Abstract
Methotrexate (MTX) is one of the most active and widely used agents for the treatment of acute lymphoblastic leukemia (ALL). To elucidate the mechanism for higher accumulation of MTX polyglutamates (MTX-PG) in hyperdiploid ALL and lower accumulation in T-lineage ALL, expression of the reduced folate carrier (RFC) was assessed by reverse transcription-polymerase chain reaction in ALL blasts isolated from newly diagnosed patients. RFC expression exhibited a 60-fold range among 29 children, with significantly higher expression in hyperdiploid B-lineage ALL (median, 11.3) compared with nonhyperdiploid ALL (median, 2.1; P < .0006), but no significant difference between nonhyperdiploid B-lineage and T-lineage ALL. Furthermore, mRNA levels of RFC (mapped by FISH to chromosome 21) were significantly related to chromosome 21 copy number (P = .0013), with the highest expression in hyperdiploid ALL blasts with 4 copies of chromosome 21. To assess the functional significance of gene copy number, MTX-PG accumulation was compared in ALL blasts isolated from 121 patients treated with either low-dose MTX (LDMTX; n = 60) or high-dose MTX (HDMTX; n = 61). After LDMTX, MTX-PG accumulation was highest in hyperdiploid B-lineage ALL with 4 copies of chromosome 21 (P = .011), but MTX-PG accumulation was not significantly related to chromosome 21 copy number after HDMTX (P = .24). These data show higher RFC expression as a mechanism for greater MTX accumulation in hyperdiploid B-lineage ALL and indicate that lineage differences in MTX-PG accumulation are not due to lower RFC expression in T-lineage ALL.
METHOTREXATE (MTX) is one of the most active and widely used medications for the treatment of childhood acute lymphoblastic leukemia (ALL).1,2 Recent studies have shown significant lineage and ploidy differences in the intracellular accumulation and metabolism of MTX in ALL blasts,3providing insights into mechanisms underlying prognostic differences in these ALL subtypes. Intracellular accumulation of MTX and MTX polyglutamate can be an important determinant of event-free survival in ALL patients.4 After entering cells via the reduced folate carrier (RFC) and by passive diffusion at higher extracellular concentrations, MTX is metabolized to polyglutamylated metabolites. These active MTX polyglutamates (MTX-PG) inhibit dihydrofolate reductase, thymidylate synthase, and enzymes involved in de novo purine synthesis and are retained in cells longer than the parent drug (MTX).1,2,5 It has been shown that B-lineage lymphoblasts accumulate higher intracellular concentrations of active MTX-PG compared with T-lineage ALL3 and that this is related in part to higher activity of folylpolyglutamate synthetase (FPGS) in B-lineage lymphoblasts.6 Among patients with B-lineage ALL, those with hyperdiploid (>50 chromosomes) ALL accumulate higher MTX-PG in their leukemia cells compared with nonhyperdiploid ALL.3 However, the mechanism of higher MTX-PG accumulation in hyperdiploid ALL has not been elucidated, although it has been shown that FPGS activity does not differ between hyperdiploid and nonhyperdiploid B-lineage ALL.6 Because the great majority (∼97%) of hyperdiploid ALL blasts have 3 or 4 copies of chromosome 21,7 we hypothesized that hyperdiploid blasts have increased expression of the RFC, which is located on human chromosome 21 (21q22.2-q22.3),8 and that chromosome 21 copy number is associated with RFC mRNA expression and MTX accumulation in hyperdiploid ALL. The present study was therefore undertaken to assess the relation between chromosome 21 copy number and (1) the level of RFC mRNA expression and (2) in vivo MTX-PG accumulation in leukemia cells of children with newly diagnosed ALL.
MATERIALS AND METHODS
Human subjects.
The diagnosis of B-lineage and T-lineage ALL was made using immunological criteria, as previously described.9 To determine MTX-PG accumulation, ALL blasts were isolated from bone marrow aspirates obtained from consecutively treated children with newly diagnosed ALL, 44 hours after MTX treatment. After providing informed consent, patients enrolled on St Jude Total-XIIIA protocol were randomized to initial single agent therapy with either low-dose (180 mg/m2 administered orally as 30 mg/m2every 6 hours 6 times) or high-dose (1,000 mg/m2 administered intravenously as a 24-hour infusion) MTX, plus leucovorin rescue, as previously described.3 ALL blasts were isolated by Ficoll gradient separation, and MTX-PG concentrations were measured in 5 × 106 blasts by a high-performance liquid chromatography (HPLC) radioenzymatic assay as previously described.3 ALL ploidy and chromosome 21 copy number were determined by cytogenetic analysis, as previously described.7
To determine RFC mRNA expression, ALL blasts were isolated from either bone marrow aspirates or peripheral blood obtained from patients enrolled on the TOTAL-XIIIB protocol, either before treatment (n = 26) or within 48 hours of starting chemotherapy (n = 3). RNA was isolated from ALL blasts within 6 hours of being obtained from patients (or cell culture) and then analyzed for RFC expression by the reverse transcription-polymerase chain reaction (RT-PCR) method described below. Informed consent was obtained from the patient’s parents or guardian according to IRB guidelines.
Human leukemia cell lines.
The CEM/MTX and K500E/MTX cell lines with impaired MTX uptake and the RFC wild-type K562/wt cell lines were generous gifts from Dr L. Matherly (Karmanos Cancer Institute, Detroit, MI).10,11 CCRF-CEM cells were purchased from ATCC (Rockville, MD). The CEM/T-cell line with impaired MTX RFC transport was a generous gift from Dr J. Bertino (Memorial Sloan Kettering, New York, NY).12 NALM6 cells were purchased from DSMZ (Braunschweig, Germany).
Total RNA preparation.
Total RNA from cell lines and patient’s lymphoblasts were isolated using TRI REAGENT from Molecular Research Center, Inc (Cincinnati, OH). Typically, 1 mL of TRI REAGENT solution was used to isolate total RNA from 5 to 10 × 106 cells. The yield of total RNA varied from 5 to 10 μg total RNA per 1 × 106 cultured cells and from 1 to 2 μg total RNA per 1 × 106 lymphoblasts from patients.
Preparation of RFC mRNA external standard.
All oligonucleotides used as primers for PCR were synthesized in the Center for Biotechnology at St Jude Children’s Research Hospital (Memphis, TN). Primers RFC617 (5′-CCAAGCGCAGCCTCTTCTTCAACC) and RFC949 (5′-CCAGCAGCGTGGAGGCAGCATCTGCC)13 were used to generate a fragment corresponding to nucleotides 617-949 of the human RFC cDNA by RT-PCR (numeration based on the complete RFC cDNA sequence10). The synthesized DNA fragment was directly cloned into pCR2.1 vector (Invitrogen, San Diego, CA) and sequenced in both directions. Acc I treatment followed by self-ligation gave a plasmid with a 58-bp deletion within the 617-949 fragment of cDNA. The resulting fragment was excised by EcoRI treatment, purified by electrophoresis in an agarose gel, isolated by QIAquick Gel extraction kit from QIAgen (Santa Clarita, CA), and cloned into the plasmid pGEM7 with a previously inserted polyadenylate stretch (A30) in the multiple cloning site. Orientation of the inserted fragment was determined by restriction analysis withAcc I and Xba I. The insert was completely sequenced in both directions, and an external standard for RT-PCR (stRNA) with 58 ribonucleotides deleted was obtained using T7 RNA polymerase from RiboMax Large Scale RNA production system from Promega (Madison, WI) and purified using Oligotex direct mRNA Midi/Maxi kit from QIAgen. stRNA concentration was measured by UV absorbance (GeneQuant; Pharmacia Biotech, Cambridge, UK).
RFC mRNA quantitation by competitive RT-PCR.
Serial 10-fold dilutions (1:1 to 1:105) of stRNA were made to obtain PCR signals comparable to those obtained for RFC using mRNA from the MTX-sensitive human leukemia cell line, CCRF-CEM/wt. An aliquot of stRNA was added to 1 μg of total RNA and isolated either from cell lines or lymphoblasts from patients, and this mixture was converted to cDNA as follows. The reaction mixture (20 μL) containing 50 mmol/L Tris-HCl (pH 8.3), 75 mmol/L KCl, 3 mmol/L MgCl2, 10 mmol/L dithiothreitol, 3 μg of random primers from GIBCO BRL Life Technology (Gaithersburg, MD), and 1 U of RQ1 RNAse-free DNAse I (Promega) was incubated at 37°C for 30 minutes and at 75°C for 5 minutes to inactivate DNAse I and then chilled for 3 minutes on ice. Two hundred units (1 μL) of Moloney murine leukemia virus reverse transcriptase (SuperScript II) from GIBCO BRL Life Technology was added and the mixture was incubated at 42°C for 50 minutes. The final PCR amplification mixture contained buffer J (PCR Optimizer kit) from Invitrogen [60 mmol/L Tris-HCl (pH 9.5), 15 mmol/L (NH4)2SO4, 0.4 mmol/L MgCl2], 0.25 μg of each primer, 1 U Taq DNA polymerase (Promega), and 2 μL cDNA template. The amplification program was initiated according to the “Hot start” protocol as suggested by the manufacturer: 5 μL 10 mmol/L dNTPs and 5 μCi [α-32P] dCTP (10 μCi/μL) from Amersham (Arlington Heights, IL) were added at 80°C, followed by the denaturation step at 94°C for 2 minutes. PCR was performed as follows: 30 cycles at 94°C for 1 minute, 55°C for 2 minutes, and 72°C for 3 minutes followed by a final cycle at 72°C for 7 minutes. The PCR products were purified by QIAquick PCR purification kit (QIAgen) and separated by gel electrophoresis at 300 V in 5% nondenaturating PAAG gel (4°C), the gel was dried under vacuum at 80°C for 1 hour and visualized using a Molecular Dynamics PhosphorImager (Molecular Dynamics, Sunnyvale, CA), and the amount of32P-labeled RFC fragments was quantified by “ImageQuaNT” software (ver 4.2a; Molecular Dynamics).
To estimate a suitable range of RNA concentrations and number of PCR cycles with good RFC signal and minimal nonspecific products, we determined conditions in which the amount of the PCR product increased linearly with the number of PCR cycles in logarithmic coordinates.14 An excellent correlation (r2 = .94) was achieved between the amount of PCR product and the number of cycles performed (data not shown).
Amplification of β-actin as an internal standard.
To amplify β-actin mRNA fragment, primers BA-67 (5′-GGGAGAGCGGGAAATCGTGCGTGACATT) and BA-68 (5′-GATGGAGTTGAAGGTAGTGGCGTG) were used as described previously.15 Because β-actin mRNA is much more abundant in cells than RFC mRNA, the cDNA template was diluted 1,000-fold to obtain a signal from β-actin comparable to that of RFC mRNA. PCR of β-actin, separation, and quantification of PCR products were performed under the conditions described above.
Northern blotting.
To validate RFC mRNA quantification by RT-PCR, RFC mRNA levels were determined by Northern analysis of human leukemia cell lines. Samples of total RNA (40 μg per lane, 8.1 μL) were incubated at 50°C for 1 hour with 8.1 μL of 6 mol/L glyoxal, 24 μL of dimethyl sulfoxide (DMSO), and 4.5 μL of 0.1 mol/L sodium phosphate buffer (pH 7.0). After adding loading buffer, samples were electrophoresed in 1.4% agarose with 10 mmol/L sodium phosphate (pH 7.0) at 60 V with buffer circulation. Separated products were transferred to the nylon membrane Hybond-N+ (Amersham), and the membrane was washed in 6× SSC, dried at room temperature for 30 minutes, and baked at 80°C for 30 minutes under vacuum. The RFC mRNA probe, 32P-labeled by Rediprime from Amersham, was a fragment of approximately 1,000 bp of the 3′ end of the RFC coding region. The probe was hybridized overnight in Rapid-hyb buffer from Amersham and then washed at 65°C with 2× SSC for 15 minutes, 2× SSC with 0.1% sodium dodecyl sulfate (SDS) for 30 minutes, and then 0.1× SSC for 30 minutes. Signals were visualized by PhosphorImager and quantified by “ImageQuaNT” software, as described above.
Fluorescence in situ hybridization (FISH).
A BAC clone containing the human RFC gene was used to perform FISH analysis of metaphase chromosomes from lymphoblasts isolated from two patients with B-lineage ALL, one containing 4 copies of chromosome 21 and the other containing 2 copies of chromosome 21, with 1 involved in a 12;21 translocation (ie, ETV6-CBFA2 fusion). To isolate the RFC probe, human genomic DNA from Molt4 cells was cloned into the PBeloBacII vector (Genome Systems, St Louis, MO). The library was screened by hybridization with the RFC EST cDNA (Genbank Accession no.R87517) containing a 2,034-bp cDNA insert. One positive clone was isolated and further characterized by hybridization with the RFC cDNA and the RFC promoter. For in situ hybridization, the RFC probe was labeled with biotin 11-dUTP by nick translation (Life Technologies, Inc, Gaithersburg, MD). As a control probe for chromosome 21, digoxigenin-labeled 21q22.3-ter DNA was used (Oncor, Gaithersburg, MD). Slides were denatured in 70% formamide for 2 minutes and dehydrated. The RFC probe was denatured for 5 minutes at 70°C and preannealed for 10 minutes at 37°C. The control probe was prewarmed to 37°C, and the probes were mixed and applied to the denatured slide. Slides were placed in a humidified chamber and hybridized at 37°C overnight. Posthybridization washes were performed at 45°C in 50% formamide three times for 5 minutes each, followed by two 2× SSC washes at room temperature. The probes were detected using fluorescein isothiocyanate (FITC)-avidin (RFC) and Rodamine-labeled antidigoxigenin (21q22.3 probe). The slides were stained with 4′, 6-diamidino-2-phenylindole (DAPI) and the cells were analyzed using an Olympus microscope and an image capturing system (Vysis Inc, Downer’s Grove, IL). For the second case, the digoxigenin-labeled Coatasome 12-chromosome probe (Oncor) was denatured for 10 minutes at 70°C and preannealed for 2 hours at 37°C. The biotin-labeled RFC probe was denatured separately and hybridized together on the denatured slide. Probes were detected using the Rhodamine-labeled antidigoxigenin and FITC-avidin, and the slide was stained with DAPI.
Statistical analysis.
Pearson coefficient was used to assess the correlation between RFC mRNA measured in the same cells by different methods. The difference in RFC mRNA between hyperdiploid and nonhyperdiploid B-lineage ALL was evaluated using the Mann-Whitney U test. The relation between in vivo MTX-PG1-7 accumulation and chromosome 21 copy number was evaluated separately in ALL patients treated with low-dose and high-dose MTX. To adjust for differences in extracellular MTX concentrations among patients treated with the same dose of MTX, the ratio of intracellular MTX-PG (picomoles per 109 blasts) to extracellular steady-state plasma MTX concentration (Cpss) was also assessed. The shape of data distribution was fitted by normal and log-normal functions and the quality of the fit was assessed by χ2 test. For log-normally distributed data, a logarithmic transformation was applied before parametric analyses (ie, all MTX-PG datasets were log-transformed for analysis). Standard deviations of log-normal data were calculated from log-transformed values and are thus symmetrical when depicted on log-scale graphs. The amount of RFC mRNA and MTX-PG accumulation among groups with different chromosome 21 copy number were compared using analysis of variance (ANOVA), followed by the Tukey multiple comparison test for unequal sample sizes. Computations were performed with STATISTICA, version 5.1 (StatSoft, Inc, Tulsa, OK), and P < .05 was considered statistically significant.
RESULTS
Estimation of RFC mRNA by RT-PCR versus Northern analysis.
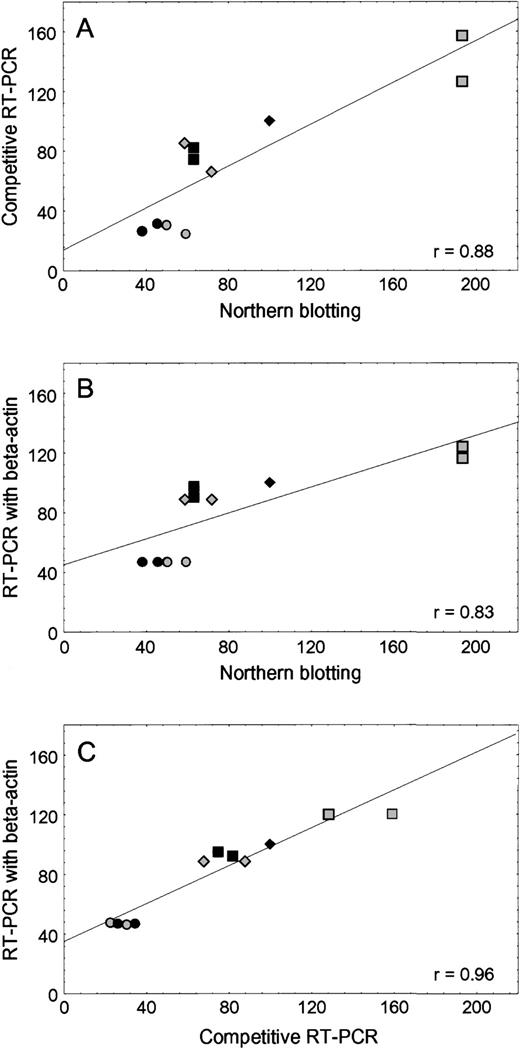
As shown in Fig 1, there was good agreement between the level of RFC mRNA in human leukemia cell lines when determined by RT-PCR and Northern analysis, using either competitive RT-PCR with external standard (r2 = .77,P = .0016) or RT-PCR with β-actin as the internal standard (r2 = .69, P = .002). With these methods, the amount of RFC mRNA in the transport-deficient CEM/T cells was approximately 30% to 60% lower than CCRF-CEM/wt, and the CEM/MTX line (with a mutant RFC gene) had a 20% to 70% higher RFC mRNA level compared with CCRF-CEM/wt. The RFC mRNA amounts in K562/wt and MTX-resistant K500E/MTX cells were comparable to each other.
Concordance among three methods of RFC mRNA quantification. (A) Comparison of Northern blotting and competitive RT-PCR using stRNA as an external standard. (B) Comparison of Northern blotting and RT-PCR using β-actin as an internal standard. (C) Comparison of competitive RT-PCR using stRNA and RT-PCR using β-actin. The amount of RFC mRNA from the CCRF-CEM/wt cell line was assigned a value of 100%. The RFC mRNA amount from other cell lines was calculated as a percentage of CCRF-CEM/wt. CCRF-CEM/wt (⧫), CEM/T (◥), CEM/MTX (░), Nalm6 (▪), K562/wt (◍), K500E/MTX (•).
Concordance among three methods of RFC mRNA quantification. (A) Comparison of Northern blotting and competitive RT-PCR using stRNA as an external standard. (B) Comparison of Northern blotting and RT-PCR using β-actin as an internal standard. (C) Comparison of competitive RT-PCR using stRNA and RT-PCR using β-actin. The amount of RFC mRNA from the CCRF-CEM/wt cell line was assigned a value of 100%. The RFC mRNA amount from other cell lines was calculated as a percentage of CCRF-CEM/wt. CCRF-CEM/wt (⧫), CEM/T (◥), CEM/MTX (░), Nalm6 (▪), K562/wt (◍), K500E/MTX (•).
RFC mRNA in lymphoblasts from patients.
RFC expression was determined in lymphoblasts isolated from 29 newly diagnosed children with ALL, 26 of them before treatment and 3 at 44 hours after treatment with MTX. In 6 patients studied both before and 44 hours after treatment, there was not a significant difference in RFC mRNA at the two time points (data not shown). The 29 patients were selected from newly diagnosed patients entered on the Total XIIIB protocol (between April 1997 and December 1997 and in June and July 1998), including all available children with T-lineage ALL (n = 8), all with hyperdiploid (>50 chromosomes) B-lineage ALL (n = 7), and a comparable number of patients with nonhyperdiploid B-lineage ALL (n = 14). None of these patients had Down’s syndrome (ie, germline trisomy 21). There was a 60-fold range in RFC mRNA expression in leukemia cells isolated from these patients. Using RFC mRNA expression in CCRF-CEM/wt cells as a reference value of 1.0, patients with hyperdiploid B-lineage ALL had significantly higher levels of RFC mRNA expression (median, 11.3) compared with those with nonhyperdiploid ALL (median, 2.1; P < .0006), with no significant difference between nonhyperdiploid B-lineage (median, 1.4) and T-lineage (median, 4.8) ALL (Fig 2A). As depicted in Fig 2B, there was a relation between the level of RFC mRNA and chromosome 21 copy number in B-lineage ALL lymphoblasts. ANOVA showed significant differences (P < .002) in the relative amounts of RFC mRNA among the three patient groups: B-lineage with 2 (median, 1.35), 3 (median, 6.7), or 4 (median, 12.8) copies of chromosome 21 (Fig 2B). Pairwise comparisons using the Tukey test showed that B-lineage ALL with 4 copies of chromosome 21 had significantly higher levels than B-lineage with 2 copies (P = .004), whereas the other groups did not differ significantly (P> .3). Seven of 10 lymphoblast samples with greater than 2 copies of chromosome 21 were hyperdiploid (>50 chromosomes). As depicted in Fig2B, the three nonhyperdiploid samples had the lowest RFC mRNA levels among those with greater than 2 copies of chromosome 21.
RFC mRNA in ALL blasts from newly diagnosed patients. (A) Relative RFC mRNA expression in nonhyperdiploid T-lineage lymphoblasts (n = 8), nonhyperdiploid B-lineage lymphoblasts (n = 14), and hyperdiploid B-lineage lymphoblasts (n = 7). Diamonds (⧫, ◥) depict lymphoblasts isolated from peripheral blood before treatment, squares (▪, ░) depict lymphoblasts from bone marrow before treatment, and circles (•) depict lymphoblasts from bone marrow 44 hours after MTX treatment. Shaded symbols depict nonhyperdiploid blasts and solid symbols depict hyperdiploid blasts (>50 chromosomes). Horizontal lines depict the median values in each group. (B) RFC mRNA expression in B-lineage ALL blasts with either 2 (n = 11), 3 (n = 5), or 4 (n = 5) copies of chromosome 21. Symbols are the same as in (A). Among lymphoblasts with 3 copies of chromosome 21, the 2 samples with the lowest RFC expression have less than 50 chromosomes, and the lowest value had a translocation involving the long arm of 1 copy of chromosome 21 and the short arm of chromosome 12 (ie, a 12;21 [p13;q22] translocation) resulting in the ETV6-CBFA2 fusion. Similarly, among those with 4 copies of chromosome 21, the sample with the lowest RFC expression had less than 50 chromosomes.
RFC mRNA in ALL blasts from newly diagnosed patients. (A) Relative RFC mRNA expression in nonhyperdiploid T-lineage lymphoblasts (n = 8), nonhyperdiploid B-lineage lymphoblasts (n = 14), and hyperdiploid B-lineage lymphoblasts (n = 7). Diamonds (⧫, ◥) depict lymphoblasts isolated from peripheral blood before treatment, squares (▪, ░) depict lymphoblasts from bone marrow before treatment, and circles (•) depict lymphoblasts from bone marrow 44 hours after MTX treatment. Shaded symbols depict nonhyperdiploid blasts and solid symbols depict hyperdiploid blasts (>50 chromosomes). Horizontal lines depict the median values in each group. (B) RFC mRNA expression in B-lineage ALL blasts with either 2 (n = 11), 3 (n = 5), or 4 (n = 5) copies of chromosome 21. Symbols are the same as in (A). Among lymphoblasts with 3 copies of chromosome 21, the 2 samples with the lowest RFC expression have less than 50 chromosomes, and the lowest value had a translocation involving the long arm of 1 copy of chromosome 21 and the short arm of chromosome 12 (ie, a 12;21 [p13;q22] translocation) resulting in the ETV6-CBFA2 fusion. Similarly, among those with 4 copies of chromosome 21, the sample with the lowest RFC expression had less than 50 chromosomes.
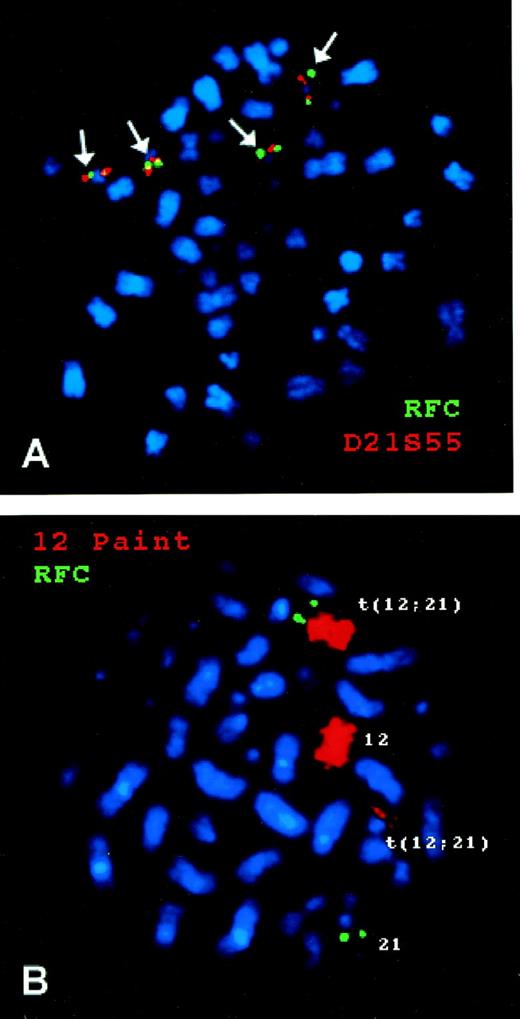
As shown in Fig 3A, FISH of chromosomes from a hyperdiploid ALL blast with 4 copies of chromosome 21 showed 4 copies of the RFC gene located telomeric to the human chromosome 21q22.3-ter probe. Figure 3B documents that the RFCgene is translocated with the ETV6 gene on the long arm of chromosome 21 in the 12;21 translocation that produces theETV6-CBFA2 fusion.
FISH of a human RFC probe with chromosomes from B-lineage ALL blasts. (A) is from a hyperdiploid ALL blast with 4 copies of chromosome 21. The green signal is from the RFC gene probe and the red signal from a chromosome 21q22.3-ter probe. (B) is from a nonhyperdiploid ALL blast with 2 copies of chromosome 21, one of which is involved in a 12;21 translocation. The green signal is from the RFC gene probe and the red signal is from a chromosome 12 probe (Coatasome 12).
FISH of a human RFC probe with chromosomes from B-lineage ALL blasts. (A) is from a hyperdiploid ALL blast with 4 copies of chromosome 21. The green signal is from the RFC gene probe and the red signal from a chromosome 21q22.3-ter probe. (B) is from a nonhyperdiploid ALL blast with 2 copies of chromosome 21, one of which is involved in a 12;21 translocation. The green signal is from the RFC gene probe and the red signal is from a chromosome 12 probe (Coatasome 12).
MTX-PG accumulation and chromosome 21 copy number.
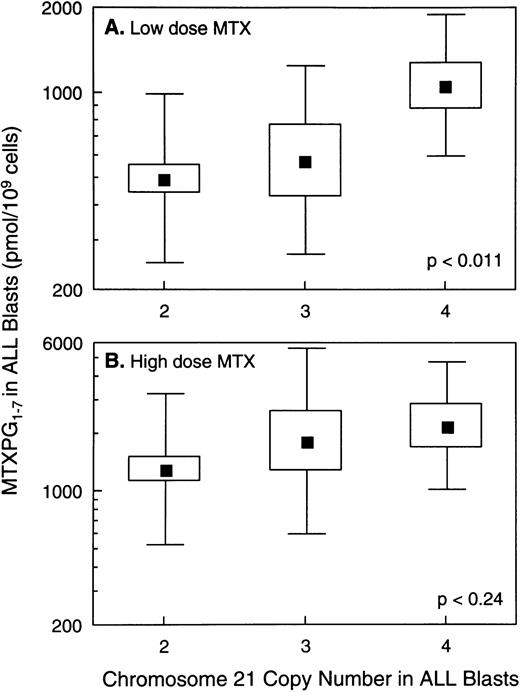
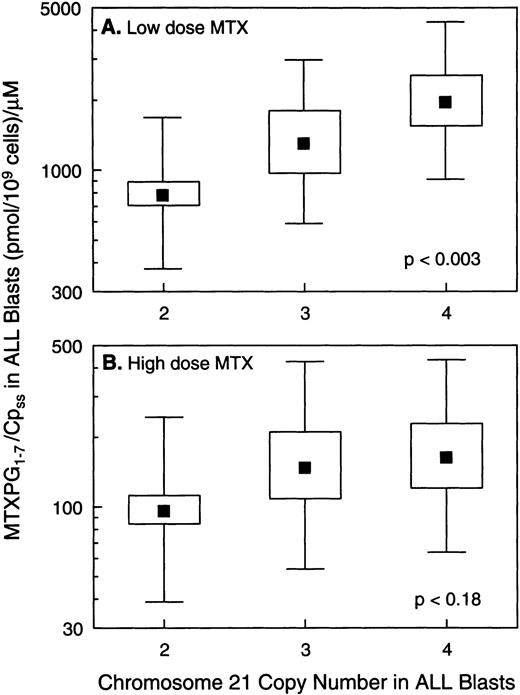
In vivo MTX-PG concentrations were measured in ALL blasts isolated from bone marrow aspirates in a total of 140 patients, 121 with B-lineage ALL and 19 with T-lineage ALL. As previously reported,3patients with T-lineage ALL had significantly lower MTX-PG accumulation when compared with nonhyperdiploid B-lineage ALL after either high-dose (P < .03) or low-dose MTX (P < .001). Because essentially all patients with T-lineage ALL have nonhyperdiploid lymphoblasts with only 2 copies of chromosome 21 (18 of 19 T-lineage ALL were nonhyperdiploid), all analyses of the relation between chromosome 21 copy number and MTX-PG accumulation were restricted to B-lineage ALL. For patients treated with low-dose MTX, there were statistically significant differences in MTX-PG among patients whose ALL blasts had 2 (mean, 499 pmol/109 cells; n = 43), 3 (mean, 581 pmol/109 cells; n = 7), or 4 (mean, 1,064 pmol/109 cells; n = 10) copies of chromosome 21 (P< .011 by ANOVA for log-transformed values; Fig 4). Likewise, differences in MTX-PG/Cpss were significant (P < .003) among patients treated with low-dose MTX (Fig 5). Pairwise comparisons using the Tukey test showed that those with 4 copies of chromosome 21 had significantly higher MTX-PG accumulation when compared with those with 2 copies (P = .044 for MTX-PG andP = .024 for MTX-PG/Cpss), whereas other pairwise comparisons among chromosome copy number groups were not significant. In contrast, after high-dose MTX treatment (Figs 4 and 5), there was not a significant difference among ALL blasts with 2 (mean, 1,309 pmol/109 cells; n = 42), 3 (mean, 1,839 pmol/109 cells; n = 10) or 4 (mean, 2,217 pmol/109 cells; n = 9) copies of chromosome 21 (P = .24 for MTX-PG and P = .18 for MTX-PG/Cpss by ANOVA). There were 7 patients with B-lineage ALL treated with low-dose MTX in whom in vivo MTX-PG accumulation in ALL blasts and RFC mRNA levels were measured in the same sample. In these patients, the Spearman rank order correlation for RFC mRNA and the relative amount of MTX-PG accumulation at 44 hours was R = .678 (P = .09), with higher RFC mRNA associated with greater MTX-PG accumulation. This association was not evident in patients treated with HDMTX (P = .6; n = 4).
Relation between MTX-PG concentrations in ALL blasts and chromosome 21 copy number. (A) Data for B-lineage ALL blasts isolated from bone marrow at 44 hours after low-dose MTX treatment of 60 children (n = 43, 7, and 10 for 2, 3, and 4 copies of chromosome 21, respectively). (B) Data for B-lineage ALL blasts isolated from 61 patients at 44 hours after treatment with high-dose MTX (n = 42, 10, and 9 for 2, 3, and 4 copies of chromosome 21, respectively). (▪) Mean values; the boxes depict the standard errors (SE) of the mean; and the bars depict the range of ±1 standard deviation (SD) in each group. The SE and SD are symmetrical because they were calculated from log-transformed values of log-normal data.
Relation between MTX-PG concentrations in ALL blasts and chromosome 21 copy number. (A) Data for B-lineage ALL blasts isolated from bone marrow at 44 hours after low-dose MTX treatment of 60 children (n = 43, 7, and 10 for 2, 3, and 4 copies of chromosome 21, respectively). (B) Data for B-lineage ALL blasts isolated from 61 patients at 44 hours after treatment with high-dose MTX (n = 42, 10, and 9 for 2, 3, and 4 copies of chromosome 21, respectively). (▪) Mean values; the boxes depict the standard errors (SE) of the mean; and the bars depict the range of ±1 standard deviation (SD) in each group. The SE and SD are symmetrical because they were calculated from log-transformed values of log-normal data.
Relation between the ratio of intracellular MTX-PG to extracellular plasma MTX concentration (Cpss) versus chromosome 21 copy number in ALL blasts. (A) Data for B-lineage ALL blasts isolated from bone marrow at 44 hours after low-dose MTX treatment of 60 children (n = 43, 7, and 10 for 2, 3, and 4 copies of chromosome 21, respectively). (B) Data from B-lineage ALL blasts isolated from bone marrow at 44 hours after high-dose MTX treatment in 61 children (n = 42, 10, and 9 for 2, 3, and 4 copies of chromosome 21, respectively). (▪) Mean values; the boxes depict the standard errors of the mean; and bars depict the range of ±1 standard deviation in each group.
Relation between the ratio of intracellular MTX-PG to extracellular plasma MTX concentration (Cpss) versus chromosome 21 copy number in ALL blasts. (A) Data for B-lineage ALL blasts isolated from bone marrow at 44 hours after low-dose MTX treatment of 60 children (n = 43, 7, and 10 for 2, 3, and 4 copies of chromosome 21, respectively). (B) Data from B-lineage ALL blasts isolated from bone marrow at 44 hours after high-dose MTX treatment in 61 children (n = 42, 10, and 9 for 2, 3, and 4 copies of chromosome 21, respectively). (▪) Mean values; the boxes depict the standard errors of the mean; and bars depict the range of ±1 standard deviation in each group.
Among patients treated with low-dose MTX, there were 2 hyperdiploid B-lineage cases whose ALL blasts had only 2 copies of chromosome 21, yet these 2 patients had significantly higher MTX-PG accumulation (989 and 2,187 pmol/109 cells) when compared with nonhyperdiploid B-lineage ALL with 2 copies of chromosome 21 (n = 41; median MTX-PG, 517 pmol/109 cells; P = .028).
DISCUSSION
Previous studies from our laboratory and others have established that MTX-PG accumulation is greater in hyperdiploid B-lineage ALL when compared with nonhyperdiploid B-lineage or T-lineage ALL, both in vivo3 and ex vivo.4 Because increased intracellular accumulation of MTX-PG has been associated with greater antileukemic effects,4,16 this may explain, in part, the favorable prognosis of children with hyperdiploid B-lineage ALL17 and offer insights for developing alternative treatment strategies for different subtypes of ALL.
The present study has identified a novel mechanism for higher intracellular concentrations of MTX-PG in hyperdiploid ALL, showing significantly higher expression of the RFC in these leukemic lymphoblasts. Furthermore, the present study showed a relation between chromosome 21 copy number and both the level of RFC mRNA expression and the level of MTX-PG accumulation in hyperdiploid ALL blasts, with the highest level of expression and MTX-PG accumulation in hyperdiploid lymphoblasts with 4 copies of chromosome 21. The human RFC gene has been mapped to chromosome 21q22.2-q22.3,8 suggesting a gene-dose effect for RFC expression in these lymphoblasts. Because hyperdiploid B-lineage ALL blasts almost always have at least 1 extra copy of chromosome 21 (97% in one large series7), this appears to be a common mechanism for increased MTX-PG accumulation in this favorable subgroup of childhood ALL. It is known that, in individuals with Down’s syndrome, trisomy 21 is associated with overexpression of a number of genes on this chromosome, including cystathionine β synthase,18,19 phosphoribosylglycinamide synthetase, and phosphoribosylaminoimidasole synthetase.20Constitutive overexpression of the RFC gene in all cells of patients with Down’s syndrome may explain why these individuals are more susceptible to MTX toxicity,21 a hypothesis that remains to be investigated. It is also interesting that, among lymphoblasts with greater than 2 copies of chromosome 21, the nonhyperdiploid samples (n = 3) had lower RFC mRNA than samples that were hyperdiploid (n = 7; medians, 3.4 v 11.3). This finding is also consistent with higher MTX-PG accumulation in patients who had hyperdiploid blasts with only 2 copies of chromosome 21 (n = 2) compared with nonhyperdiploid blasts with two chromosomes 21 (n = 41; P = .028).
One nonhyperdiploid sample with 3 copies of chromosome 21 had a 12;21 translocation involving 1 copy of chromosome 21. This is a reciprocal translocation that fuses the long arm of chromosome 21 (q22) to the short arm of chromosome 12 (p13), resulting in the ETV6-CBFA2fusion. It is not known whether translocation of the RFC gene (22q22.2-q22.3), which maps close to the CBFA2 gene, alters its expression, but these cells had the lowest level of RFC mRNA among all samples with greater than 2 copies of chromosome 21.
Given the low abundance of RFC mRNA, we developed a PCR-based technique that allows quantitation of RFC mRNA in a relatively small number of leukemia cells (1 × 106) that differs from published methods.13 22 This method permits assessment of RFC expression using an aliquot of patient cells that is not sufficient to quantitate RFC mRNA by Northern analysis. To enhance the accuracy of RFC mRNA measurements, we prepared an artificial RFC RNA fragment with a deletion of 58 ribonucleotides and used it as an external standard. This competitive RT-PCR method permitted estimation of RFC mRNA in both cultured cell lines and patient lymphoblasts with good precision (coefficient of variation of 19.9% within day and 26.5% between days).
The current studies with cultured human leukemia cell lines indicate that MTX-transport–deficient CEM/T cells are resistant to MTX, at least in part due to decreased RFC mRNA (30% to 60% lower compared with CCRF-CEM/wt). In contrast, the transport-deficient CEM/MTX cells had a high level of RFC mRNA, which is not unexpected, because the RFC gene in this MTX-resistant cell line contains inactivating mutations, resulting in substitution of Ser-127 by Asn, or a 4-bp (CATG) insertion at position 191, generating a frame shift and a premature stop codon.23 It was previously reported that, in the transport-defective L1210 murine leukemia cell line, the RFC gene contains a G429→C429 mutation, resulting in an amino acid substitution (Ala-130 → Pro).24 In both CEM/MTX and transport-deficient L1210 cells, these mutations are located in a very homologous and highly conserved region in the predicted fourth transmembrane domain of the protein. We found no difference in RFC mRNA level in the MTX-resistant K500E/MTX cells compared with the parent MTX-sensitive K562/wt cells, suggesting that the RFC gene in these cells may also contain inactivating mutations. These data indicate that either decreased RFC expression or inactivating mutations in the RFC gene are potential mechanisms for MTX resistance, although neither has yet been identified in primary leukemia cells isolated from patients. In contrast, this is the first report of increased RFC expression as a mechanism for enhanced sensitivity to MTX in a genetically defined subtype of ALL (ie, hyperdiploid ALL).
It is recognized that mechanisms other than RFC expression may also contribute to greater MTX-PG accumulation in hyperdiploid ALL, such as increased FPGS activity and (or) decreased γ glutamyl hydrolase (GGH) activity.25,26 However, in contrast to lineage differences in FPGS activity,6,25 we did not find a difference in FPGS activity in hyperdiploid versus nonhyperdiploid B-lineage ALL,3 whereas the activity of GGH has not been investigated in these subtypes of childhood ALL. The human FPGS gene has been mapped to chromosome 9q34.127 and the human GGH gene to chromosome 8q12.23-13.1,28 and these chromosomes are not commonly present in increased (or decreased) copy number in hyperdiploid ALL (ie, 20% have an extra chromosome 9 and 34% an extra chromosome 8).7 Two lines of evidence from the current study indicate that at least one mechanism in addition to an RFC gene-dose effect contributes to ploidy differences in MTX-PG accumulation. First, in patients treated with low-dose MTX, MTX-PG concentrations were significantly higher (P = .028) in hyperdiploid blasts with only 2 copies of chromosome 21 compared with nonhyperdiploid blasts with 2 copies of chromosome 21, although the small number of patients in the former group limits the certainty of this finding. Second, RFC mRNA levels were higher in the 7 hyperdiploid samples with greater than 2 copies of chromosome 21 compared with the 3 nonhyperdiploid samples with greater than 2 copies of chromosome 21 (Fig 2B). It is plausible that hyperdiploid blasts have greater expression of selected transcription factors, leading to overexpression of genes such as RFC, a hypothesis requiring further investigation.
It is interesting that the relation between in vivo MTX-PG accumulation and chromosome 21 copy number was statistically significant after treatment with low-dose MTX, but did not reach statistical significance (P < .24) after treatment with high-dose MTX (Figs 4 and 5). With the doses of MTX evaluated in the current study, the mean steady-state MTX plasma concentrations were approximately 0.9 μmol/L with low-dose MTX treatment and approximately 12 μmol/L with high-dose MTX.3 It is known that MTX accumulation at lower plasma concentrations is more dependent on the level of RFC expression and function, whereas MTX entry into lymphoblasts occurs by additional mechanisms (eg, passive diffusion) at high extracellular MTX concentrations.29,30 It is also possible that intracellular metabolism to MTX-PG via FPGS is saturated at the higher intracellular MTX concentrations produced by high-dose MTX, providing another explanation for why lymphoblasts with extra copies of chromosome 21 do not accumulate significantly higher MTX-PG after high-dose MTX, in contrast to low-dose MTX (Figs 4 and 5). Because MTX-PG accumulation in B-lineage lymphoblasts with either 2, 3, or 4 copies of chromosome 21 was higher after high-dose MTX compared with low-dose MTX, there appears to be a rationale for using high-dose MTX in all children with B-lineage ALL. However, the current results indicate that hyperdiploid B-lineage ALL with extra copies of chromosome 21 may be adequately treated with relatively lower doses than nonhyperdiploid B-lineage or T-lineage ALL. Because the mechanisms responsible for lineage and ploidy differences in MTX-PG accumulation are not the same, it is probable that the optimal dose of high-dose MTX will differ for specific subtypes of childhood ALL,31 a hypothesis currently under investigation.
ACKNOWLEDGMENT
The authors thank Drs G. Rivera, R. Riberio, J.T. Sandlund, J. Rubnitz, and F. Behm and all other individuals involved in the treatment of these patients; N. Kornegay for her expertise in database management and quality control; E.T. Melton, M. Needham, M. Chung, L. McNinch, E. Su, E. Ye, Y. Chu, and A. Atkinson for excellent technical assistance; our research nurses, S. Ring, L. Walters, T. Kuehner, and M. Edwards; Drs Joseph Bertino and Larry Matherly for providing cell lines; and most importantly, the patients and parents who volunteered to participate in this study.
Supported in part by National Institutes of Health Grants No. R37 CA36401 and R01 CA78224, by Cancer Center Grant No. CA21765, by a Center of Excellence grant from the State of Tennessee, and by American Lebanese Syrian Associated Charities.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to William E. Evans, PharmD, St Jude Children’s Research Hospital, 332 N Lauderdale St, Memphis, TN 38105; e-mail: william.evans@stjude.org.

![Fig. 2. RFC mRNA in ALL blasts from newly diagnosed patients. (A) Relative RFC mRNA expression in nonhyperdiploid T-lineage lymphoblasts (n = 8), nonhyperdiploid B-lineage lymphoblasts (n = 14), and hyperdiploid B-lineage lymphoblasts (n = 7). Diamonds (⧫, ◥) depict lymphoblasts isolated from peripheral blood before treatment, squares (▪, ░) depict lymphoblasts from bone marrow before treatment, and circles (•) depict lymphoblasts from bone marrow 44 hours after MTX treatment. Shaded symbols depict nonhyperdiploid blasts and solid symbols depict hyperdiploid blasts (>50 chromosomes). Horizontal lines depict the median values in each group. (B) RFC mRNA expression in B-lineage ALL blasts with either 2 (n = 11), 3 (n = 5), or 4 (n = 5) copies of chromosome 21. Symbols are the same as in (A). Among lymphoblasts with 3 copies of chromosome 21, the 2 samples with the lowest RFC expression have less than 50 chromosomes, and the lowest value had a translocation involving the long arm of 1 copy of chromosome 21 and the short arm of chromosome 12 (ie, a 12;21 [p13;q22] translocation) resulting in the ETV6-CBFA2 fusion. Similarly, among those with 4 copies of chromosome 21, the sample with the lowest RFC expression had less than 50 chromosomes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/5/10.1182_blood.v93.5.1643/4/m_blod40506002x.jpeg?Expires=1769105682&Signature=M5PqUUpl27R2frBrgBdtw3sLl-sN106ZL9bNqozwaZXoK-zdnkKcFOfBWVtfFeqjW8gI4Ow-Z8wtBMEQUOXwlP86rWbuosCMz06tbYuuSjTpTOMCDXFhcmi7yJzj0pyXq-k~rEVktEKdtq35olfa53gJWbF27RuLihY1IsbTsqLgGe0ReESQg1S4-IxFqnF-GU7OSMV2~3EXA57rtKNzn~xgs1jWxpDigzeLZH~9tIQDW6imVfhZZ0242aAI9WlXYNtrwEJzX8I3r2hsGsf-h92oASR5ptodCf7VrWeckVJlvwSROjVPMJIW8OXmm0CIVSjJajmT2h1zQVlwuPcAig__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal