is upstream of CDX2 exon 1.
DISCUSSION
We have found a fusion between ETV6 and a new partner gene, CDX2, in a patient with AML M1 with a t(12;13)(p13;q12). Chimeric ETV6-CDX2 fusion transcripts were not detected in two other patients with AML M0 and a t(12;13) who have been shown previously to have breakpoints within the ETV6 gene,25 indicating that this translocation is heterogeneous at the molecular level. Several different types of chimeric mRNA were identified in patient no. 1, of which two were predominant. The first of these was a direct in-frame fusion between ETV6 exon 2 and CDX2 exon 2 and is predicted to encode a 187-amino acid fusion protein consisting of a short ETV6-derived amino-terminal domain of unknown function joined to the DNA-binding domain of CDX2. This protein would lack the ETV6 helix-loop-helix domain, implicated in the leukemogenic activity of the fusions between ETV6 and ABL, PDGFRB, and JAK2,26-28 and also the ETV6 DNA-binding domain. The second major transcript had an additional 547 bp spliced between ETV6 exon 2 and CDX2 exon 2. This transcript would not be translated into a fusion protein because the additional sequence introduces an in-frame stop codon 57 bp after the end of ETV6 exon 2. Potentially it could encode a 74-amino acid truncated ETV6 protein. In addition, there are two possible AUG start codons within the inserted sequence that are in frame with CDX2 and it is conceivable that these could initiate translation, for example via an internal ribosome entry site (IRES), a mechanism of internal ribosome binding found in some viruses and several cellular genes.29
Unexpectedly, we also detected apparently normal CDX2 transcripts in patient no. 1 using primers designed to amplify between CDX2 exons 1 and 2. In adults, expression of this gene is usually tightly restricted to the intestinal epithelium.30 However, analysis of a further 20 patients with acute leukemia showed similar transcripts in one other case, indicating that ectopic expression of CDX2 is not restricted to patients with a t(12;13) or other abnormalities of chromosome 13. Taken together, these data suggest that the leukemogenic event in patient no. 1 may have been the ectopic expression of CDX2, and that deregulation of this gene by a mechanism that remains to be defined may be a contributory factor in other leukemias. This situation is reminiscent of the EVI-1 gene: overexpression of EVI-1 was initially found in association with the t(3;3)(q21;q26) or the inv(3)(q21q26). Subsequently, however, EVI-1 overexpression was found in other cases of acute leukemia.31 Furthermore, a related translocation, the t(3;12)(q26;p13), has been described in which a fusion was found between ETV6 exon 2 and MDS1 exon 2, which in turn was fused to EVI1.15 The investigators suggested that the function of ETV6 in these cases was to drive the inappropriate expression of MDS1/EVI1.
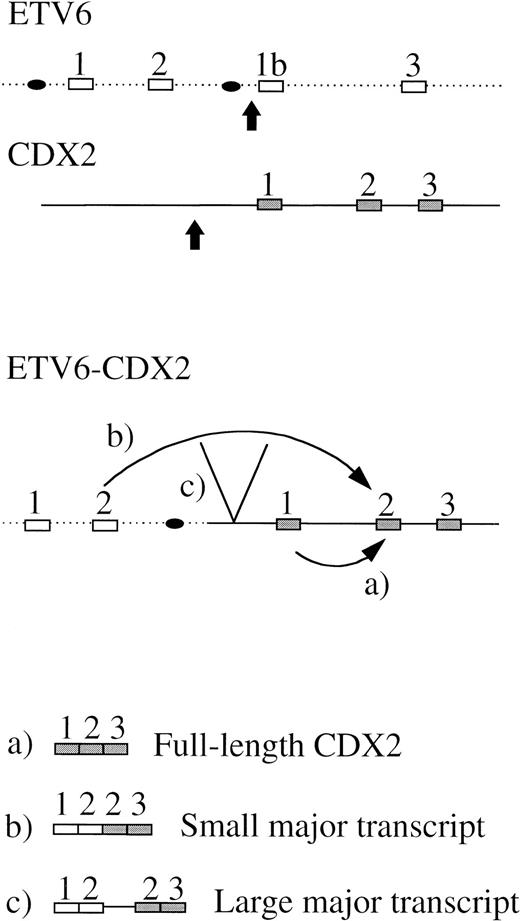
We have shown that the chromosome 13 breakpoint lies upstream of the CDX2 gene. One possible explanation for the aberrant expression of apparently normal CDX2 transcripts could be that this gene is translocated downstream of one of the two alternative first ETV6 promoters that are located between ETV6 exons 2 and 3,15 32and that this promoter then drives the expression of a CDX2 mRNA that would be predicted to be translated into normal CDX2 protein (Fig5, transcript a). The fusion transcripts would be initiated at the normal ETV6 promoter, from which CDX2 exon 1 would be spliced out due to the lack of a 3′ splice acceptor sequence, resulting in the observed ETV6 exon 2-CDX2 exon 2 chimeric mRNAs or various minor products (Fig 5, transcripts b and c). However, alternative explanations for the ectopic expression of CDX2 are possible. The ETV6-CDX2 fusion protein may dysregulate the normal and/or the translocated CDX2 allele. Alternatively, translocation of CDX2 and its promoter into the active ETV6 locus may relieve chromatin-mediated transcriptional suppression of CDX2.
Schematic diagram to illustrate a possible mechanism for expression of an apparently normal full-length CDX2 transcript (a) and ETV6-CDX2 fusion transcripts (b and c). (▪, exon; •, promoter; , breakpoint).
Schematic diagram to illustrate a possible mechanism for expression of an apparently normal full-length CDX2 transcript (a) and ETV6-CDX2 fusion transcripts (b and c). (▪, exon; •, promoter; , breakpoint).
CDX1 and CDX2 in humans, Cdx1, Cdx2, and Cdx4 in mice, and also genes identified in chicken, Xenopus, zebra fish, silk worm, andCaenorhabditis elegans are related to the Drosophila homeobox gene caudal. Caudal family members share a highly conserved DNA-binding homeodomain that is similar to, but distinct from, the homeodomains of the class I homeobox genes. The caudal protein forms an anterior-posterior gradient in the Drosophilaoocyte, which plays an important role in embryo segmentation.33 Later in embryogenesis caudal is involved in formation of the gut, Malpighian tubules, and other posterior structures. Similarly in mice, in early embryogenesis Cdx2 is expressed in posterior regions, but in later embryos and adults Cdx2 is reported as restricted to the intestinal epithelium.22,30,34 InDrosophila, mutations of caudal produce severe disruptions to body segmentation.35 In mice, Cdx2 homozygous null mutants are not viable beyond 3 to 5 days postcoitum, but heterozygotes show an anterior homeotic shift of the vertebrae. Surprisingly, these mice developed intestinal neoplasia and metaplasia, suggesting that abnormalities of CDX2 may be relevant to human intestinal malignancies.36 Indeed, a case of colorectal cancer with biallelic mutations of CDX2 has recently been described.18 The fundamental segment disruption in caudal-null Drosophila and the homeotic changes in Cdx2 heterozygous mice suggest that CDX2 influences the expression of other homeobox genes and its action may be one of the early steps in the hierarchy of control of homeobox gene expression. In support of this hypothesis, CDX2 consensus DNA binding sequences have been identified in the promoter regions of 13 murine homeobox genes37 and 6 human homeobox genes.36 38
Disruption or aberrant regulation of homeobox gene have been implicated in the pathogenesis of distinct leukemias. HOXA9 is fused to the nucleoporin gene NUP98 by the t(7;11) in AML.39,40 HOX11 is juxtaposed to the T-cell receptor by the t(10;14) in T-ALL41 and PBX1 is fused to E2A by the t(1;19) in pre-B ALL. The MLL gene, involved in translocations with many different partners in both lymphoblastic and myeloblastic leukemias, is a human homologue of the Drosophila gene trithorax, a positive regulator of homeobox gene expression. In addition, CBFA2 and CBFB, which are disrupted by the t(8;21) and inv(16), respectively, in AML, contain domains homologous to runt, a Drosophila pair-rule gene involved in defining expression domains of the Drosophila class I homeobox genes. Finally, retinoic acid is a potent modulator of homeobox gene expression42 and this may be relevant to the mechanism of transformation in AML induced by the fusion between PML and the α-subunit of the retinoic acid receptor in the t(15;17). Our findings provide a further example of a deregulated homeobox gene in leukemia and reinforce the notion that alterations in the expression patterns of this family may critically alter the balance between hematopoietic cell differentiation and proliferation.
ACKNOWLEDGMENT
We thank the MRC HGMP Resource Centre (Hinxton, UK) and the RZPD (Berlin, Germany) for providing the YAC and cosmid clones used in this study, and also Dr P. Marynen (Leuven, Belgium) for providing the ETV6 cosmid clones.
Supported by the Leukaemia Research Fund, the Dr. Mildred Scheel Stiftung, the Associazione Italiana per la Ricerca sul Cancro, and the Fondazione Tettamanti.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Andrew Chase, Department of Haematology, Imperial College School of Medicine, Hammersmith Hospital, Du Cane Rd, London W12 ONN, UK; e-mail: achase@rpms.ac.uk.