Abstract
Analysis of peptide binding to human neutrophils (PMN) using phage display techniques has revealed cell-specific motifs reactive with the PMN surface. Phage libraries displaying either linear 9-mer or cyclic 10-mer and 6-mer peptides were incubated with normal human neutrophils followed by elution of bound phage with low pH (pH 2.2) and non-ionic detergent. Three rounds of selection generated several related peptide sequences that bound with high avidity to PMN. Using the linear 9-mer library, PMN-binding phage expressed peptides with the motif (G/A)PNLTGRW. The binding of phage bearing this motif was highly specific since no binding was observed on lymphocytes, fibroblasts, epithelial, or endothelial cells. Functional assays revealed that phage bearing the sequence FGPNLTGRW induced a pertussis toxin-sensitive increase in PMN cytosolic calcium analogous to that observed with Gi coupled receptors. Other prominent motifs identified included phage bearing the consensus DLXTSK(M/L)X(V/I/L), where X represents a non-conserved position. Phage with this motif bound exclusively to a sub population of human PMN that comprised approximately 50% of the total and did not elicit a calcium response. The binding of such phage to PMN was prevented by co-incubation with competing peptides displaying identical or similar sequences (IC50 range from 0.6 μmol/L to 50 μmol/L for DLXTSK and GPNLTG, respectively). We speculate that these techniques will be useful in identifying functional cell-specific binding motifs and contribute to the development of new therapeutic and diagnostic strategies in human disease.
THE IDENTIFICATION of specific cell types has become a crucial component in the diagnosis and treatment of disease and in experimental work requiring strict definition of cell populations. In general, specific cell types have been defined through the identification of individual or groups of protein/carbohydrate(s) that reside on the cell surface through the use of monoclonal antibodies (MoAbs). The ability to specifically identify individual or groups of specific cell surface structures has greatly increased our understanding of both normal and pathologic processes. For example, by labeling specific cell surface structures, it has been possible to hypothesize pathways of differentiation in both normal and malignant cells. It has also been possible to define states of activation in a variety of cell types through the identification and quantification of specific cell surface structures. In neutrophils, for example, this approach has demonstrated that cellular activation with physiologic agonists results in the mobilization of internal stores of CD11b/CD18 to the cell surface along with shedding of cell surface L-selectin.1,2 Furthermore, cellular activation, as dictated by conformational changes in cell surface proteins such as β2 integrins3 is readily detectable by this general approach.
Recently, others have taken a new approach to identify ligands on cells and whole organs by using phage display techniques.4-9Through these observations, it has become apparent that cell-type specific recognition might be achieved by scanning whole cells with phage display peptide libraries. To date, however, few cell-type specific binding peptide sequences have been identified using phage display techniques. Furthermore, little information exists on library constructs that preferentially interact with cellular targets and the selection procedures used to identify cell-specific phage.
Here we show the identification of specific, cell-binding peptide sequences using phage display libraries. We demonstrate that affinity purified phage-bearing these peptide sequences bind specifically to the membrane surface of human neutrophils (PMN) or monocytes. We also show that phage bearing these peptide sequences can induce receptor-mediated functional responses in PMN as indicated by intracellular calcium measurements. Our results demonstrate that the use of both linear and structurally constrained libraries is complementary and may be crucial for the identification of high affinity ligands. Finally, we show that numerous, unique cell-binding motifs can be identified through the use of different phage affinity purification procedures. Taken together, these data indicate that the phage display approach is likely to have broad applications in diagnosis and management of human disease.
MATERIALS AND METHODS
Reagents and buffers.
Oligonucleotides for library construction as well as synthetic peptides were purchased from Macromolecular Resources (Fort Collins, CO). Peptides were synthesized by solid phase synthesis (Macromolecular Resources) and analyzed by high performance liquid chromatography (>90% purity) and mass spectrometry. HBSS consisted of (in g/L): 0.185 CaCl2, 0.094 MgSO4, 0.4 KCl, 0,06 KH2PO4, 8 NaCl, 0.048 Na2HPO4, 1 glucose, and HEPES added to 10 mmol/L (pH 7.4). HBSS(−) was prepared as HBSS but without CaCl2 or MgSO4. Saline HEPES consisted of 150 mmol/L NaCl and 10 mmol/L HEPES (pH 7.4). TBS buffer consisted of 150 mmol/L NaCl and 50 mmol/L Tris/HCl pH 7.4.
Random peptide phage display libraries.
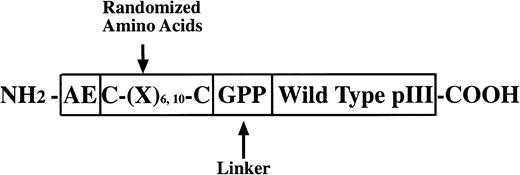
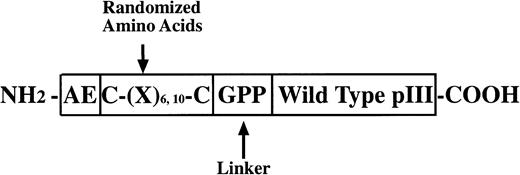
Three random peptide phage display libraries were used in these studies. J404, a linear random nonapeptide M13 phage library with kanamycin resistance, has been successfully used in the identification of several epitopes recognized by MoAbs and interactive regions of proteins.10-13 As detailed below, two structurally constrained libraries displaying hexa- and decapeptide loops, respectively, were constructed essentially as previously described14 using a vector, M13KBstX,15 which has kanamycin resistance. A schematic diagram of the modified pIII coat protein for the constrained library construction is shown in Fig1.
PIII protein construct for phage libraries. The glycine and two proline residues at the carboxyl end of the random regions function as a flexible linker and may serve to discourage association of the random peptide with the native pIII protein. Structural constraints in the CL10 and CL6 libraries were imposed by disulfide bonds between the cysteine residues flanking the variable region. CL10 and CL6 also include at the amino terminus of the mutated pIII protein wild-type residues (glutamic acid and alanine) and may help to preserve the signal peptidase cleavage site allowing cleavage of the peptide backbone at the correct site on the pIII pre-protein.
PIII protein construct for phage libraries. The glycine and two proline residues at the carboxyl end of the random regions function as a flexible linker and may serve to discourage association of the random peptide with the native pIII protein. Structural constraints in the CL10 and CL6 libraries were imposed by disulfide bonds between the cysteine residues flanking the variable region. CL10 and CL6 also include at the amino terminus of the mutated pIII protein wild-type residues (glutamic acid and alanine) and may help to preserve the signal peptidase cleavage site allowing cleavage of the peptide backbone at the correct site on the pIII pre-protein.
M13KBstX replicative form (RF) DNA was extracted from infected K91Escherichia coli, purified on a equilibrium CsCl gradient,16 cleaved with BstXI, further purified on a potassium acetate gradient,15,17 followed by dephosphorylation with calf intestinal phosphatase (Pharmacia Biotech, Piscataway, NJ) as described by the manufacturer. A synthetic DNA construct containing the ligation-compatibleBstXI ends and the random inserts with the designed flanking residues was prepared by annealing a collection of degenerate oligonucleotides with two “half-site” oligonucleotides as described.14 The coding strand of this DNA construct contained (NNK)6 or (NNK)10 codons, where N corresponds to equimolar amounts of A, T, G, or C and where K corresponds to equimolar amounts of G or T. Following ligation and fill-in reaction, the DNA product was transfected into electrocompetent MC 1061 cells18 with a Bio-Rad Gene Pulser. The decapeptide library termed “CL10” was produced by 108 separate electroporations using a total of 440 μg of ligated vector DNA resulting in a diversity of approximately 1.04 × 109unique clones. To produce the hexapeptide library termed “CL6,” a total of 136 μg of ligated vector DNA was transfected in 34 separate electroporations yielding a library containing 9 × 108unique clones. Libraries were propagated in LB containing kanamycin, harvested and resuspended in TBS buffer containing 0.03% sodium azide at a final concentration of 1 × 1013 Pfu/mL as previously described.19 To replenish depleted stocks of the J404 linear library, an additional 9-mer linear library was constructed using the original degenerate oligonucleotide constructs10as above. For this library, termed LL9, 450 μg of ligated vector DNA was transfected in 106 separate electroporations yielding a library containing 3.66 × 109 unique clones. Analysis of the LL9 library revealed a random distribution of the frequency of codons in the insert with no duplicate occurrences of peptide sequences as has been described for the J404 library10 (data not shown).
PMN isolation.
PMN were isolated from whole blood (anticoagulated with citrate/dextrose) obtained from healthy volunteers, using a gelatin sedimentation technique previously described.20 PMN were resuspended in modified HBSS devoid of Ca2+ and Mg2+ (HBSS(−)) at a concentration of 4 × 107 cells/mL (4°C) and used for subsequent experiments.
Selection of PMN-binding phage.
Freshly isolated PMN (2 × 107) were incubated with 20-μL aliquots of phage library (∼5 × 1010 phage) in 1 mL HBSS containing 0.1% bovine serum albumin (phage buffer) for 2 hours at 4°C or for 1 hour at 20°C in a 1.5-mL microcentrifuge tube. PMN were then washed five times with phage buffer and bound phage were eluted for 5 minutes in 2 mL of 0.1 mol/L glycine (pH 2.2) followed by addition of phage buffer containing 0.5% Tween 20 to the remaining cell pellet. After neutralization with Tris buffer pH 8.1, the titer of phage was determined in each fraction by plaque assay according to standard procedures.21 Phage eluates were then amplified in “starved” K91 E. coli19 on solid LB agar containing 75 μg/mL kanamycin,10 purified by precipitation with polyethylene glycol, and resuspended in 600 μL saline/HEPES buffer. An aliquot (20 μL) of purified phage was subsequently re-applied to newly isolated PMN for a total of three affinity purification and two amplification steps. Individual phage clones were then isolated, amplified, and DNA prepared (Sequenase version 2.0 kit; Amersham, Cleveland OH). The random peptide sequences were deduced after sequencing the unique nucleotide region of the pIII protein by the Sanger method. As a control for PMN-specific selection, parallel phage affinity purification steps were carried out in a 1.5-mL microcentrifuge tube in the absence of PMN, and a sample of the recovered phage was analyzed by sequencing of the DNA insert.
ELISA for detecting phage binding to PMN.
Suspensions of 2.5 × 105 PMN in 100 μL HBSS were allowed to settle and bind to wells of a microtiter plate (Limbro-Titertek; Flow Laboratories, Irvine, CA) for 30 minutes (20°C). After gentle washing, plate wells were blocked with 0.5% bovine serum albumin (BSA) in HBSS for 30 minutes (blocking buffer). Amplified phage clones (2.5 × 1010 phage particles) were added to each well and allowed to bind for 20 minutes. Unbound phage were subsequently removed by gentle washing. Bound phage were fixed to PMN by addition of paraformaldehyde (3.7%) for 10 minutes followed by washing and incubation in blocking buffer for 45 minutes. Bound phage were assayed after incubation of wells with a 1:1,000 dilution of biotin-sheep anti-phage antibody (5prime-3prime, Boulder, CO) followed by incubation with alkaline phosphatase-streptavidin (Pierce). After substrate addition, wells were analyzed in a microtiter plate reader at 405 nm.
Immunofluorescence.
For immunofluorescence studies, suspensions of 1 × 105PMN in HBSS were allowed to spread on glass coverslips for 30 minutes at room temperature followed by washing and blocking with 0.5% BSA in HBSS for 30 minutes. PMN were then incubated with individual phage clones for 20 minutes (109 phage/coverslip). PMN were then washed, fixed with 3.7% paraformaldehyde, and permeabilized with 0.5% Triton X-100 in HBSS for 30 minutes at 20°C. After blocking reactive groups with 0.5% BSA in HBSS, PMN were incubated with biotinylated sheep anti-phage antibody as described above and followed by incubation with FITC-Streptavidin (1:1,000; Jackson Laboratories, Bar Harbor, ME). PMN were viewed with a BioRad MRC-600 confocal fluorescence microscope.
Flow cytometry studies.
Suspensions of PMN (5 × 106) in 1 mL HBSS containing 0.5% BSA were incubated with individual phage clones for 2 hours at 4°C or for 1 hour at 20°C followed by washing and fixation with 3.7% paraformaldehyde. PMN were then stained with anti-phage antibody and FITC-streptavidin as described above. Cells were analyzed for bound phage with a FACSscan flow (Becton-Dickinson Immunocytometry Systems, Mountain View, CA). Identical experiments were performed with other human cell types including monocytic cells (U937), epithelial cells (T84 and HT29), an endothelial cell line (ECV 304), fibroblasts (VA2), and peripheral blood lymphocytes. The latter were isolated using Ficoll density purification from heparinized blood of healthy volunteers. Isolated lymphocytes were then cultured with Phytohemagglutinin (GIBCO-BRL, Gaithersburg, MD) in RPMI 1640 medium supplemented with 10% fetal bovine serum and 2 nmol/L recombinant human IL-2 (Boehringer Mannheim) and used after 1 week of culture.
PMN binding assays with selected clones were also performed in the presence of competing peptides or of control scrambled peptides. The inhibitory effect of added peptide was quantified by evaluating the percentage of maximal phage binding as defined by flow cytometry studies above.
Measurement of cytosolic [Ca++] in PMN.
PMN were loaded with the calcium indicator Indo-1 and cytoplasmic [Ca++] measured as previously described22,23using a Hitatchi F-4500 spectrofluorimeter. PMN, 2 × 106, were suspended in 960 μL of HBSS and stimulated by the adding 40 μL of purified phage (4 × 1010 PFU) from a stock solution containing 1012 PFU/mL in saline/HEPES buffer. Controls included buffer vehicle alone, irrelevant phage, and the well characterized agonists fMLF (100 nmol/L) and IL-8 (14 nmol/L). In a subset of experiments, PMN were pre-incubated with 1 μg/mL pertussis toxin (Sigma, St Louis, MO) for 2 hours (37°C) in HBSS(−) before calcium experiments.22 23
RESULTS
Library construction.
Two random peptide phage libraries displaying cyclic hexapeptides or decapeptides were produced with the M13KBst vector and used in addition to a previously characterized linear library (J404) with the intent to characterize PMN-binding phage peptides. The two circular libraries, CL10 and CL6, were analyzed by nucleotide sequencing of the 5′ end of the pIII protein. All of the 112 different phage clones sequenced were found to contain the random insert flanked by the correct invariant sequences. Approximately 3% of the sequences coded for random peptides shorter than the intended 10 or 6 residues, indicating rearrangements occurring during the assembly of the DNA construct.
The amino acid sequences of random peptides deduced from the nucleotide sequence of 48 clones from the CL10 and 64 clones from the CL6 library indicated a random distribution of the frequency of codons in the insert. Duplicate occurrences of peptide sequences were not found. Codons for Lys and Ala were found to be slightly over-represented, whereas those for Tyr and Cys were somewhat under-represented. Cysteine residues in the displayed peptides were almost exclusively found in pairs. Phage with odd numbers of cysteine residues were not found suggesting negative biological selection. Thus, no relevant bias in the CL10 and CL6 libraries was observed, in agreement with other reports of phage library construction.14,15 24
Affinity purification of PMN-binding phage bearing linear motifs.
Experiments were performed to affinity isolate phage binding to human PMN isolated from whole blood obtained from normal human volunteers. In parallel experiments, PMN were resuspended in HBSS containing BSA and coincubated with phage library at either 20°C or 4°C. The two different binding temperatures were used to evaluate the effects of temperature and potential phage internalization on binding motif patterns. Phage binding to human PMN were initially selected with the linear nonpeptide phage library. Bound phage were first eluted under acidic conditions (pH 2.2) followed by PMN incubation in detergent-containing buffer to recover any remaining cell-associated phage. The acid-eluted and cell-associated phage fractions were then amplified separately and re-applied to fresh PMN. After three rounds of such selection, an amplification of approximately 104 was achieved for affinity purification’s either at 4°C or at 20°C (data not shown). No significant increase of nonspecific phage binding was observed.
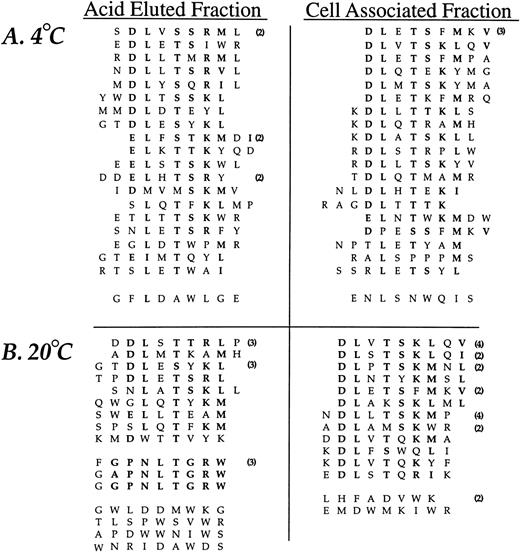
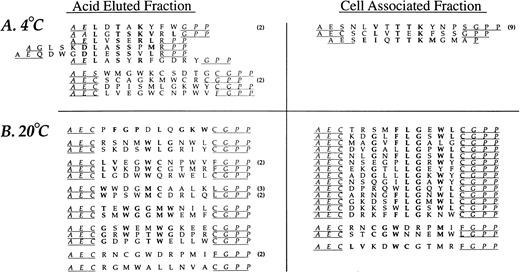
The random insert region was sequenced from 44 phage selected in the third round of affinity purification from experiments performed at 4°C and 47 phage derived from experiments performed at 20°C (Fig2). Sequence similarity was present in all but two sequences (95%) from phage selected at 4°C and 35 of the 47 sequences (74%) from phage recovered at 20°C. A consensus, DLXTSK(M/L)X(V/I/L), where X represents a non-conserved position, was particularly apparent in the cell-associated (detergent lysis) fractions (Fig 2). Here, powerful selective pressure of the PMN-phage peptide interaction is apparent with exact matches observed in seven residue sequences in some of the recovered phage (peptide sequences such as DLVTSKLQV and DLSTSKLQI). The most conserved residues displayed at the amino terminus of peptide inserts were Asp and Leu followed by Thr and Lys. Similarity between phage peptides was increased by considering conservative substitutions between amino acids.
Neutrophil-binding phage peptide sequences isolated from the linear (X9) phage display library. The unique region of phage DNA recovered from the third round of affinity purification was sequenced and the motifs aligned to form the consensus DLXTSK(M/L)X(V/I/L) and GPNLTGRW, respectively. Identical residues and conservative substitutions are shown in bold. Following sets are considered to contain homologous amino acids (I, L, V), (K, R), (D, E), (S, T), (Y, F). The number of clones encoding the same peptide is shown in parentheses. The linker sequence at the amino terminus (GPP, not shown) was correctly displayed in all clones. Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Neutrophil-binding phage peptide sequences isolated from the linear (X9) phage display library. The unique region of phage DNA recovered from the third round of affinity purification was sequenced and the motifs aligned to form the consensus DLXTSK(M/L)X(V/I/L) and GPNLTGRW, respectively. Identical residues and conservative substitutions are shown in bold. Following sets are considered to contain homologous amino acids (I, L, V), (K, R), (D, E), (S, T), (Y, F). The number of clones encoding the same peptide is shown in parentheses. The linker sequence at the amino terminus (GPP, not shown) was correctly displayed in all clones. Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
The analysis of phage eluted with low pH buffer at 20°C revealed an additional consensus sequence, GPNLTGRW. While this motif was present in only three different phage clones, there was an eight residue match between sequences, indicating a very strong selection. The DNA insert corresponding to the random peptides displayed by the three consensus-bearing phage showed different triplets encoding for Pro, Leu, and Arg, indicating uniqueness of each selected phage. Such eight residue matches provide an alternative estimate of the true amino acid sequence diversity of the library which is much larger than the initial estimate of ∼3.6 × 108. Finally, a small group of phage peptides binding to PMN at 20°C and recovered by acid elution showed a relative abundance of Tyr residues without displaying an obvious consensus.
Affinity purification PMN-binding phage bearing circular motifs.
Affinity isolation experiments using two circular libraries, CL6 and CL10, were performed as above with the linear library. An amplification of approximately 103 to 104 was achieved after three rounds of affinity purifications for both cyclic libraries at either 4°C or 20°C (data not shown). The deduced amino acid sequences of phage selected from the CL10 library after three rounds of affinity purification are shown in Fig 3. Nine of the 13 (69%) distinct phage clones identified in the 4°C experiments had the same linear consensus DLXTSK(M/L)X(I/V/L) that was identified with the J404 library. Conservative substitutions indicated an even greater homology of the phage peptides to this consensus sequence. However, as can be seen in Fig 3, none of these clones displayed the linker sequences as designed by the library construction (Fig 1). In particular, the Cysteine residues flanking the random region were absent, suggesting a linear conformation instead of a constrained loop. Phage clones bearing a Cys residue in either the 5′ or 3′ portion of the linker sequence always displayed an additional Cys within the random region, suggesting the formation of a short, structurally constrained sequence within the displayed peptide. Conversely, the analysis of the PMN binding phage selected after three rounds of affinity purification performed at 20°C revealed different consensus motifs that were mostly cyclic. In particular, 14 of the 17 phage clones (82%) isolated from the cell-associated fraction displayed peptide sequences related to the consensus CXXGXFLGXWLC. The residues Leu and Gly were absolutely conserved and occupied same position within the random region in all of the phage clones. Analysis of phage clones recovered from the acid eluted fraction showed additional consensus motifs that are grouped in Fig 3. Interestingly, two phage clones, CLVEGWCNPWV and CLVKDWCGTMR, contain a short loop displaying the consensus CLVXXWC and were identical to phage clones recovered from the acid eluted fraction at 4°C and from the cell-associated fraction at 20°C, respectively. Finally, phage bearing the cyclic peptide CPFGPDLQGKWC showed a 6 residue match with the peptide FGPNLTGRW selected with the linear J404 library under the same conditions.
Neutrophil-binding phage peptide sequences isolated from the cyclic (CL10) phage display library. Selection and sequencing of phage was performed as described in the text. The peptides displayed by the phage recovered from the experiments performed at 4°C are aligned to the consensus motif DLXTSK(M/L)X(V/I/L) that was previously identified with the linear nonapeptide library. Identical residues and conservative substitutions are shown in bold. Underlined residues represent the putative linker sequences which, in several clones, diverge from the linker sequences designed by the library construction (see Fig 1) due to the absence of the cysteine residues. The phage clones recovered from the cell-associated fraction at 20°C are aligned to form the consensus motif CXXGXFLGXWLC. One phage in the eluate fraction at 20°C (first from the top) shows a high degree of homology with the FGPNLTGRW displaying phage recovered with the J404 library under the same experimental conditions, representing a structurally related sequence. Other possible consensus motifs are tentatively grouped and shown in bold.
Neutrophil-binding phage peptide sequences isolated from the cyclic (CL10) phage display library. Selection and sequencing of phage was performed as described in the text. The peptides displayed by the phage recovered from the experiments performed at 4°C are aligned to the consensus motif DLXTSK(M/L)X(V/I/L) that was previously identified with the linear nonapeptide library. Identical residues and conservative substitutions are shown in bold. Underlined residues represent the putative linker sequences which, in several clones, diverge from the linker sequences designed by the library construction (see Fig 1) due to the absence of the cysteine residues. The phage clones recovered from the cell-associated fraction at 20°C are aligned to form the consensus motif CXXGXFLGXWLC. One phage in the eluate fraction at 20°C (first from the top) shows a high degree of homology with the FGPNLTGRW displaying phage recovered with the J404 library under the same experimental conditions, representing a structurally related sequence. Other possible consensus motifs are tentatively grouped and shown in bold.
Analysis of the random region of PMN-binding phage selected from the CL6 library revealed similarities to the motifs obtained from the libraries with longer random inserts. Twenty-eight phage clones sequenced from the acid eluted and cell associated fractions affinity purified at 4°C displayed 12 distinct peptides. Eight of them (66%) confirmed the strong selection for a phage subpopulation displaying an unconstrained linear form similar to the main consensus identified with the J404 library. In particular, the phage clone bearingAESDLLTNRLGPP, recovered nine times in the acid eluted fraction, did not contain Cys residues in the linker sequences (underlined) and displayed a four residue match (bold) to the DLXTSK(M/L)X(V/I/L) motif. Analysis of 41 phage affinity purified at 20°C showed, in the acid eluted and in the cell-associated fractions, 19 distinct phage clones. Eleven of them (58%) contained homologies to the consensus WLGXW, which is similar to the motif CXXGXFLGXWLC identified with the CL10 library.
Binding studies with affinity purified phage.
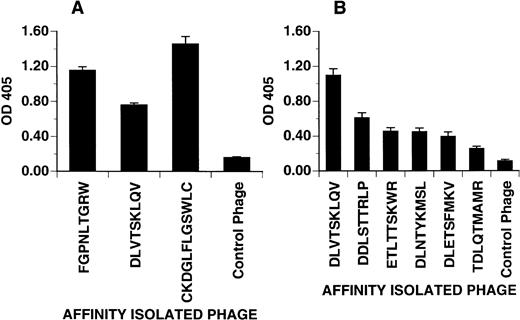
Phage bearing consensus-related sequences were tested for their ability to bind to adherent PMN in a microtiter plate. As shown in Fig4A, three phage clones bearing different consensus-related sequences bound to PMN when compared with an irrelevant control phage. OD values for phage displaying the peptides FGPNLTGRW and CKDGLFLGSWLC were consistently higher than that for the phage clone bearing the peptide DLVTSKLQV suggesting either higher affinity or more binding sites per PMN. In Fig 4B, the effect of peptide sequence variability on phage binding to PMN was explored. Here, related phage displaying different grades of homology to the consensus DLXTSK(M/L)X(V/I/L) were tested for binding to adherent PMN. As expected, peptide sequences with fewer residue matches showed diminished binding.
Phage attachment assay. Purified phage clones were incubated in microtiter plates coated with PMN freshly isolated from peripheral venous blood. Bound phage were detected with polyclonal anti-phage antibody and quantified by light absorbance as described in Materials and Methods. (A) Distinct PMN binding phage clones selected from different experiments. The DLVTSKLQV phage consistently showed a weaker detection signal as compared with the other binding phage. (B) The binding affinity of phage clones displaying different grades of homology to the consensus motif DLXTSK(M/L)X(V/I/L). Compared with the DLVTSKLQV-displaying phage, clones with fewer residues matching to the consensus motif show a decreased binding affinity. Control phage displayed irrelevant sequences (AQPQVRPIG or GPRPGPPKL). Optical density at 405 represents phage incubated wells after subtraction of the wells without phage incubation. Phage clones incubated on BSA coated wells (not shown) yielded OD value between 0.1 and 0.2. Values representative of one of two experiments showing the mean ± SD of quadruplicate determinations.
Phage attachment assay. Purified phage clones were incubated in microtiter plates coated with PMN freshly isolated from peripheral venous blood. Bound phage were detected with polyclonal anti-phage antibody and quantified by light absorbance as described in Materials and Methods. (A) Distinct PMN binding phage clones selected from different experiments. The DLVTSKLQV phage consistently showed a weaker detection signal as compared with the other binding phage. (B) The binding affinity of phage clones displaying different grades of homology to the consensus motif DLXTSK(M/L)X(V/I/L). Compared with the DLVTSKLQV-displaying phage, clones with fewer residues matching to the consensus motif show a decreased binding affinity. Control phage displayed irrelevant sequences (AQPQVRPIG or GPRPGPPKL). Optical density at 405 represents phage incubated wells after subtraction of the wells without phage incubation. Phage clones incubated on BSA coated wells (not shown) yielded OD value between 0.1 and 0.2. Values representative of one of two experiments showing the mean ± SD of quadruplicate determinations.
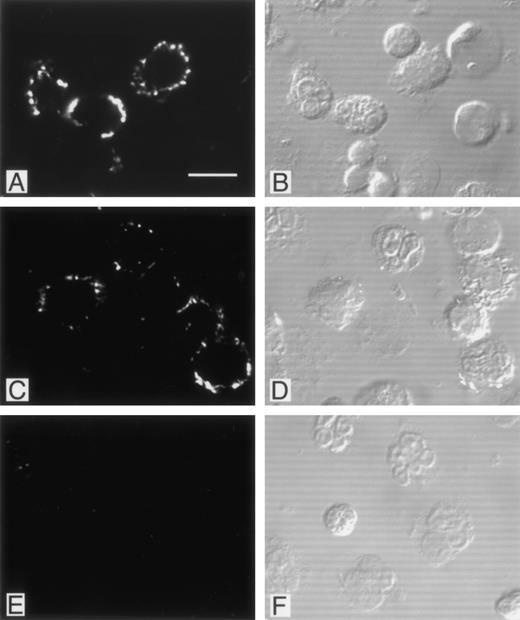
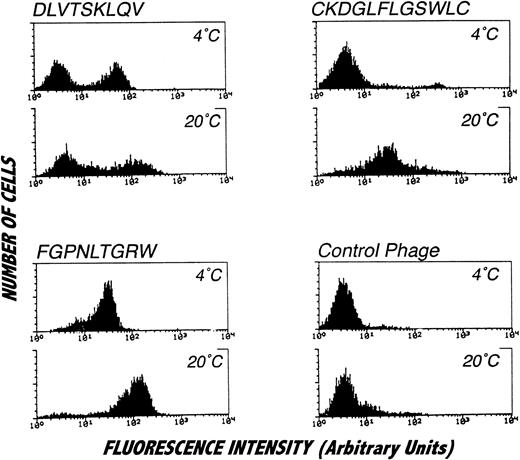
Immunofluorescence studies were performed in an attempt to localize PMN-bound phage. Adherent phage were visualized by confocal microscopy after staining with biotinylated anti-phage antibody and FITC-avidin (Fig 5). As shown in the figure, phage bearing DLVTSKLQV or FGPNLTGRW motifs showed a comparable multifocal staining pattern restricted to the PMN surface, whereas control phage failed to label. Intracellular fluorescence staining patterns indicative of internalized phage particles were not observed in the permeabilized PMN preparations. Interestingly, a considerable proportion of the PMN did not show any detectable fluorescent signal (approximately 50%). No morphological difference between labeled and unlabeled PMN were observed to account for the difference in labeling. Additional fluorescence studies with the CKDGLFLGWLC bearing phage clone yielded a similar focal staining pattern (data not shown).
Localization of PMN-binding phage clones by immunofluorescence. PMN isolated from peripheral blood were incubated with phage clones, fixed with paraformaldehyde, permeabilized with Triton X-100, and labeled with biotinylated polyclonal antibodies as described in Materials and Methods. There was a bright membrane staining after incubation with the FGPNLTGRW (A) or DLVTSKLQI (C) displaying phage. Incubation of PMN with an irrelevant phage resulted in no detectable immunofluorescence (E). Cytoplasmic staining suggestive of phage internalization was not detected. For orientation, the corresponding Nomarski images are shown in (B), (D), and (F). As can be seen, phage incubation labeled only part of the neutrophils. Incubation with the CKDGLFLGSWLC-displaying phage yielded staining patterns comparable with those found with the other PMN-binding phage (data not shown).
Localization of PMN-binding phage clones by immunofluorescence. PMN isolated from peripheral blood were incubated with phage clones, fixed with paraformaldehyde, permeabilized with Triton X-100, and labeled with biotinylated polyclonal antibodies as described in Materials and Methods. There was a bright membrane staining after incubation with the FGPNLTGRW (A) or DLVTSKLQI (C) displaying phage. Incubation of PMN with an irrelevant phage resulted in no detectable immunofluorescence (E). Cytoplasmic staining suggestive of phage internalization was not detected. For orientation, the corresponding Nomarski images are shown in (B), (D), and (F). As can be seen, phage incubation labeled only part of the neutrophils. Incubation with the CKDGLFLGSWLC-displaying phage yielded staining patterns comparable with those found with the other PMN-binding phage (data not shown).
Because of the focal nature of the phage staining observed by confocal microscopy, the PMN binding properties of phage bearing consensus-related sequences were examined by flow cytometry. Interestingly, as shown in Fig 6, flow cytometric analysis revealed binding of phage bearing consensus-related sequences to a subpopulation of PMN. In particular, phage with the DLVTSKLQV peptide, as well as other phage clones bearing the same consensus motif (data not shown), exclusively stained a subpopulation of PMN comprising approximately 50% of the cells. While an identical pattern of staining was observed at 4°C and 20°C, the fluorescence signal from cells labeled at 20°C was substantially brighter than that obtained at 4°C. In contrast to this bimodal staining pattern, phage displaying the FGPNLTGRW motif labeled all PMN and produced a particularly bright signal when the binding reactions were performed at 20°C. Temperature sensitive labeling was observed for one of the cyclic motifs identified. As shown in the figure, there was no detectable labeling of PMN at 4°C with phage bearing the CKDGLFLGSWLC peptide sequence. However, all of PMN were labeled with the phage when binding was performed at 20°C. This finding is consistent with the affinity purification results, since phage bearing CKDGLFLGSWLC-related sequences were not recovered from selection procedures performed at 4°C. As can be seen in Fig 6, labeling PMN at 4°C with phage bearing the peptide sequence CKDGLFLGSWLC did result in a small population of highly fluorescent cells (approximately 4%). However, analysis of orthogonal light scatter revealed that this small population of cells most likely represented contaminating monocytes. Additionally, such binding profiles were not affected by stimulation with phorbol myristate acetate or by chelation of extracellular calcium (data not shown).
Staining of isolated PMN with affinity purified phage clones and analysis by flow cytometry. PMN were incubated with affinity purified phage clones or irrelevant control phage followed by biotinylated anti-phage antibody and labeled with FITC conjugated to streptavidin. The distinct staining pattern of the analyzed phage suggests binding to different target molecules. All phage clones, but not the control phage, showed brighter signals by incubation at 20°C. The DLVTSKLQV displaying phage (or other homologous phage, data not shown) consistently labeled approximately 50% of the cells. Incubation of cells with the CKDGLFLGSWLC-displaying phage at 4°C resulted in staining of a small cell subpopulation that could be identified as monocytes on the orthogonal light scatter (data not shown) but not in labeling of the main cell population consisting of PMN. Histograms represent specifically stained cell numbers on the vertical axis (labeled counts) plotted against fluorescence on a log scale from 5,000 cells per condition. These results were obtained in four separate analyses.
Staining of isolated PMN with affinity purified phage clones and analysis by flow cytometry. PMN were incubated with affinity purified phage clones or irrelevant control phage followed by biotinylated anti-phage antibody and labeled with FITC conjugated to streptavidin. The distinct staining pattern of the analyzed phage suggests binding to different target molecules. All phage clones, but not the control phage, showed brighter signals by incubation at 20°C. The DLVTSKLQV displaying phage (or other homologous phage, data not shown) consistently labeled approximately 50% of the cells. Incubation of cells with the CKDGLFLGSWLC-displaying phage at 4°C resulted in staining of a small cell subpopulation that could be identified as monocytes on the orthogonal light scatter (data not shown) but not in labeling of the main cell population consisting of PMN. Histograms represent specifically stained cell numbers on the vertical axis (labeled counts) plotted against fluorescence on a log scale from 5,000 cells per condition. These results were obtained in four separate analyses.
To examine cell specificity of phage clones bearing consensus-related sequences, the binding of selected phage clones to various cell types was examined. In Table 1 is a summary of the results of phage binding to a number of different cell lines analyzed by flow cytometry. As summarized in the table, DLVTSKLQV-displaying phage bound exclusively to PMN, whereas the FGPNLTGRW and the CKDGLFLGSWLC bound to both PMN and monocytic (U937) cells. Interestingly, phage bearing a sequence related to the former monocyte-reactive one, CPFGPDLQGKWC, retained reactivity with PMN but failed to label U937 monocytic cells. No phage binding was observed on fibroblasts, endothelial cells, epithelial cells, and peripheral blood lymphocytes.
Synthetic peptide binding and inhibition of phage–PMN interactions.
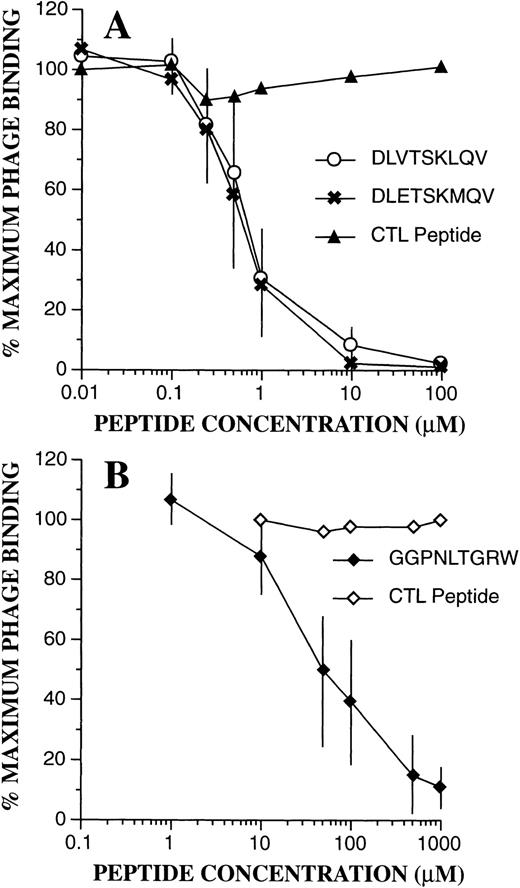
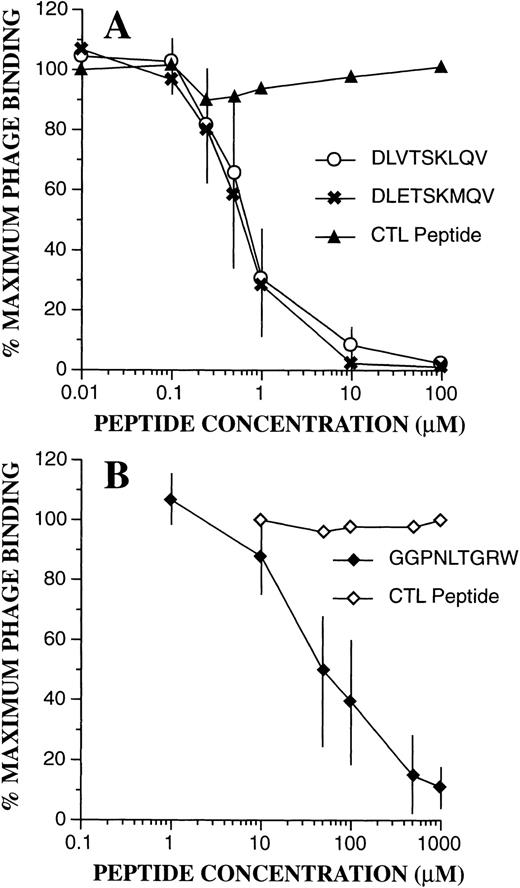
To confirm the binding specificity of phage-bearing consensus-related peptide sequences, competition assays were performed with synthetic peptides (Fig 7). Bound phage were then quantitated by flow cytometric techniques as above. As can be seen, binding of DLVTSKLQV-displaying phage to PMN was markedly inhibited in the presence of synthetic peptides containing identical or similar sequences of DLVTSKLQV and DLETSKMQV. A control peptide with a scrambled sequence KQLSEMVTD had no effect. Half maximal inhibition (IC50) for both peptides was seen at 0.6 μmol/L. Similarly, as shown in Fig 7B, addition of the peptide GGPNLTGRW resulted in marked inhibition of binding of FGPNLTGRW containing phage to PMN. Here, the IC50 was estimated at 50 μmol/L, and phage binding was unaffected by a control scrambled peptide (RPGNGWLGT). Interestingly, even high concentrations of the GGPNLTGRW peptide (1 mmol/L) had no effect on the PMN binding of the phage displaying the cyclic CPFGPDLQGKWC motif. These results suggest that the cyclic motif may either recognize a different receptor/receptor configuration, or that it binds with higher affinity to the putative receptor for the linear nonpeptide.
Phage binding competition assays. Effect of the synthetic peptides was evaluated as described in Materials and Methods. (A) Inhibition of PMN binding of the DLVTSKLQV displaying phage by the DLVTSKLQV and DLETSKMQV peptides. The scramble peptide KQLSEMVTD had no effect. (B) Inhibition of PMN binding of the FGPNLTGRW displaying phage by the GGPNLTGRW synthetic peptide. Phage binding was unaffected by the control scramble peptide RPGNGWLGT. The data points represent the mean ± SD from three experiments.
Phage binding competition assays. Effect of the synthetic peptides was evaluated as described in Materials and Methods. (A) Inhibition of PMN binding of the DLVTSKLQV displaying phage by the DLVTSKLQV and DLETSKMQV peptides. The scramble peptide KQLSEMVTD had no effect. (B) Inhibition of PMN binding of the FGPNLTGRW displaying phage by the GGPNLTGRW synthetic peptide. Phage binding was unaffected by the control scramble peptide RPGNGWLGT. The data points represent the mean ± SD from three experiments.
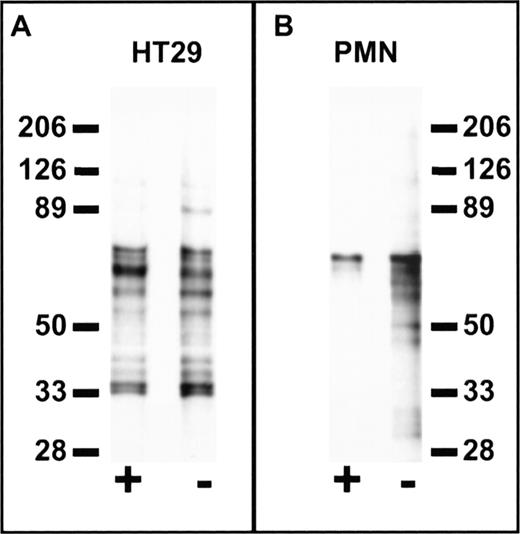
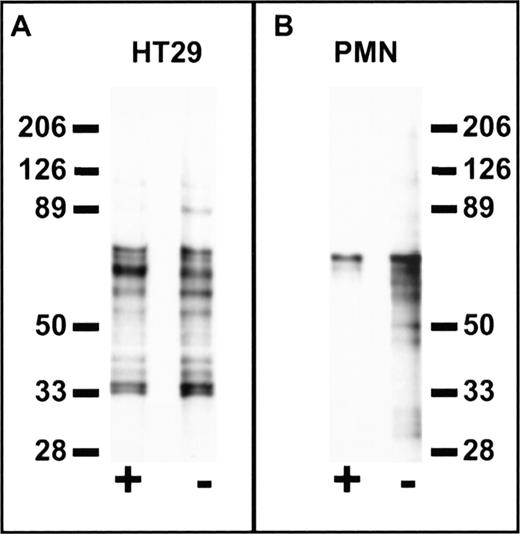
Cell specificity of peptide binding was further confirmed in experiments using a photo-activatable derivative of FGPNLTGRW. Using the biotin-conjugated photo-activatable reagent, Biotin-GF(Benzoyl)GPNLTGRW, we compared the labeled protein profiles of PMN and HT29 epithelial cells (Fig 8). Cells were incubated with 100 μmol/L labeled peptide alone or in the presence of excess (1 mmol/L) unlabeled peptide followed by photolysis. As shown in the figure, probing Western blots of the biotinylated cell extracts with avidin-peroxidase revealed significant differences between PMN and HT29 cells. In particular, labeled protein bands from HT29 cells were not eliminated by excess unlabeled peptide, whereas biotin labeling of PMN protein bands in the 30 to 60 kD range was inhibited by coincubation with unlabeled peptide. These results suggest that peptides derived from cell binding phage retain cell-type binding specificity at the protein level. Furthermore, such reagents may be useful in the characterization of receptors for phage bearing specific cell binding sequences.
Cell-specific labeling by a synthetic peptide derived from phage containing a PMN-binding peptide sequence. Suspensions of 1 × 107 HT29 epithelial cells (A) or PMN (B) in HBSS containing 0.5% BSA were incubated at 4°C for 1 hour in the presence (+) or absence (−) of 1 mmol/L FGPNLTGRW followed by the addition of Biotin-GF(Benzoyl)GPNLTGRW to a concentration of 100 μmol/L. After a 30-minute incubation in the dark, the samples were placed on ice and photolyzed for 10 minutes under UV light. The washed cell pellets were solubilized in 150 mmol/L NaCl, 100 mmol/L KCl, 2 mmol/L EDTA, and 10 mmol/L Hepes pH 7.4 containing 1% N-octylglucoside and the protease inhibitors DFP, PMSF, aprotinin, bestatin, chymostatin, and pepstatin. Cell lysates were then incubated with avidin-Sepharose for 2 hours followed by washing and denaturation of the avidin beads with reduced sodium dodecyl sulfate (SDS) sample buffer. Samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 6% to 16% polyacrylamide gradient gels followed by western blot, probed with peroxidase-conjugated streptavidin, and developed by enhanced chemiluminescence (Amersham Inc). Each lane represents the biotin-labeled protein profiles of ∼3 × 106 cells. Molecular weights in kiloDaltons are shown to the side. As can be seen, specific labeling of PMN protein bands in the 30 to 60 kD range was inhibited by coincubation with unlabeled peptide, whereas no specific labeling is observed for HT29 cells.
Cell-specific labeling by a synthetic peptide derived from phage containing a PMN-binding peptide sequence. Suspensions of 1 × 107 HT29 epithelial cells (A) or PMN (B) in HBSS containing 0.5% BSA were incubated at 4°C for 1 hour in the presence (+) or absence (−) of 1 mmol/L FGPNLTGRW followed by the addition of Biotin-GF(Benzoyl)GPNLTGRW to a concentration of 100 μmol/L. After a 30-minute incubation in the dark, the samples were placed on ice and photolyzed for 10 minutes under UV light. The washed cell pellets were solubilized in 150 mmol/L NaCl, 100 mmol/L KCl, 2 mmol/L EDTA, and 10 mmol/L Hepes pH 7.4 containing 1% N-octylglucoside and the protease inhibitors DFP, PMSF, aprotinin, bestatin, chymostatin, and pepstatin. Cell lysates were then incubated with avidin-Sepharose for 2 hours followed by washing and denaturation of the avidin beads with reduced sodium dodecyl sulfate (SDS) sample buffer. Samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 6% to 16% polyacrylamide gradient gels followed by western blot, probed with peroxidase-conjugated streptavidin, and developed by enhanced chemiluminescence (Amersham Inc). Each lane represents the biotin-labeled protein profiles of ∼3 × 106 cells. Molecular weights in kiloDaltons are shown to the side. As can be seen, specific labeling of PMN protein bands in the 30 to 60 kD range was inhibited by coincubation with unlabeled peptide, whereas no specific labeling is observed for HT29 cells.
Phage effects on PMN calcium signaling.
To test for phage effects on PMN function, experiments were performed to determine whether motif-bearing phage could induce changes in cytosolic [Ca++] in PMN. As shown in Fig 9A, addition of phage bearing the sequence FGPNLTGRW induced a significant change in cytosolic [Ca++] when compared with the addition of control irrelevant phage. In contrast, phage bearing the binding sequence DLVTSKLQV had absolutely no effect. The magnitude of the increase in cytosolic [Ca++] induced by the FGPNLTGRW-displaying phage was approximately one third of that induced by a saturating dose (10−7 mol/L) of the well-characterized PMN agonist fmet-leu-phe (fMLF). To test whether the phage-induced calcium signal might be G-protein mediated, calcium experiments were performed on pertussis toxin-treated PMN. As shown in Fig 9B, incubation of PMN for 2 hours with pertussis toxin completely blocked the phage-induced response, as well as the fMLF-induced response (positive control, not shown), indicating that the FGPNLTGRW-displaying phage is indeed activating through a Gαi-mediated pathway. As a negative control, demonstrating that pertussis toxin had not simply blocked all PMN signal transduction, stimulation with immune complexes that are known to activate PMN by a pertussis toxin-insensitive Fcγ receptor-mediated pathway,22,23 25 still exhibited a large cytosolic [Ca++] increase (data not shown).
DISCUSSION
Currently, there are limited approaches that can be taken to identify peptide binding domains on cells or proteins without prior specific functional or structural information. While antibody approaches have proven successful in this regard, extreme variability in the antigenic response has remained problematic. Recently, phage libraries containing vast repertoires of random peptide inserts of various lengths and conformations have been used in the identification of interactive regions of proteins and other molecules. In addition, with phage techniques it is possible to identify peptide binding domains on cells or proteins without having any preexisting information on such domains or the biology of the target cells. We have used three such phage display libraries to identify peptides that bind preferentially to human neutrophils. These include groups of peptide sequences isolated under different affinity purification procedures that contained both the linear and structurally constrained consensus motifs DLXTSK(M/L)X(V/I/L), GPNLTGRW, and CXXGXFLGXWLC, respectively. We have also shown that one of these peptide motifs, GPNLTGRW, has functional effects as defined by a Gαi-linked rise in intracellular calcium. These results define, for the first time, functional cell-specific binding peptides identified with phage display library technology.
A dramatic finding in these studies was the identification of distinct PMN-binding peptides that were exclusively displayed in a linear or in a cyclic form by the bearing phage. In general, it has been assumed that libraries displaying cyclic peptides are more capable of interacting with targets at higher affinities than libraries displaying unconstrained oligopeptides free to assume different structural conformations in solution.9,26-31 In particular, previous reports have demonstrated that peptides containing cysteine pairs can be selected in high affinity screens of linear peptide libraries.26 In our studies, however, we were able to affinity purify PMN-binding phage containing linear sequences homologous to the unconstrained DLXTSK(M/L)X(V/I/L) motif even by screening phage libraries constructed to display the random region with a cyclic configuration. These latter findings suggest an extremely strong selection from the CL10 and CL6 libraries for a phage subpopulation displaying the random peptide without the intended structural constraints, ie, missing one or both Cys residues in the linker sequences intended to be conserved throughout all of the library members. These findings are consistent with previous studies on MoAbs that demonstrated an unpredictable preference for a specific type of constraint32 but also raise the possibility of nonrandom selection of particular clones in the unscreened libraries. Fortunately, such phage variants have never been identified in randomly selected clones from our library stock solutions or in phage selected from epitope mapping studies we performed with a number of different MoAbs (L.M. and C.A.P., unpublished observations, 1996, 1997).
Most likely, these biases result from errors in the DNA encoding for the random region and the linker sequences. Events such as single base changes and rearrangements undoubtedly occur during oligonucleotide synthesis and assembly of the DNA construct. While the majority of these errors probably result in defective phage particle formation, our data suggests that the CL10 and CL6 phage libraries contain only a very small percentage (<1 in 105 or 106) of members displaying an unconstrained peptide. However, given the large diversity of these libraries, even this small percentage results in a considerable phage subpopulation.
In addition to the unusual selection for structural variants contained in cyclic phage libraries, several other findings indicate that at least part of the selected peptides bind to PMN with high affinity. For instance, the selection of certain motifs in the detergent fractions after low pH treatment implies high affinity binding. We excluded the trivial explanation of internalization by low temperature experiments (4°C) and by the lack of evidence by confocal fluorescence microscopy on permeabilized cells. The specificity of such binding was confirmed by demonstrating that phage binding to PMN could be competitively inhibited by coincubation with relatively low concentrations of synthetic peptides displaying identical or similar sequences. This latter finding is particularly significant since the competing synthetic peptides interact with PMN in a monovalent fashion, whereas the phage binding is likely to occur in a multivalent fashion, and since each phage carries five copies of the modified pIII protein.
While the nature of the PMN receptors for the peptide sequences identified by these phage display library studies are unknown, the results of the calcium signaling experiments indicate that FGPNLTGRW-displaying phage induce a transient intracellular calcium increase that is indistinguishable from those observed with pertussis toxin-sensitive Gαi-coupled receptors. While the seven membrane spanning chemoattractant receptors are classical examples of this class, we have not detected chemotactic activity in experiments with the synthetic peptide FGPNLTGRW. As shown in Fig 8, cell labeling experiments using photoaffinity derivatives of this peptide may be useful in the identification and characterization of the membrane receptor(s).
Analyses of GPNLTGRW, DLXTSK(M/L)X(V/I/L), and CXXGXFLGXWLC using Gen-Bank/EMBL did not reveal any significant homology with known membrane proteins/receptors. This is not surprising since phage may also recognize complex, discontinuous epitopes11 or non-proteinacious determinants, such as sugar groups. Interestingly, several of the analyzed peptide motifs showed homology with various viral coat proteins. In addition, some homology was identified between the DLXTSK(M/L)X(V/I/L) motif and integrins. Specifically, the Asp and Leu residues displayed at the amino terminus of this motif are also present in a number of putative integrin ligand binding sites33 and integrin recognition sequences.27In addition, the TSK motif is present in the 10th type-III repeats of the cell binding region of fibronectin.34 However, the binding of phage bearing consensus-related sequences to PMN is not reduced under conditions that inhibit integrin function such as low temperature and in the absence of divalent cations.
A notable finding in our report was that phage bearing the DLXTSK(M/L)X(V/I/L) motif recognized approximately 50% of the PMN in flow cytometry studies and suggests heterogeneity in peripheral blood PMN. Indeed, there are other reports of MoAbs that label subpopulations of human PMN.35 36 Interestingly, flow cytometry studies with these antibodies showed a labeling pattern indistinguishable from those observed here with DLVTSKLQV-bearing phage. While the protein antigen for one of these antibodies was identified as a 59-kD membrane protein, it remains uncharacterized at the molecular level.
The identification of only a few distinct peptide sequences by scanning whole cells (this report) or even organs7,9 is unexpected. However, important variables such as membrane receptor number and affinity might prevent the selection of weakly binding peptides. Phage particles are relatively large and might be sterically hindered from interacting with potential target molecules on the PMN membrane that are of lower affinity or lower density. Additionally, because of the relatively large size of the phage, higher affinity interactions might tend to “coat” the cell with phage and effectively form a barrier for phage bearing lower affinity peptides. In support of this hypothesis, we found that affinity selections performed in the presence of competing synthetic peptides (1 mmol/L of DLETSKMQV and GGPNLTGRW) resulted in total inhibition of recovery of phage with related sequences. In these experiments, new phage populations were selected that were enriched in tryptophan residues (L.M. and C.A.P., unpublished results, 1996, 1997). Thus, it is likely that additional cell binding sequences can be identified by inclusion of competing peptides derived from sequence information obtained from phage with high binding affinity. Additional factors that might influence the recovery of specific motifs include nonspecific cell binding and selection bias for particular sequence-bearing phage. Other groups have indicated that absorption steps in which the library is incubated with absorber cells may be very effective to prevent the amplification of “undesired” sequences.5 37 Lastly, it is unlikely that the motifs identified here are secondary to nonspecific phage selection due to some particular property of the mutated pIII protein. We tested phage displaying DLVTSKLQV, FGPNLTGRW, and CKDGLFLGWLC motifs for their ability to infect E coli, and differences between these clones and control phage were not found (data not shown).
Phage bearing a specific PMN-binding peptide sequence induce a pertussis toxin-sensitive calcium signal in PMN. (A) As described in Materials and Methods, INDO-1 loaded PMN were suspended for 2 minute at 37°C in a Ca++-containing buffer and cytosolic [Ca++] measured by spectrofluorimetry before and after the addition of 4 × 1010 of phage bearing the sequences indicated. (B) PMN were treated for 2 hours with pertussis toxin or buffer (control), loaded with INDO-1 and treated with FGDNLTGRW-displaying phage as in (A). Positive and negative controls, respectively (not shown), included stimulation with 10−7 mol/L fMLF and immune complexes.
Phage bearing a specific PMN-binding peptide sequence induce a pertussis toxin-sensitive calcium signal in PMN. (A) As described in Materials and Methods, INDO-1 loaded PMN were suspended for 2 minute at 37°C in a Ca++-containing buffer and cytosolic [Ca++] measured by spectrofluorimetry before and after the addition of 4 × 1010 of phage bearing the sequences indicated. (B) PMN were treated for 2 hours with pertussis toxin or buffer (control), loaded with INDO-1 and treated with FGDNLTGRW-displaying phage as in (A). Positive and negative controls, respectively (not shown), included stimulation with 10−7 mol/L fMLF and immune complexes.
In addition to the prominent consensus motifs reported, our affinity selection procedures yielded single clones that were not investigated further but may be relevant. For instance, no matching clones were found for the partially constrained sequence CRNCGWDRPMIF that was recovered from the CL10 library in the acid eluted and cell-associated fraction at 20°C. In this case, the substitution of the Cys residue in the linker sequence at the carboxy terminus with a Phe strongly suggests that the recovering of this phage represents a genuine selection.
In conclusion, we report herein on the identification of neutrophil-specific binding peptides using random phage display techniques. Peptide sequences identified by these techniques provide new information to assist in the identification and characterization of functional surface receptors that are cell-type specific. While the selection of relatively few high affinity peptide ligands is favored with these methods, the number of peptide sequences identified can be considerably expanded through modification of affinity purification procedures, thus increasing the probability of identifying motifs with potential biological and medical importance.
ACKNOWLEDGMENT
Thanks to Claudia Baggi and Lily Hong for expert technical assistance and James Madara for helpful suggestions and critical review of this manuscript.
Supported by grants from the Swiss National Science Foundation, the Swiss Cancer League, and the Bern Cancer League (L.M.) and by grants from the National Institutes of Health—HL54229, HL60540 (C.A.P.), AI26711, and AI22735 (A.J.J.), and a Biomedical Science grant from the Arthritis Foundation (C.A.P.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Charles A. Parkos, MD, PhD, Department of Pathology and Laboratory Medicine, Emory University, Woodruff Memorial Research Bldg, Room 2309, Atlanta, GA 30322.




![Fig. 9. Phage bearing a specific PMN-binding peptide sequence induce a pertussis toxin-sensitive calcium signal in PMN. (A) As described in Materials and Methods, INDO-1 loaded PMN were suspended for 2 minute at 37°C in a Ca++-containing buffer and cytosolic [Ca++] measured by spectrofluorimetry before and after the addition of 4 × 1010 of phage bearing the sequences indicated. (B) PMN were treated for 2 hours with pertussis toxin or buffer (control), loaded with INDO-1 and treated with FGDNLTGRW-displaying phage as in (A). Positive and negative controls, respectively (not shown), included stimulation with 10−7 mol/L fMLF and immune complexes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/5/10.1182_blood.v93.5.1738/4/m_blod40518009x.jpeg?Expires=1767834703&Signature=XFB7H7Sv5RkIhnrTmjFT1SEpSbKxUA73yubmTnjpo~JPrSY74LNsTkHvii1YshN7jWs8fOlntnAtX0gYJ2HunfQt1mVkIGcQzq5REHKSN5Fk81ySBblLbPSLoz3anhL236DfHOPGQL-wqM2Pck9D6YiAldb7sWMZYv-74o0lt3PEucC4tPRqXwep~~MavAT6y1Bo0pJoGaJ5hEDgB3EDas7kxSi951bjbrYKZjsPcrbqbXcBvssT9AjRKHuMO-IUzenwNhQnO~n5XzTRoOU3l7UkPdkQVP8QtYuM3HC1YvjuHdISsJSASKyamfFoSKv-tTDtVGOtwqzLyPdj-tifoA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Fig. 9. Phage bearing a specific PMN-binding peptide sequence induce a pertussis toxin-sensitive calcium signal in PMN. (A) As described in Materials and Methods, INDO-1 loaded PMN were suspended for 2 minute at 37°C in a Ca++-containing buffer and cytosolic [Ca++] measured by spectrofluorimetry before and after the addition of 4 × 1010 of phage bearing the sequences indicated. (B) PMN were treated for 2 hours with pertussis toxin or buffer (control), loaded with INDO-1 and treated with FGDNLTGRW-displaying phage as in (A). Positive and negative controls, respectively (not shown), included stimulation with 10−7 mol/L fMLF and immune complexes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/5/10.1182_blood.v93.5.1738/4/m_blod40518009x.jpeg?Expires=1767932700&Signature=TyGgPcDXuTBImTQCcpT6H1FDQMHKCCN8qKALpLBVJegq~5UC6y4sls93iaqdbp1TtYraYEadsEwLiEIP4jQUB4DoZAl8wz3VXsy-XhZ06qWIZtFVOnlj6gOQOenrzxlsS6XGSzZnZCz~9NY9dQ03-6pfrV4bhySSfalGcP2yULD68Z8t~gx6yrjQB3IVqCT2IJGB1pfbG97W56J6j-Q4iPYsYONYzXa21rmone5N1GKkmUU8RYFh1TA6y5LCGFVtAMyfEIru1VuPNWVDqyfJSet13aJ3vreYmxhSoc~9EZDKNpXq1ZRBnGKkZVXksRiB9MrDtSgOIzFazVOrs23fxA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)