Abstract
Chromosome translocations involving band 12p13 are known to be involved in a variety of hematologic malignancies, some of them resulting in rearrangement of the ETV6/TEL gene. Applying the fluorescence in situ hybridization (FISH) method, we found a cryptic translocation t(12;15)(p13;q25) in an adult acute myeloid leukemia (AML) patient. Hybridization with cosmid probes showed that the ETV6 gene was rearranged in this translocation. A patient-specific cDNA library was screened with ETV6 cDNA, and a novel fusion transcript was identified between the ETV6 andTRKC/NTRK3 gene located on 15q25. TRKC is a receptor tyrosine kinase that is activated by neurotrophin-3 (NT-3). It is known to be expressed broadly in neural tissues but not in hematologic cells, so far. ETV6-TRKC chimeric transcript encoded the pointed (PNT) domain of the ETV6 gene that fused to the protein-tyrosine kinase (PTK) domain of the TRKC gene. Two types of fusion transcript were determined, one that included the entire PTK domain of TRKC and the other in which the 3′-terminal 462 bp of TRKC was truncated within the PTK domain. Western blot analysis showed the expression of both chimeric proteins of 52 and 38 kD in size. Our results suggest that chimeric PTK expressed in the leukemic cells may contribute to cellular transformation by abnormally activating TRK signaling pathways. Moreover, this is the first report on truncated neurotrophin receptors associated in leukemia.
SPECIFIC CHROMOSOME abnormalities found in hematologic and nonhematologic malignancies have given many important insights into the pathogenesis of malignant transformation. Recent molecular biological studies have shown that important genes associated with cell proliferation and/or differentiation are often involved.1 Cytogenetic abnormalities involving the short arm of chromosome 12 have been observed in a wide variety of hematologic malignancies, including acute myeloid leukemia (AML), acute lymphoid leukemia (ALL), and myelodysplastic syndrome (MDS). TheETV6/TEL gene, cloned from chronic myelomonocytic leukemia harboring t(5;12)(q33;p13) as the translocation partner of platelet-derived growth factor receptor β (PDGFRβ),2 is now known to be disrupted in some of these 12p13 abnormalities.3ETV6 is a member of the ETS family of transcriptional regulators that are defined by the ETS domain responsible for specific DNA binding activities. It is also distinctive in having the pointed (PNT) domain (also referred to as helix-loop-helix oligomerization domain) that is suggested to play a role in the protein-protein interaction of ETS factors.2Besides t(5;12), the ETV6 gene is reported to form fusion transcripts in some other 12p13-associated translocations, ie,ETV6-JAK2 in myeloproliferative disorders (MPD) or ALL with t(9;12)(p24;p13)4,5; ETV6-MN1 in MPD with t(12;22)(p13;q11)6; ETV6-MDS1/EVI1 in MPD with t(3;12)(q26;p13)7; and ETV6-AML1,8,9ETV6-ABL,10,11 and ETV6-STL12in childhood B-precursor ALL with t(12;21)(p13;q22), t(9;12)(q34;p13), or t(6;12)(q23;p13), respectively.
Several members of the ETS family are known to be associated with various malignancies as the result of specific chromosome translocations. These include t(11;22)(q24;q12) and t(7;22)(p22;q12) in Ewing’s sarcoma13,14 and t(21;22)(q22;q12) in clear cell sarcoma.15 In these translocations, the transactivation motif of EWS fuses to the ETS binding motif of FLI1,ETV1, or ERG, respectively, which are suspected to be oncogenic. In contrast to these EWS-associated translocations, the ETV6 fusion gene mentioned above, with exception ofETV6-MN1 and ETV6-STL, loses its ETS binding domain but retains the PNT domain in the 5′ ETV6 fusion transcript, indicating some different underlying mechanism in transformation.
Translocation involving ETV6 is often subtle and therefore overlooked by conventional cytogenetic analysis. We report here a novelETV6-associated translocation, t(12;15)(p13;q25), detected by fluorescence in situ hybridization (FISH) analysis in an adult AML patient. A patient-specific cDNA library was screened with ETV6cDNA, and a fusion transcript between the ETV6 andTRKC/NTRK3 gene was identified. The fusion transcript encoded the PNT domain of ETV6 that fused to the protein-tyrosine kinase (PTK) domain of TRKC. Abnormal protein expression was detected by Western blot analysis, which was suspected to be oncogenic.
MATERIALS AND METHODS
Patient material.
Bone marrow samples obtained from a 59-year-old AML (FAB M2) patient with chromosome abnormalities of 48,XX,add(6)(q27),+8, inv(12)(p13q15),add(15)(q25),+add(15)(q25) were used in this study. The clinical and cytogenetic data of the patient have been described previously.16 Despite aggressive chemotherapy, she died of multiple organ failure associated with massive infiltration of leukemic cells in the systemic organs, 5 months after the onset of the leukemia.
Cytogenetic studies.
The metaphase samples were prepared from bone marrow cells, which were fixed after unstimulated short-term cultures. G-banded karyotypes were determined according to the International System for Human Cytogenetic Nomenclature (ISCN 199517).
FISH analysis.
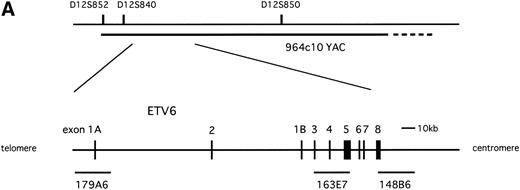
Metaphase sample that was applied to cytogenetic studies was also used for FISH analysis. Probes used were 964c10 YAC probe covering theETV6 locus on chromosome 12p13 (kindly provided by Dr Janet D. Rowley, Section of Hematology/Oncology, University of Chicago, Chicago, IL3), LL12NCO1 cosmid probes (179A6, 163E7, and 148B6, which are located within the ETV6 gene; kindly provided by Dr Peter Marynen, Center for Human Genetics, Flanders Interuniversity Institute for Biotechnology, Leuven, Belgium18), c-FES genomic probe (provided by HSRRB, Osaka, Japan), 170I1 BAC clone located on 15q25 (Research Genetics, Huntsville, AL), and D12Z3 (Oncor, Gaithersburg, MD), which hybridizes to the α satellite region of chromosome 12. The probes were labeled with biotin-11-dUTP or digoxigenin-11-dUTP (Boehringer Mannheim, Mannheim, Germany) using polymerase chain reaction (PCR) labeling after sequence-independent amplification,19 and were hybridized to metaphase samples as previously described.20 The probes were detected with avidin fluorescein (Vector Laboratories, Burlingame, CA) or anti-digoxigenin rhodamine (Boehringer Mannheim) and then counterstained with 4′ 6-diamidino-2-phenylindole dihydrochloride (DAPI). Images of the hybridized signals were captured under fluorescence microscopy (Olympus Optical Co, Tokyo, Japan).
Molecular cloning of the fusion mRNA.
Poly(A)+ RNA was extracted from leukemic cells with a Fast Track mRNA Isolation Kit (Invitrogen, San Diego, CA). Five micrograms of the extracted mRNA was reverse transcribed using cDNA synthesis module by random hexamer (Amersham, Buckinghamshire, UK), ligated withEcoRI-Not I-BamHI adaptor (Takara, Kyoto, Japan), and subcloned into EcoRI-digested λZap II vector (Stratagene, La Jolla, CA). The constructed cDNA library was screened with ETV6 full-length cDNA (kindly provided by Dr Todd R. Golub, Division of Pediatric Oncology, Dana-Farber Cancer Institute, Boston, MA2). The positive clones were isolated and subcloned into pBluescript phagemids according to the manufacturer’s instructions. Nucleotide sequences were determined using fluorescently labeled dideoxy terminators on 373A Applied Biosystems automated sequencer (Applied Biosystems, Urayasu, Japan). The sequence of nucleotides was compared with those in databases on the NCBI Blast server (Bethesda, MD).
Northern blot analysis.
Total RNA of the leukemic cells was isolated by the acid guanidinium thiocyanate-phenol-chloroform method. Ten micrograms of the RNA was separated by electrophoresis on 1.2% agarose gels containing 0.7 mol/L formaldehyde and transferred to Hybond-N membranes (Amersham). Probes used for hybridization were full-length ETV6 cDNA andTRKC cDNA fragment (at nucleotide position [nt] 1556-197021) prepared by PCR from fetal brain cDNA library (Clontech, Palo Alto, CA). Primers used were TRKC-1556f (5′-CGTGGCTGTCATCAGTGGTGAGG-3′) and TRKC-1947r (5′-CCATAGAACTTGACAATGTGCTC-3′). The same filter was rehybridized with a β-actin–specific probe to check the amount of RNA loaded in each lane.
Reverse transcriptase-mediated PCR (RT-PCR).
One microgram of the total RNA was reverse transcribed with 10 U of Moloney murine leukemia virus reverse trancriptase (MMLV-RT; Stratagene) using a random hexamer. One tenth of the synthesized cDNA was directed to PCR analysis using a PCR kit (Takara). The PCR conditions were as follows: 94°C for 3 minutes, followed by 30 cycles of 94°C for 1 minute, 55°C for 1 minute, 72°C for 1 minute and, finally, extension at 72°C for 10 minutes. The products were electrophoresed on 3% agarose gels and stained by ethidium bromide. The primers used were ETV6-116f (5′-CGATTCCGCCACTTCATGTTCCA-3′), ETV6-575r (5′-CGGTGATTTGTCGTGATAGGTG-3′), and TRKC primers mentioned above. Total RNA extracted from NBTM cell line, which was derived from a neuroblastoma (kindly provided by Dr Akira Nakagawara, Division of Biochemistry, Chiba Cancer Center, Chiba, Japan), was used as the positive control for TRKC expression.
Anchored PCR analysis.
Poly(A)+ RNA was extracted from the leukemic cells, and 500 ng was reverse transcribed and ligated with EcoRI-NotI-BamHI adaptor as mentioned above. To detect the 3′ETV6 mRNA, PCR analysis was performed with adaptor primer (5′-AATTCGGCGGCCGCGGATCC-3′) as the sense primer andETV6-541rE (5′-CGAATTCGCAACAGTTCAATGGTGGGAGG-3′) as the antisense primer. To detect the 5′ TRKC mRNA, PCR was performed with TRKC-1602fE (5′-CGAATTCCACATCAACCACGGCATCACC-3′) as the sense primer and adaptor primer as the antisense primer. The PCR conditions were the same as previously described. The resulting PCR product was subcloned into EcoRI/BamHI-digested into pBluescript phagemid (Stratagene) and sequenced.
Western blot analysis.
Leukemic cells (2 × 107) were washed in PBS; suspended in RIPA that contained 1% NP-40, 0.5% deoxycholate, and 0.5 mmol/L phenylmethyl sulfonyl fluoride (PMSF); homogenized with a Dounce homogenizer; and kept on ice for 30 minutes. The samples were centrifuged at 12,000g to remove any insoluble particles. Twenty-five micrograms of protein was separated on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels and transblotted onto Hybond-ECL membranes (Amersham). After appropriate blocking with 5% bovine serum albumin (BSA; Sigma Co, St Louis, MO), immunoblot analysis was performed with anti-TRK (C-14; Santa Cruz Biotechnology, Santa Cruz, CA), anti–N-TEL (kindly provided by Dr Olivier Bernard, U434 INSERM, Paris, France), and anti–β-actin antibodies (Santa Cruz). The signal was detected by incubating with horseradish peroxidase (HRP)-conjugated second antibody according to the manufacturer’s instructions and was exposed on FUJI RX film (FUJI Photo Film, Tokyo, Japan).
RESULTS
Rearrangement of the ETV6 gene in complex translocations between 12p13, 12q15, and 15q25.
To know whether the ETV6 gene was involved in inv(12)(p13q15), we applied FISH analysis using the 964c10 YAC probe that covers theETV6 gene. The signals turned out to be located on normal 12p13, on der(12)(q15), and on two der(15)(q25), but not on der(12)(p13) (Fig 1A and B).16This result caused us to hybridize the metaphase with cosmid probes (179A6, 163E7, and 148B6) that locate within the ETV6 gene. As a result, 179A6 hybridized on two der(15)(q25), 148B6 on der(12)(q15), and 163E7, which spans exon 3 to exon 5 of the ETV6 gene, was detected as split signals between der(12)(q15) and two der(15)(q25) (Figs 1A and B and 2A). These results indicated that two events, t(12;15)(p13;q25) and inv(12)(p13q15), occurred within the ETV6 gene in the leukemic cells.
(A) Physical map surrounding ETV6. The cosmid and YAC probes used in FISH analysis are shown underneath. (B) Schematic representation of t(12;15)(p13;q25) and inv(12)(p13q15) detected in the leukemic cells. Circles on the right indicate localization of probes detected in FISH analysis.
(A) Physical map surrounding ETV6. The cosmid and YAC probes used in FISH analysis are shown underneath. (B) Schematic representation of t(12;15)(p13;q25) and inv(12)(p13q15) detected in the leukemic cells. Circles on the right indicate localization of probes detected in FISH analysis.
FISH analysis of ETV6 and TRKC gene rearranged in the metaphase chromosomes. (A) Split signals of cosmid 163E7 (red) are observed on der(12)(q15) and on der(15)(q25). Large arrow indicates der(12), which is identified by chromosome 12 centromeric probe (D12Z3; green). The small arrows indicate der(15). Note that two der(15)s are detected in most metaphases. Large arrowhead indicates normal chromosome 12. (B) 170I1 BAC probe hybridized with 964c10 YAC probe. The 170I1 BAC (red) that encompasses the breakpoint of TRKC shows split signals on der(15)(q25) and on der(12)(p13). 964c10 YAC (green) covering ETV6 locus splits between der(12)(q15) and der(15)(q25), which results in fusion with 170I1 BAC signal on der(15)(q25). Normal chromosome 15 is indicated by a small arrowhead.
FISH analysis of ETV6 and TRKC gene rearranged in the metaphase chromosomes. (A) Split signals of cosmid 163E7 (red) are observed on der(12)(q15) and on der(15)(q25). Large arrow indicates der(12), which is identified by chromosome 12 centromeric probe (D12Z3; green). The small arrows indicate der(15). Note that two der(15)s are detected in most metaphases. Large arrowhead indicates normal chromosome 12. (B) 170I1 BAC probe hybridized with 964c10 YAC probe. The 170I1 BAC (red) that encompasses the breakpoint of TRKC shows split signals on der(15)(q25) and on der(12)(p13). 964c10 YAC (green) covering ETV6 locus splits between der(12)(q15) and der(15)(q25), which results in fusion with 170I1 BAC signal on der(15)(q25). Normal chromosome 15 is indicated by a small arrowhead.
Cloning and identification of a novel ETV6 fusion product.
To identify the fusion partner of the ETV6 gene, we constructed a cDNA library from leukemic cells that consisted of 1.25 × 105 independent clones and was screened with the full-length ETV6 cDNA probe. Five positive clones were obtained and were subcloned in pBluescript phagemid. Sequence analysis showed that two clones included a full-length ETV6 sequence, and the other three clones included ETV6 exon 1 to exon 4, followed by an unknown sequence, which was proven to be TRKC by BLAST database searching (ETV6 was referred to GenBank accession no.U11732 and human TRKC to U05012). In one clone (C-1), 487 bp of the ETV6 sequence, including the entire PNT domain, fused in-frame to the 3′-terminal 1,010 bp of the human TRKCgene. The fusion point of ETV6 was after nt 487, which is the end of exon 4, followed by nt 1741 of TRKC, including the entire PTK domain of TRKC (Fig 3). In the other two clones (C-2 and C-3), 487 bp of ETV6 fused toTRKC nt 1741 just the same as the C-1 clone, but the 3′-terminal 462 bp of TRKC was truncated at TRKCnt 2288 within the PTK domain, followed by 2 unrelated amino acids and a TGA stop codon (see Fig 7). This unreported sequence afterTRKC nt 2288 was confirmed to originate in the read-through intron sequence of normal TRKC by PCR for genomic DNA (data not shown). This short-form TRKC mRNA seems to be one of the splice variants of TRKC, because it is detected by RT-PCR in neuroblastoma samples that express TRKC (data not shown). Sequence analysis showed a deduced 450 amino acid open reading frame in C-1 that was suspected to code 52-kD protein, and a 339 amino acid open reading frame in C-2 and C-3 that were suspected to code 38-kD protein. The cDNA library was rehybridized with theTRKC cDNA probe spanning the breakpoint, but no positive clone including the 5′-portion of TRKC was obtained.
Schematic representation of ETV6, TRKC, and predicted ETV6-TRKC protein. Extracellular and transmembrane domain of TRKC is replaced by PNT domain of ETV6 as the result of t(12;15). C-1 chimeric transcript encodes the complete sequence of the PTK domain, but in C-2 and C-3, the 3′-terminal 462 bp of TRKC is truncated within the PTK domain. Nucleic acid and amino acid sequences surrounding the breakpoint are shown beneath. Vertical arrows indicate the breakpoint. Primers used for RT-PCR are indicated with small arrows. SP, putative signal peptide; TM, transmembrane domain; PTK, protein-tyrosine kinase domain; PNT, pointed domain; ETS, ETS binding domain.
Schematic representation of ETV6, TRKC, and predicted ETV6-TRKC protein. Extracellular and transmembrane domain of TRKC is replaced by PNT domain of ETV6 as the result of t(12;15). C-1 chimeric transcript encodes the complete sequence of the PTK domain, but in C-2 and C-3, the 3′-terminal 462 bp of TRKC is truncated within the PTK domain. Nucleic acid and amino acid sequences surrounding the breakpoint are shown beneath. Vertical arrows indicate the breakpoint. Primers used for RT-PCR are indicated with small arrows. SP, putative signal peptide; TM, transmembrane domain; PTK, protein-tyrosine kinase domain; PNT, pointed domain; ETS, ETS binding domain.
To know whether TRKC was involved in inv(12), we screened the human genome bacterial artificial library (BAC; Research Genetics) with a TRKC cDNA probe and obtained one positive clone, 170I1. FISH analysis was applied to identify the location of the probe on the metaphase of the leukemic cells, which resulted in split signals between der(15)(q25) and der(12)(p13) (Figs 1B and 2B). The c-FES probe located telomeric to the TRKC gene hybridized to normal chromosome 15 and to der(12)(p13). Finally, the karyotypes corrected by FISH analysis were 48,XX,add(6)(q27),+8, der(12)t(12;15)(p13;q25)inv(12)(p13q15),der(15)t(12;15)(p13;q25),+der(15)t(12;15)(p13;q25).
ETV6-TRKC but not TRKC-ETV6 fusion transcript is expressed.
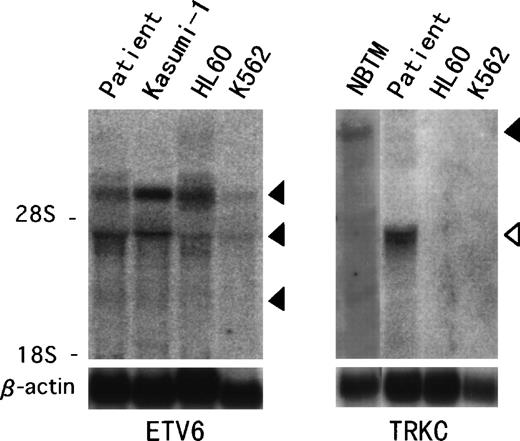
Northern blot analysis hybridized with ETV6 full-length cDNA showed previously described transcripts of 6.2, 4.3, and 2.4 kb in the t(12;15) leukemic cells and in other leukemic cell lines (Fig 4).18 On the other hand, aTRKC cDNA probe detected an abnormal 4.3-kb band only in the t(12;15) leukemic cells, compared with the 14-kb wild-type message detected in the NBTM cell line. RT-PCR analysis was performed to confirm the fusion transcript between the ETV6 and TRKCgene. We detected an amplified fragment of expected size using sense primer on ETV6 exon 2 (ETV6-116f) and antisense primer on the PTK domain of TRKC (TRKC-1947r) (Fig 5). On the other hand, we could not detect any reciprocal products of TRKC-ETV6 using the sense primer on the transmembrane domain of TRKC (TRKC-1556f) and the antisense primer on ETV6 exon 5 (ETV6-575r). A normal TRKC message was also undetectable.
Northern hybridization of t(12;15) leukemic cells.ETV6 full-length cDNA detects 6.2-, 4.3-, and 2.4-kb transcripts (solid arrowhead) in the t(12;15) leukemic cells and Kasumi-1, HL60, and K562 leukemic cell lines (left panel). TRKCprobe spanning the breakpoint detects a wild-type 14-kb transcript (solid arrowhead) in the NBTM cell line, whereas a 4.3-kb abnormal band (open arrowhead) is detected in the t(12;15) leukemic cells (right panel). No band is detected in HL60 or K562.
Northern hybridization of t(12;15) leukemic cells.ETV6 full-length cDNA detects 6.2-, 4.3-, and 2.4-kb transcripts (solid arrowhead) in the t(12;15) leukemic cells and Kasumi-1, HL60, and K562 leukemic cell lines (left panel). TRKCprobe spanning the breakpoint detects a wild-type 14-kb transcript (solid arrowhead) in the NBTM cell line, whereas a 4.3-kb abnormal band (open arrowhead) is detected in the t(12;15) leukemic cells (right panel). No band is detected in HL60 or K562.
RT-PCR analysis of ETV6-TRKC fusion products. Total RNA from t(12;15) leukemic cells (lanes 1 through 4) or NBTM cell line (lane 5) was reverse transcribed. PCR amplification was performed using primers ETV6-116f and ETV6-575r (lane 1),ETV6-116f and TRKC-1947r (lane 2), TRKC-1556f and ETV6-575r (lane 3), and TRKC-1556f andTRKC-1947r (lanes 4 and 5), respectively.
RT-PCR analysis of ETV6-TRKC fusion products. Total RNA from t(12;15) leukemic cells (lanes 1 through 4) or NBTM cell line (lane 5) was reverse transcribed. PCR amplification was performed using primers ETV6-116f and ETV6-575r (lane 1),ETV6-116f and TRKC-1947r (lane 2), TRKC-1556f and ETV6-575r (lane 3), and TRKC-1556f andTRKC-1947r (lanes 4 and 5), respectively.
Anchored-PCR was performed to confirm whether the reciprocal chimericETV6 mRNA exists in the cDNA library. An adaptor primer at the end of cDNA was used as the anchored sense primer andETV6-541rE primer was used as the antisense primer. The resulting PCR products were subcloned into pBluescript phagemid and 30 clones were sequenced, but all the clones encoded only the normalETV6 sequence. To isolate the reciprocal chimeric TRKCmRNA, the cDNA library was also amplified by the anchored-PCR withTRKC-1602fE primer as the sense primer and an adaptor primer as the antisense primer, but no PCR product was obtained. Although different types of chimeric mRNA, TRKC-ETV6, and/orTRKC–an unknown gene on 12q15, were suspected to express in the same leukemic cells, any reciprocal chimeric transcripts associating with the 3′-portion of the ETV6 gene or the 5′-portion of the TRKC gene have not been isolated by conventional cDNA screening, RT-PCR, and anchored PCR experiments.
Expression of ETV6-TRKC fusion protein.
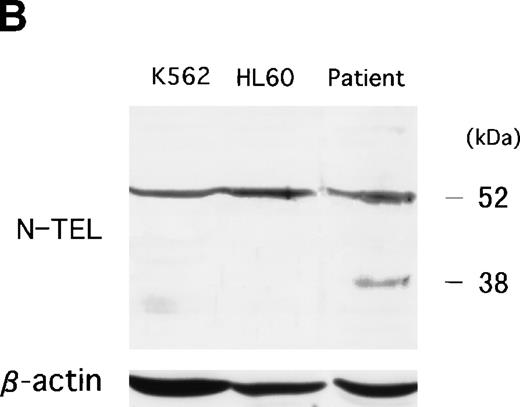
To examine whether the chimeric protein was expressed in the leukemic cells, Western blot analysis was performed with an anti-TRK antibody that recognizes the carboxy terminal of the TRK family (TRKA, TRKB, and TRKC). In the NBTM cell line, which expresses TRKA and TRKC, a positive band was detected at 140 to 145 kD in a normal size. On the other hand, an abnormal 52-kD band was detected in the t(12;15) leukemic cells as the expected size of a C-1 fusion transcript (Fig 6A). Because the carboxy terminal of TRK protein containing the epitope of anti-TRK antibody was deleted in C-2 and C-3 protein, the expected 38-kD band of C-2 and C-3 protein could not be detected. A 140-kD weak band was detected by an anti-TRK antibody in K562 cell line. However, the positive band in K562 was derived from TRKA22 but not from TRKC expression, which was confirmed by RT-PCR analysis (data not shown). No TRK band was detected in other leukemic cell lines such as HL60, Kasumi-1, Kasumi-3, and RS4;11 (data not shown). To determine whether small-sized fusion protein (C-2 and C-3) are also translated in the leukemic cells, anti–N-TEL antibody was used for Western blot analysis. A 52-kD band, which is the expected size of endogenous ETV623 and C-1 protein, was detected in all leukemic cells examined. On the other hand, the 38-kD band that reflects the size of the C-2 and C-3 protein was only detected in the leukemic cells with t(12;15) (Fig 6B). Therefore, both types of ETV6-TRKC were expressed in the leukemic cells.
(A) Abnormal expression of TRKC protein in t(12;15) leukemic cells. Total cell lysates were prepared from NBTM, t(12;15) leukemic cells, HL60 and K562. Wild-type TRK expression is detected in NBTM and weakly in K562. On the other hand, 52-kD abnormal protein is detected only in the t(12;15) leukemic cells. Anti-TRK antibody detects carboxy terminal of the TRK gene family. (B) ETV6/TEL expression in leukemic cells. Western blotting analysis using anti–N-TEL antibody detects a 52-kD band corresponding to the normal ETV6 protein in t(12;15) leukemic cells, HL60, and K562. C-1 ETV6-TRKC protein in t(12;15) leukemic cells is also detected as the 52-kD band. The t(12;15) leukemic cells express a 38-kD protein corresponding to the C-2 and C-3 ETV6-TRKC protein.
(A) Abnormal expression of TRKC protein in t(12;15) leukemic cells. Total cell lysates were prepared from NBTM, t(12;15) leukemic cells, HL60 and K562. Wild-type TRK expression is detected in NBTM and weakly in K562. On the other hand, 52-kD abnormal protein is detected only in the t(12;15) leukemic cells. Anti-TRK antibody detects carboxy terminal of the TRK gene family. (B) ETV6/TEL expression in leukemic cells. Western blotting analysis using anti–N-TEL antibody detects a 52-kD band corresponding to the normal ETV6 protein in t(12;15) leukemic cells, HL60, and K562. C-1 ETV6-TRKC protein in t(12;15) leukemic cells is also detected as the 52-kD band. The t(12;15) leukemic cells express a 38-kD protein corresponding to the C-2 and C-3 ETV6-TRKC protein.
DISCUSSION
We report here an ETV6-associated cryptic translocation t(12;15)(p13;q25) found in an adult patient with AML (M2). The molecular cloning of the ETV6 cDNA showed a novel fusion partner gene, TRKC, which for the first time is reported to be associated with leukemia. TRKC is a receptor tyrosine kinase that is activated by NT-3, a member of the nerve growth factors.21The TRKC gene product encodes three leucine-rich motifs flanked by conserved cysteine residues and two Ig-like loops in the extracellular domain. The intracellular domain encodes the well-conserved receptor tyrosine kinase domain, which is a common structure in the TRK subfamily members, TRKA, TRKB, and TRKC. TRKC is known to be essential in the development of the central and peripheral nervous systems.24 Expression of TRKC is linked to a favorable outcome in both neuroblastoma and medulloblastoma.25,26 Activation of TRKA as a result of recombination with various genes has been reported in some solid tumors such as colon carcinoma and thyroid carcinoma.27 28 Because abnormality of the TRKC gene in oncogenesis has not been reported so far, this is the first report that TRKC activation occurred in a type of leukemia.
Receptor and nonreceptor tyrosine kinase is known to play an important role in cell growth and differentiation. On the other hand, activation of these genes by amplification, mutation, or recombination results in sustained activation of their kinase domain, which leads to cellular transformation. The BCR-ABL fusion gene, which is observed in t(9;22)-positive chronic myeloid leukemia (CML) or ALL, is one of the well-known models in hematologic malignancies.29 Recent studies have shown that someETV6-associated fusion genes, ETV6-PDGFRβ,ETV6-ABL, and ETV6-JAK2, also have oncogenic properties due to their activated tyrosine kinase activities.5,11,30,31 These fusion transcripts contain the PNT domain of ETV6 that fused to the catalytic domain of the partner PTK gene. It has been suggested that the PNT domain derived from ETV6 contributes to the oncogenic properties of these chimeric proteins by forming PNT domain-dependent homo-oligomers that lead to ligand-independent tyrosine kinase activation.30 31TRKC is preferentially expressed in neural tissues and in some nonneural tissues, but it has not yet been detected in hematopoietic cells. Taking these findings into consideration, theETV6-TRKC chimeric transcript, composed of the PNT domain of ETV6 that fused to the PTK domain of TRKC, may possess transforming properties by aberrantly activated PTK expression in the leukemic cells induced by oligodimerization. In this case, two types of chimeric transcripts were detected: one included the entire PTK domain of TRKC (C-1) and the other had the truncated PTK domain of TRKC (C-2 and C-3). These chimeric transcripts may possess different potentials in transforming activities. Further experiments are needed for addressing the status of these ETV6-TRKC chimeric proteins in cells to show the activation mechanism of this kinase.
In t(12;15), the reciprocal transcript of ETV6-TRKC was not detected and neither was any kind of chimeric mRNA including the 3′-portion of ETV6 gene. In t(5;12) and t(9;12), the reciprocal transcripts of ETV6-PDGFRβ, ETV6-ABL, andETV6-JAK2 were not detected either.2,4 11 Two kinds of genetic alteration, t(12;15) and inv(12)(p13q15), were found in the leukemic cells. However, the ETV6-TRKC chimeric transcript was only isolated from the breakpoints, suggesting that theETV6-TRKC transcripts seem to have some role in leukemogenesis in this type of leukemia.
The leukemic cells had very aggressive clinical characteristics. The patient’s bone marrow was accompanied by severe fibrosis at the time of diagnosis, and the leukemic cells had invaded multiple organs in a short period.16 It is not yet clear whether this distinct phenotype is due to t(12;15). An accumulation of reported cases would be needed to discuss this point. Because this translocation is too cryptic to be detected by conventional cytogenetic analysis, FISH or RT-PCR analysis is recommended to study the presence of this translocation. The combination of 964c10 YAC and 170I1 BAC probes is an appropriate tool to detect t(12;15) by FISH analysis. One hundred AML patients were screened by the FISH method to see whether cytogenetically undetected t(12;15) exists, but it has not yet been found. t(12;15) seems to be a rather rare event.
While this report was in preparation, the sameETV6-TRKC fusion gene was reported in cases of congenital fibrosarcoma.32 The reported chimeric transcript encoded exon 1 to exon 5 of the ETV6 gene that fused to nt 1741 of the TRKC gene, resulting in a fusion protein functionally resembling the one in our study. There were only two differences between the chimeric transcript defined in the leukemic cells and in congenital fibrosarcoma. The first is the existence of ETV6exon 5 in the chimeric transcript in the latter case. The second is an additional 14 amino acid insertion which was observed within the catalytic kinase domain of TRKC (nt 2288) in the case of congenital fibrosarcoma (Fig 7). The amino acid insertion occurred just downstream from the TDYYR motif that encompasses the putative autophosphorylation site of the TRK receptor family.33,34 The biological properties of this TRKCreceptor isoform were previously studied.33,35 Upon interaction with NT-3, both isoforms became rapidly phosphorylated on the tyrosine residues and induced DNA synthesis in quiescent cells. However, only TRKC without amino acid insertion had mitogenic activity in NIH3T3 cells and also induced neuronal differentiation of pheochromocytoma-derived PC12 cells. Moreover, only noninsertion type TRKC phosphorylated phospholipase Cγ1 or phosphatidylinositol-3 kinase, which is one of the TRKC substrates. These results suggest that different trophic activities are mediated by TRKC isoforms, which may contribute to the clinical and pathological differences between a very aggressive leukemia and rather benign congenital fibrosarcoma. There are some examples of genes that affect both leukemia and solid tumors such as ERG and ALL-1,36 37 but no identical fusion transcript has been described in either case so far.ETV6-TRKC gives a new entity of the fusion gene that may have the potential to transform both hematologic and nonhematologic cells.
Nucleotide and deduced amino acid sequence of the PTK domain of ETV6-TRKC chimeric genes detected in t(12;15)-positive AML (C-1, C-2, and C-3) and congenital fibrosarcoma (CFS). In C-2 and C-3, 3′-terminal 462 bp was truncated within the PTK domain at nucleotide position (nt) 2288 of TRKC, followed by 2 unrelated amino acids and TGA stop codon originating from the intron sequence of TRKC. The number shown above is the amino acid residue in C-1. The nucleotide and deduced amino acid sequence of the 42-bp sequence inserted in the kinase domain in CFS are also indicated. The solid box shows the tyrosine residues (YY) corresponding to the putative major autophosphorylation sites ofTRKC.
Nucleotide and deduced amino acid sequence of the PTK domain of ETV6-TRKC chimeric genes detected in t(12;15)-positive AML (C-1, C-2, and C-3) and congenital fibrosarcoma (CFS). In C-2 and C-3, 3′-terminal 462 bp was truncated within the PTK domain at nucleotide position (nt) 2288 of TRKC, followed by 2 unrelated amino acids and TGA stop codon originating from the intron sequence of TRKC. The number shown above is the amino acid residue in C-1. The nucleotide and deduced amino acid sequence of the 42-bp sequence inserted in the kinase domain in CFS are also indicated. The solid box shows the tyrosine residues (YY) corresponding to the putative major autophosphorylation sites ofTRKC.
ACKNOWLEDGMENT
The authors thank Dr Janet D. Rowley for providing the 964c10 YAC probe, Dr Peter Marynen for providing the LL12NCO1 cosmid probes, Dr Todd R. Golub for providing TEL cDNA probe, and Dr Olivier Bernard for providing anti–N-TEL antibody. We also thank Dr Akira Nakagawara for providing NBTM cell line and Dr Eiso Hiyama (Department of General Medicine, Hiroshima University, School of Medicine, Hiroshima, Japan) for providing neuroblastoma samples.
Supported by a Grant-in-Aid for Scientific Research of Cancer (08266109) from the Ministry of Education, Science, Sports and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Nanao Kamada, MD, Department of Cancer Cytogenetics, Research Institute for Radiation Biology and Medicine, Hiroshima University, 1-2-3, Kasumi, Minami-ku, Hiroshima 734-8553, Japan.