Abstract
The idiopathic hypereosinophilic syndrome (IHES) is a rare disorder characterized by unexplained, persistent eosinophilia associated with multiple organ dysfunction due to eosinophilic tissue infiltration. In the absence of karyotypic abnormalities, there is no specific test to detect clonal eosinophilia in IHES. Analysis of X-chromosome inactivation patterns can be used to determine whether proliferative disorders are clonal in origin. Methylation of HpaII andHha I sites near the polymorphic trinucleotide repeat of the human androgen receptor gene (HUMARA) has been shown to correlate with X-inactivation. In this study, we have used the polymerase chain reaction (PCR) with nested primers to analyze X-inactivation patterns of the HUMARA loci in purified eosinophils from female patients with eosinophilia. Peripheral blood eosinophils were isolated by their autofluoresence using flow cytometric sorting. Eosinophils purified from a female patient presenting with IHES were found to show a clonal pattern of X-inactivation. Eosinophil-depleted leukocytes from this patient were polyclonal by HUMARA analysis, thus excluding skewedness of random X-inactivation. After corticosteroid suppression of her blood eosinophilia, a clonal population of eosinophils could no longer be detected in purified eosinophils. In contrast, eosinophils purified from a patient with Churg-Strauss syndrome and from six patients with reactive eosinophilias attributed to allergy, parasitic infection, or drug reaction showed a polyclonal pattern of X-inactivation by HUMARA analysis. The finding of clonal eosinophilia in a patient presenting with IHES indicates that such patients may have, in reality, a low-grade clonal disorder that can be distinguished from reactive eosinophilias by HUMARA analysis. Further, the method described can be used to monitor disease progression.
THE IDIOPATHIC hypereosinophilic syndrome (IHES) is a rare disorder characterized by persistent eosinophilia of unknown origin often associated with multiple organ dysfunction as a result of infiltration by tissue eosinophils and the toxic effects of their released granule contents.1 There is no specific test that is diagnostic for IHES; rather the syndrome is defined as the combination of unexplained prolonged eosinophilia and evidence of organ involvement.2 It must be distinguished from reactive, clinically benign eosinophilia associated with parasitic, allergic, and other unexplained causes of eosinophilia,3 and also from eosinophilic leukemias associated with increased blast cells and cytogenetic abnormalities. Moreover, some patients with IHES may show features usually associated with myeloproliferative syndromes such as splenomegaly and thrombocytopenia.1,4,5 Cases of clonal eosinophilia as evidenced by karyotypic abnormalities in bone marrow cells can usually be classified as chronic eosinophilic leukemia, myelodysplastic syndromes, or myeloproliferative disorders.6 However, in the absence of karyotypic abnormalities, the distinction between these entities and IHES may be difficult. A method that can detect clonal eosinophilia could allow more accurate diagnosis and treatment of cases presenting as IHES and help to clarify its relationship to the myeloproliferative disorders.
Analysis of maternal and paternal X-chromosome activation status has been used as a means of determining whether certain proliferative disorders are clonal in origin.7-10 These methods are based on the assumption that random inactivation of one X-chromosome occurs in each somatic cell during early embryogenic development, and that this is passed on to the progeny of the cell in a stable fashion.11-14 Therefore, some cells from a normal female carry active maternal X-chromosome, while other cells carry active paternal X-chromosome. Differential methylation patterns between the active and inactive X-chromosome have been documented at several loci such as the phosphoglycerate kinase (PGK), hypoxanthine phosphoribosyl transferase (HPRT), and the hypervariable DXS255 (M27β) loci.15 Recently, Allen et al9have shown that methylation of HpaII and Hha I sites near the polymorphic trinucleotide CAG repeat of the human androgen receptor gene (HUMARA) correlates with X-inactivation. Digestion of DNA with a methylation-sensitive restriction endonuclease, such asHpaII, permits the distinction of the active (unmethylated) from the inactive (methylated) X-chromosome. Therefore, one strategy to determine whether eosinophils from female patients with IHES are clonal would be to analyze the X-inactivation patterns of the HUMARA loci in purified eosinophils.
In a recent report, Luppi et al16 analyzed the methylation status of the PGK gene in granulocytes from a female IHES patient with 70% eosinophilia and found this to show a clonal pattern by Southern hybridization. However, PGK gene analysis is restricted by a low incidence of constitutional heterozygosity of approximately 40% in the general female population.10 By contrast, the HUMARA loci have been reported to show a much higher incidence of constitutional heterozygosity of approximately 90%.9,17 Further, it would be an advantage to extend such studies to patients with lesser proportions of eosinophils in the peripheral blood by isolating eosinophils for clonality analysis. Human eosinophils emit marked fluorescence at 520 nm when excited at 450 nm due to their fluorescent granule contents, and this property may be used to purify eosinophils for study.18 19 Here, we have purified peripheral blood eosinophils by their autofluorescence using flow cytometric sorting and investigated the clonality of purified eosinophils from female patients by analysis of X-inactivation patterns of the HUMARA loci using polymerase chain reaction (PCR) amplification with nested primers.
MATERIALS AND METHODS
Sample Preparation
Peripheral blood samples in EDTA were collected from the patients studied. Informed consent was obtained from all patients in this study, which was approved by the local Institutional Review Board (Ethics). Red blood cells were lysed with buffer containing 15.5 mmol/L NH4Cl, 1 mmol/L KHCO3, and 10 μmol/L Na2 EDTA. White blood cells were collected by centrifugation at 2,000 rpm for 10 minutes. After centrifugation, the samples were resuspended in 5 mL phosphate-buffered saline (PBS, pH 7.4) and washed twice. Each sample was then separated by flow cytometry into a purified eosinophil fraction and an eosinophil-depleted leukocyte fraction.
Isolation of eosinophils from peripheral blood leukocytes by flow cytometry and cell sorting.
Eosinophils were separated by flow cytometry based on their autofluorescence and granularity18 19 to greater than 90% purity (Figs 1 and2). Flow cytometric analysis and cell sorting was performed on a FACStarPLUS flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Cells were excited with a 488-nm argon ion laser and fluorescence was measured through a BP 525 nm (FL1 detector) and 575 nm (FL2) filters. Results were analyzed using the LYSYS II version 1.1 software (Becton Dickinson). Cells were sorted at a cell flow rate of 800 events/s. A small sample of sorted cells were cytospun onto glass slides for Giemsa staining to confirm either eosinophil purity (Fig 2) or eosinophil depletion by morphology. The remainder of the sorted cells were processed for DNA extraction.
Flow cytometric sorting of eosinophils using autofluorescence and light scatter patterns. (A) Peripheral blood leukocytes from case 1 were obtained from whole blood after removal of erythrocytes by lysis and analyzed on a FACStarPLUS flow cytometer. (AI) The populations of leukocytes were differentiated by their side scatter and autofluorescence patterns: eosinophils (R1, 57.9%), granulocytes (R2, 26.5%), and lymphocytes (R3, 12.8%). Cells of the region R1 exhibited marked autofluorescence that was detected on FL1, and to a lesser extent, on FL2. Cells in R1 were sorted into sterile tubes containing 2 mL of sterile PBS (pH 7.4). (AII) Resultant cell populations contained 98.5% eosinophils (R1), 1.0% granulocytes (R2), and 0.4% lymphocytes (R3). (B) Twenty-six months later, peripheral blood leukocytes from case 1 were obtained and again analyzed on a FACStarPLUS flow cytometer. (BI) Eosinophil (R1, 6.0%), granulocyte (R2, 74.0%), and lymphocyte (R3, 17.3%) populations were noted among the leukocytes. Cells in R1 were purified by flow cytometric sorting. (BII) The resultant cell populations contained 92.0% eosinophils (R1), 4.0% granulocytes (R2), and 4.0% lymphocytes (R3).
Flow cytometric sorting of eosinophils using autofluorescence and light scatter patterns. (A) Peripheral blood leukocytes from case 1 were obtained from whole blood after removal of erythrocytes by lysis and analyzed on a FACStarPLUS flow cytometer. (AI) The populations of leukocytes were differentiated by their side scatter and autofluorescence patterns: eosinophils (R1, 57.9%), granulocytes (R2, 26.5%), and lymphocytes (R3, 12.8%). Cells of the region R1 exhibited marked autofluorescence that was detected on FL1, and to a lesser extent, on FL2. Cells in R1 were sorted into sterile tubes containing 2 mL of sterile PBS (pH 7.4). (AII) Resultant cell populations contained 98.5% eosinophils (R1), 1.0% granulocytes (R2), and 0.4% lymphocytes (R3). (B) Twenty-six months later, peripheral blood leukocytes from case 1 were obtained and again analyzed on a FACStarPLUS flow cytometer. (BI) Eosinophil (R1, 6.0%), granulocyte (R2, 74.0%), and lymphocyte (R3, 17.3%) populations were noted among the leukocytes. Cells in R1 were purified by flow cytometric sorting. (BII) The resultant cell populations contained 92.0% eosinophils (R1), 4.0% granulocytes (R2), and 4.0% lymphocytes (R3).
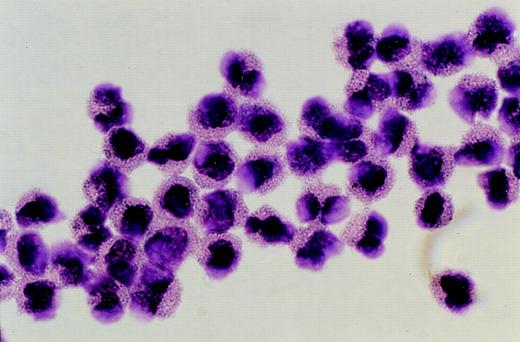
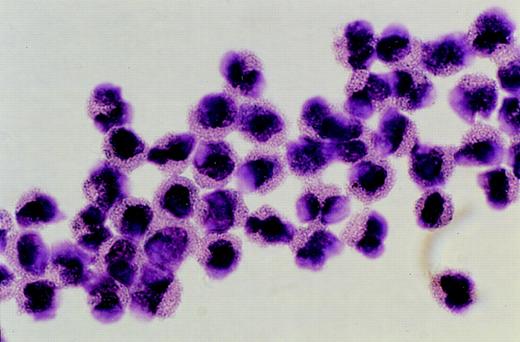
Purified eosinophils isolated by flow cytometric sorting. Photomicrograph showing eosinophils purified from peripheral blood by flow cytometric sorting to greater than 90% purity. (Giemsa, × 1,300 original magnification).
Purified eosinophils isolated by flow cytometric sorting. Photomicrograph showing eosinophils purified from peripheral blood by flow cytometric sorting to greater than 90% purity. (Giemsa, × 1,300 original magnification).
HUMARA Analysis
Preparation of DNA from peripheral blood samples was performed as described elsewhere.20 21 Briefly, about 600 μL of solution containing 50 mmol/L Tris (pH 8.0), 0.5% sodium dodecyl sulfate, 1 mmol/L EDTA, and 10 mg/mL proteinase K was added to a final concentration of 100 μg/mL and incubated at 55°C for 15 hours. The samples were then extracted once with phenol, twice with phenol-chloroform, and precipitated by 2.5 vol of ethanol. After centrifugation, the DNA was resuspended in 200 μL 10 mmol/L Tris (pH 8.0) with 1 mmol/L EDTA.
Clonality studies at the HUMARA locus were performed as described previously,9 except that internal (“nested”) primers were included (Fig 3). Briefly, about 200 ng of DNA was either digested overnight with 10 U of HpaII or not digested. The reaction was terminated by heating at 95°C for 5 minutes. The PCR reaction was performed in a total volume of 20 μL containing 0.5 μmol/L oligomers, 150 μmol/L of each dNTP (Dupont, Wilmington, DE), 2.5 mmol/L MgCl2, 50 mmol/L KCl, 10 mmol/L Tris-HCl, 5% dimethyl sulfoxide, 0.8 U of Taq polymerase, and 200 ng of DNA. The sequences of the primers were: 5′ TCCAGAATCTGTTCCAGAGCGTGC 3′ (primer A) and 5′ GCTGTGAAGGTTGCTGTTCCTCAT 3′ (primer B). Samples were amplified for 28 cycles comprising 1 minute at 95°C, 45 seconds at 60°C, and 45 seconds at 72°C with initial denaturation at 95°C for 5 minutes. One-tenth volume of this PCR mixture was added into another reaction mixture. The following internal primers were then added: 5′ GTGCGCGAAGTGATCCAGGA 3′ (primer A1) and 5′ TTCCTCATCCAGGACCAGGT 3′ (primer B1). The amplification was repeated as described above except that the PCR reaction was performed in a total vol of 10 μL for 20 amplification cycles and 50 to 100 nmol/L γ-33P-adenosine triphosphate (ATP) end-labeled primer A1 (>1,000 Ci/mmol; Dupont) was added. Five microliters of sequencing gel-loading buffer (95% formamide, 10 mmol/L EDTA, 0.025% bromophenol blue) were added to the reaction mixture. Four microliters of this mixture were loaded onto a denaturing 6% (39:1 acrylamide/bis with 6 mol/L urea) gel and electrophoresed at 80 W for 3 hours. The gel was dried and exposed.
Diagram of the region amplified in the HUMARA gene. TwoHpaII sites are adjacent to the polymorphic CAG repeats.
Diagram of the region amplified in the HUMARA gene. TwoHpaII sites are adjacent to the polymorphic CAG repeats.
HUMARA clonality analyses of all the samples in this study were repeated at least twice to ensure that the results were reproducible.
To obtain the proportional expression of the maternal and paternal alleles in HUMARA analysis, band intensities from the autoradiographs were analyzed using Molecular Analyst software (Bio-Rad Laboratories, Hercules, CA). The proportional expression of the two alleles was obtained as described by Gale et al,17 except that the final ratio was expressed as percentages of the total signal from the two alleles. Briefly, the intensity of each band after HpaII digestion was first corrected by dividing it by the signal obtained for that allele in the undigested sample. The intensity of the signals from the two alleles was then expressed as a ratio. The ratio was finally expressed as a percentage of the total signal for the two alleles. Therefore, a ratio of 50:50 would represent equal (balanced) expression of the two alleles.
RESULTS
Patients
Studies were performed on eight female patients (mean age, 48.5 years; range, 41 to 65 years) with eosinophilia (Table1). The diagnosis in one patient (case 1) was consistent with IHES, and in another (case 2), with the Churg-Strauss syndrome. Eosinophilia in the remaining patients (cases 3 to 8) was judged clinically to be reactive in nature based on a likely cause for eosinophilia, such as drug reaction due to phenytoin (case 3), parasitic infection due to Strongyloidiaisis (case 4), atopic eczema secondary to house dustmite allergy (case 5), asthma secondary to house dustmite allergy (case 6), and extrinsic allergic asthma (cases 7 and 8). The patients with reactive eosinophilia did not manifest tissue damage attributable to eosinophilia, and the duration of eosinophilia was transient in cases 3 and 4. Details of the patients presenting as IHES or Churg-Strauss syndrome were as follows.
Case 1.
A50-year-old housewife was admitted with progressive exertional dyspnea, orthopnea, and bilateral ankle swelling for several days. She gave a history of chronic nonproductive cough for several months and weight loss of 10 kg in the preceding 10 months. Examination showed physical signs of severe congestive cardiac failure. The total white cell count was 13.5 × 109/L with 40% eosinophils with an absolute eosinophil count (AEC) of 5.4 × 109/L and a mild normochromic anemia (hemoglobin, 10.2 g/dL) with occasional macrocytes in the blood film. Liver and renal biochemical screening tests showed mildly elevated serum alanine and aspartate aminotransferases and lactate dehydrogenase, but were otherwise normal. Chest x-ray showed cardiac enlargement with pulmonary edema. Cardiac doppler echocardiography confirmed a dilated cardiomyopathy with poor left ventricular function and a small pericardial effusion. Serum folate was low at 6.6 nmol/L (normal range [NR] = 10.0 to 40.0 nmol/L). Serum vitamin B12 and the neutrophil alkaline phosphatase score were in the normal range. Polyclonal IgE was increased to 1,107 U/mL (NR = 10.0 to 180 U/mL). Rheumatoid factor, antinuclear factor, and anti-DNA antibodies were negative. Stool examinations for ova, cysts, and parasites were repeatedly negative. The bone marrow aspirate showed moderately hypercellular fragments with a marked increase in eosinophils and eosinophilic precursors. Cytogenetic analysis of bone marrow cells showed a normal karyotype. She was treated with diuretics and folic acid and started on prednisolone 1 mg/kg/d. Her symptoms improved dramatically with a fall in the AEC to 0.4 × 109/L after 3 days of treatment. During the first 12 months after initial presentation, her AEC rose above 1.5 × 109/L on several occasions, accompanied by symptoms and signs of congestive heart failure. As she refused α-interferon therapy, control was reestablished by an increase in prednisolone dosage. Twelve months after initial presentation, due to increasing side effects of corticosteroid therapy, hydroxyurea was substituted for prednisolone, but had to be discontinued 20 months after initial presentation due to worsening macrocytic anemia. From that time until the present, the patient has been maintained on a small dose of prednisolone to suppress her eosinophil count. During treatment, her serum IgE level fell, but remained mildly elevated at 356 U/mL, 26 months after initial presentation. HUMARA analysis of purified blood eosinophils was performed at initial presentation and at 26 months after initial presentation.
Case 2.
A 43-year-old Caucasian woman presented with rashes on her hands and paraesthesia and swelling of her feet for 10 days. She gave a history of asthma for about 15 years, controlled with intermittent courses of corticosteroids and salbutamol. Fifteen years ago, in a hospital abroad, she had been diagnosed with “pulmonary eosinophilia” and 2 years previously, she had suffered a severe angioneurotic orbital cellulitis. On examination she had vasculitic lesions on her hands and sensory loss to pinprick in a “glove and stocking” distribution of her hands and feet and bilateral pitting edema up to the midcalves. Chest radiography showed old scarring and small areas of “ground glass” appearance consistent with pulmonary allergic granulomatosis. The total white cell count was 26.5 × 109/L, with 43% eosinophils. Biopsy of the skin lesions showed vasculitis and tissue eosinophilia. IgE levels were elevated (895 U/mL). Nerve conduction studies showed a mild sensory neuropathy of the lower limbs. A large pericardial effusion was confirmed by doppler echocardiography. Antineutrophil cytoplasmic antibodies, anti-DNA antibodies, rheumatoid factor, and antifilarial antibody were negative. A diagnosis of Churg-Strauss syndrome with peripheral neuropathy and myositis was made. Her symptoms are presently well-controlled with prednisolone and salbutamol.
HUMARA Analysis
Amplification of the HUMARA gene by nested primers gives a 240-bp fragment that spans two HpaII cutting sites and the variable CAG tandem repeats (Fig 3). Analysis of the HUMARA gene is based on the rationale that amplification by PCR cannot take place if the active (unmethylated) X-chromosome is predigested with a methylation sensitive enzyme (HpaII). Methylation at these sites has been shown to correlate with X-inactivation,9 so that a product will only be obtained if the X-chromosome is inactive. In a polyclonal cell population in which random X-inactivation has taken place, both alleles persist after HpaII predigestion so that in a high resolution polyacrylamide gel, two bands are seen, as the maternal inactive X-allele is distinct from paternal inactive X-allele because of a different number of CAG repeats. By contrast, in a monoclonal cell population in which nonrandom X-inactivation has taken place, the active allele disappears after HpaII predigestion, as all cells contain the same inactive X-chromosome,9 so that only one band from either parental inactive X-allele is shown by gel electrophoresis. The band representing an allele is sometimes accompanied by a minor band one trinucleotide repeat smaller, which is due to slippage of Taq polymerase along the repeated sequences.17
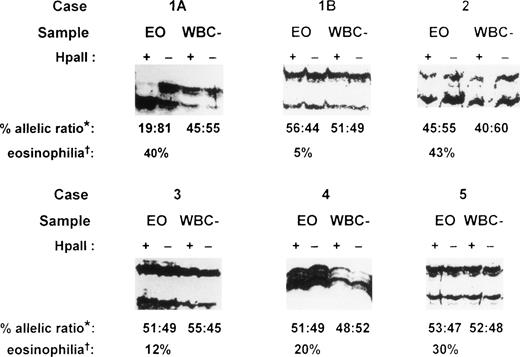
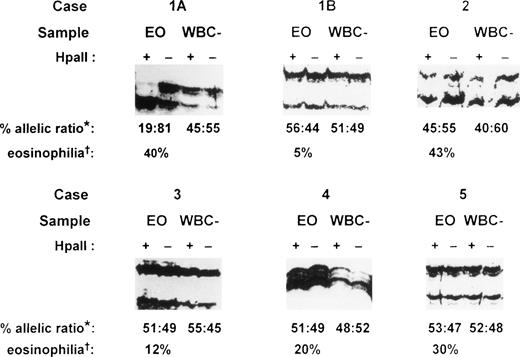
PCR amplification of the hypervariable HUMARA target gene was performed on both purified eosinophil fractions and eosinophil-depleted leukocyte fractions from each patient. Eosinophil-depleted leukocyte fractions consisted of variable proportions of a mixed population of peripheral blood neutrophils, lymphocytes, and monocytes, with fewer than 10% eosinophils. These fractions served as polyclonal controls to exclude skewedness of X-inactivation that is found in a small proportion of females in HUMARA analysis.9 17 In the patient presenting with IHES (case 1), peripheral blood eosinophils were purified to 98.5% purity by flow cytometry at initial presentation (Fig 1A). Gel electrophoresis under denaturing conditions of HpaII-digested samples of eosinophil-depleted leukocytes showed a band originating from each parental X-chromosome (Fig 4, cases 1A and 1B, WBC–). Eosinophil-depleted leukocyte fractions from case 1, therefore, showed a random (polyclonal) pattern of X-inactivation, thus excluding extreme skewedness of X-inactivation in this individual. By contrast, HUMARA analysis of the purified eosinophil fraction from this patient at initial presentation (Fig 4, case 1A, EO) showed only a band from one parental allele afterHpaII digestion, consistent with a nonrandom (clonal) pattern of X-inactivation in the purified eosinophils.
HUMARA analysis of patients with eosinophilia. Case 1, idiopathic hypereosinophilic syndrome; case 2, Churg-Strauss syndrome; case 3, eosinophilia secondary to phenytoin ingestion; case 4, eosinophilia secondary to Strongyloidiasis infection; and case 5, eosinophilia secondary to house dustmite allergy causing atopic eczema. Case 1A, sample obtained at initial presentation; case 1B, sample obtained 26 months after initial presentation; EO, purified eosinophil fraction; WBC–, eosinophil-depleted leukocyte fraction; +, DNA amplified after HpaII precutting; –, DNA amplified without HpaII precutting; *, percent allelic ratio: this is the ratio of the band intensity of the upper allele to the lower allele after HpaII digestion as a percentage of the total intensity of the two alleles. The intensity of each band after HpaII digestion was first corrected by dividing it by the signal obtained for that allele in the undigested sample (for details, see Materials and Methods). †, eosinophilia: percentage eosinophils of the peripheral blood total white cell count before eosinophil purification.
HUMARA analysis of patients with eosinophilia. Case 1, idiopathic hypereosinophilic syndrome; case 2, Churg-Strauss syndrome; case 3, eosinophilia secondary to phenytoin ingestion; case 4, eosinophilia secondary to Strongyloidiasis infection; and case 5, eosinophilia secondary to house dustmite allergy causing atopic eczema. Case 1A, sample obtained at initial presentation; case 1B, sample obtained 26 months after initial presentation; EO, purified eosinophil fraction; WBC–, eosinophil-depleted leukocyte fraction; +, DNA amplified after HpaII precutting; –, DNA amplified without HpaII precutting; *, percent allelic ratio: this is the ratio of the band intensity of the upper allele to the lower allele after HpaII digestion as a percentage of the total intensity of the two alleles. The intensity of each band after HpaII digestion was first corrected by dividing it by the signal obtained for that allele in the undigested sample (for details, see Materials and Methods). †, eosinophilia: percentage eosinophils of the peripheral blood total white cell count before eosinophil purification.
Twenty-six months after initial presentation when the patient’s symptoms and blood eosinophil count (AEC = 0.5 × 109/L) were controlled with a small dose of prednisolone, eosinophils were once again purified by flow cytometry (Fig 1B) for HUMARA reanalysis (Fig 4, case 1B). The eosinophil content of the purified eosinophil fraction was 92% (Fig 1B). By HUMARA analysis, a polyclonal pattern of X-inactivation was seen (Fig 4, case 1B, EO), indicating either that the clonal eosinophilic population was absent or that it was insufficient, as a proportion of the total eosinophil population, to give rise to a clonal pattern.
Samples from case 1 were also assayed for clonality using PGK gene analysis by PCR,22 but the patient was not heterozygous at the PGK locus, which was therefore uninformative (not shown).
In case 2, a patient with the Churg-Strauss syndrome, HUMARA analysis of both purified eosinophil and eosinophil-depleted leukocyte fractions showed a similar polyclonal pattern (Fig 4, case 2), indicating that the purified eosinophils were of polyclonal origin in this condition. In all of the other six patients with reactive eosinophilia, HUMARA analyses of both purified eosinophil and eosinophil-depleted leukocyte fractions showed a polyclonal pattern of X-inactivation, confirming that the eosinophil populations in these patients were polyclonal in origin. Representative analyses are shown (Fig 4, cases 3 to 5). HUMARA results in all samples in this study were found to be consistent on repeated analyses.
DISCUSSION
The distinction between a malignant and nonmalignant process in IHES can be extremely difficult, as there are no specific markers that can be applied to ascertain the clonality of eosinophils.2 We have attempted to solve this problem by analysis of the X-inactivation status of the HUMARA loci in purified eosinophils from female patients with eosinophilia. One patient presented with IHES, as evidenced by recurrent elevated AEC and organ damage involving the heart. A second patient presented with vasculitic lesions, asthma, pulmonary eosinophilia, peripheral neuropathy, pericarditis, and myositis and clinically fulfilled the criteria for the Churg-Strauss syndrome.23 24 The remaining six patients with reactive eosinophilia showed either transient or persistent eosinophilia that was attributable to drug reactions, allergies, or parasitic infection. These cases were clinically benign and did not show evidence of organ damage associated with eosinophilia. By X-inactivation analysis, we found clonal eosinophilia in the patient with IHES, whereas eosinophils were polyclonal in the patient with Churg-Strauss syndrome, and also in the six other patients with reactive eosinophilia. Further, in the patient with IHES, analysis of eosinophil-depleted leukocyte fractions showed a polyclonal pattern of X-inactivation. This indicated that the clonal pattern observed in purified eosinophils at initial presentation was not due to skewedness of random X-inactivation that could, in principle, result in an imbalanced expression of one allele in a population of cells.
Our patient presenting with IHES showed similarities to the case described by Luppi et al.16 Both patients showed evidence of organ damage associated with sustained eosinophilia, an absence of features associated with myeloproliferative disorders, a normal bone marrow karyotype, a clonal eosinophilia by X-inactivation analysis, and corticosteroid-responsiveness with a favorable outcome. In our patient (case 1), a very high IgE level at presentation was also found, which is associated with a good prognosis in IHES.2 After therapy, the IgE level fell, but remained mildly elevated, although clonal eosinophilia could no longer be demonstrated. In a previous study, 38% of IHES patients were found to have markedly elevated IgE levels, suggesting an association between an IgE-mediated mechanism and eosinophilia in this subgroup.25 These patients required no therapy or responded with a complete remission when treated with corticosteroids.25,26 This interesting association raises the possibility that the eosinophilia and perhaps the organ damage in this subgroup of patients with IHES is associated with an IgE-mediated hypersensitivity response to an as yet unidentified antigen or antigens.25
One advantage of HUMARA analysis compared with other X-inactivation markers is its high degree of informativeness, as the HUMARA loci show a high incidence of constitutional heterozygosity of approximately 90% in the female population.9,17 By contrast, the incidence of constitutional heterozygosity reported for other polymorphic X-linked markers, such as the PGK and HPRT loci, are significantly lower.15 In practice, X-inactivation data may be corroborated by further analyses of other polymorphic X-linked loci. We attempted to verify clonal eosinophilia in the patient presenting with IHES by PGK analysis, but the patient was found to be not heterozygous at the PGK locus. It should be noted that clonality analysis using X-linked polymorphisms is only useful in females and, therefore, only in a minority of patients with IHES, as the majority (approximately 90%2) of these patients are male.
Eosinophil purification allows HUMARA analysis to be extended to detect clonal eosinophilia when the proportion of peripheral blood eosinophils is low. This should, in principle, permit detection of clonal relapse in IHES patients and facilitate the assessment of therapeutic responses. However, it has to be noted that an absence of clonal eosinophilia by this method of analysis cannot completely exclude the possibilty that a minor clonal population of eosinophils may persist. Long-term follow-up of our patient presenting with IHES is necessary to determine whether continued suppression of clonal eosinophilia by a low dose of corticosteroids can be maintained.
Although vasculitis is not a prominent feature of IHES, individual patients with IHES may exhibit pathologic evidence of vasculitis.1,2 The major vasculitis that is associated with eosinophilia is the Churg-Strauss syndrome.23 A history of asthma, nonfixed pulmonary infiltrates, blood eosinophilia greater than 10%, paranasal sinus abnormalities, mononeuropathy or polyneuropathy, and a biopsied blood vessel demonstrating extravascular eosinophils are features of this syndrome.23 24 In some patients, clear distinction between IHES and Churg-Strauss syndrome may not be possible. However, our patient (case 2) showed the characteristic features of this syndrome. The finding that eosinophils were polyclonal in this patient provides definitive evidence that eosinophilia is reactive in nature in the Churg-Strauss syndrome.
There are several possible explanations for the underlying pathogenesis of a clonal eosinophilia in IHES. Eosinophilic proliferation and differentiation is promoted by several cytokines, including interleukin (IL)-3, granulocyte-macrophage colony-stimulating factor, and IL-5, which is the most important and, in humans, is restricted to stimulating eosinophil production.27 Reactive eosinophilias such as infections, allergies, drug reactions, skin diseases, connective tissue diseases, and malignancies are associated with eosinophilia through stimulation by IL-5 secreted by T cells.2,6 Recently, there have been reports of a clonal population of T cells with an unusual phenotype, CD3−, CD4+ or CD3+, CD4−, CD8−, in the peripheral blood of several cases of eosinophilia, including IHES.28,29 In one report,28 a patient with IHES who had excessive production of IgE was found to have a CD3−, CD4+ Type 2 helper T-cell clone that secreted high levels of IL-5 and IL-4, which stimulates IgE production. In patients with IHES who have high levels of IgE, it has been proposed that hypereosinophilia may result from an IgE-mediated hypersensitivity response to an as yet unidentified antigen or antigens.25 This subgroup of patients is characterized by responsiveness to corticosteroid therapy and a good prognosis,25,26 as seen in the case that we have described. It is possible that hypereosinophilia in these patients may be due to a dysregulated, autonomous replication of a clonal population of eosinophils after stimulation by increased levels of IL-5. Other hypotheses for the pathogenesis of IHES have been suggested, such as defects in cytokine receptors, or in their signal transduction or suppressor regulatory pathways,30,31 but these remain speculative. Alternatively, the finding of clonal eosinophilia in a patient with IHES could imply that somatic mutation affecting the eosinophil lineage is present in the patient, in the absence of karyotypic abnormalities, and that the patient has, in reality, a low-grade clonal myeloproliferative disorder. Cases of IHES that show a clonal cytogenetic abnormality should be classified as eosinophilic leukemia,32 and it has been argued that all cases of clonal eosinophilia should, by definition, be excluded from IHES.6The method of analysis that we have described would allow clonal eosinophilias to be distinguished from reactive eosinophilias in female patients presenting with IHES. In summary, in the absence of karyotypic abnormalities, HUMARA analysis of isolated eosinophils can be used to detect clonal eosinophilias and can provide a marker to assess disease progression.
Supported by National Medical Research Council Grant No 0040/94. H.-W.C. was a Medical Research Scientist Award recipient of the National Medical Research Council, Singapore.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Szu-Hee Lee, MD, PhD, Division of Haematology, National University Hospital, Singapore 119074.