DESPITE ITS well-known sensitivity to anthracyclines,1-5 acute promyelocytic leukemia (APL) was catalogued until the late 1980s as one of the most rapidly fatal human tumors, with the majority of patients dying early of intractable hemorrhage or disease relapse.6 Two major research advances—the cloning of the promyelocytic leukemia/retinoic acid receptor α (PML/RARα) fusion gene underlying the t(15;17) specific aberration7-9 and the advent of differentiation therapy with all-trans retinoic acid (ATRA)10-12—paved the way in the early 1990s for the design of modern diagnostic strategies and tailored treatment of the disease. Together with a better control of the coagulopathy, these advances contributed in the following years to a remarkable improvement in patients’ outcome.13-27 Recent data from large clinical trials conducted in Europe,19-22 the United States,23-25 Japan,26 and China27show that front-line treatments combining ATRA and anthracycline-containing chemotherapy result in long-term survival and potential cure in nearly 70% of patients. In addition to fostering basic and clinical investigation on differentiation therapy, APL may serve today as a paradigm for the development of tailored, genetically targeted treatment of human leukemia.
Among hematologic malignancies, APL is one of the best-characterized genetic-clinical entities. In fact, (1) a unique molecular lesion (the PML/RARα fusion gene) segregates with a single phenotype (hyper- or micro-granular French-American-British [FAB] M3 acute myeloid leukemia [AML]) being only sporadically reported outside this subset13-18; (2) the effectiveness of a specific therapy (ATRA) is strictly conditioned by the presence of this genetic abnormality in leukemic cells, and its detection identifies virtually 100% of ATRA-responsive patients.19,21-23,28 Besides highlighting the pathogenetic role of PML/RARα, recent studies have shown that the same hybrid protein is targeted by ATRA.29-40 In view of these findings, and for the purpose of correct therapeutic intervention, definition of APL requires diagnostic identification of the specific genetic lesion or its protein product.18
Together with conventional karyotyping, reverse transcriptase-polymerase chain reaction (RT-PCR) for PML/RARα is currently the most widely used method for the genetic diagnosis of APL.18,41 Other recently developed and equally specific techniques include fluorescence in situ hybridization (FISH) analysis and PML immunostaining with specific antibodies.42-44Compared with the latter, RT-PCR offers the additional advantage of defining the PML/RARα isoform type and enables sensitive detection of residual disease (MRD) during treatment and follow-up.28,41,45-51 In some instances, the information derived from prospective MRD monitoring of APL has already been translated into operational therapeutic decisions, such as the institution of salvage treatment at the time of PCR-detected minimal disease recurrence.52-54 Nonetheless, this technique is not devoid of drawbacks and difficulties, requiring skillful expertise as well as appropriate inter-laboratory standardization for correct interpretation of results, especially in the setting of multicenter clinical trials.
After a brief update on the role of PML/RARα in APL pathogenesis and response to ATRA, we discuss here the advantages and pitfalls of the main procedures used for genetic diagnosis in this disease, with special emphasis on RT-PCR amplification of the fusion gene. Further, we review the clinical significance of PML/RARα isoforms and the utility of RT-PCR studies to assess response to therapy and MRD monitoring.
MOLECULAR PATHOGENESIS AND RESPONSE TO ATRA IN APL
The APL phenotype is associated with chromosomal translocations disrupting the RARα locus and resulting in fusion products with other nuclear proteins (PML/RARα, PLZF/RARα, NuMA/RARα, or NPM/RARα13-18).
Despite clinical similarity, ATRA induces differentiation of leukemic blasts and disease remission only in PML/RARα APLs, whereas PLZF/RARα APLs are ATRA resistant.13-18 RARα-fusion proteins interfere with the program of terminal differentiation and are probably involved in both disease pathogenesis and response to ATRA.13-18 In particular, (1) in vitro, ectopic expression of PML/RARα, but not that of PLZF/RARα, increases ATRA-sensitivity of hematopoietic cell lines and restores ATRA-sensitivity of resistant cells29,55,56; (2) in vivo, PML/RARα and PLZF/RARα transgenic mice develop leukemias, but only leukemias from PML/RARα mice are ATRA-sensitive.30-33 40
Degradation of PML/RARα by proteaosomal pathways and activated caspases has been proposed as a critical mechanism for ATRA-response of APL blasts.34,35 However, inhibition of the PML/RARα proteolysis does not impair ATRA-dependent differentiation, further indicating a direct role for PML/RARα in this event.36
PML/RARα and PLZF/RARα retain the ability of RARα to regulate transcription of ATRA-target genes and to recruit the N-CoR/histone-deacetylase (HD) complex, which lead to a repressive chromatin conformation.37-40,57-59 High doses of ATRA release HD activity from PML/RARα but not from PLZF/RARα, which contains a second N-CoR/HD binding site in the PLZF moiety. Mutation of the N-CoR binding site abrogates the ability of PML/RARα to block differentiation, whereas inhibition of HD activity switches PLZF/RARα from an inhibitor to an activator of the ATRA-signaling pathway.38 Therefore, recruitment of the N-CoR/HD and regulation of ATRA-target genes are crucial to the transforming potential of RARα-fusion proteins.
RARα fusion proteins’ biological activity may also interfere with cell survival, and this may contribute to their leukemogenic potential. Expression of PML/RARα in hematopoietic cells inhibits programmed cell death, whereas in nonhematopoietic cells it induces apoptosis.29,60,61 Indeed, forced expression of PML in a variety of cell lines induces growth arrest through apoptotic pathways62-65 while targeted disruption of the PML locus in mice increases the rate of spontaneous or induced carcinogenesis.66 The molecular mechanisms by which PML/RARα deregulates PML intracellular pathways are not clear. PML localizes within distinct nuclear compartments (nuclear bodies; NB).67-69 The PML-NB structure is destroyed in APL cells and is restored after ATRA treatment, as a direct consequence of PML/RARα degradation.36 Also, PLZF is a growth suppressor that localizes within nuclear bodies.70 Thus, nuclear bodies could be involved in negative growth control and their deregulation by RARα-fusion proteins might represent a critical step during promyelocytic leukemogenesis.
TECHNICAL APPROACHES FOR GENETIC DIAGNOSIS OF APL
Compared with diagnosis of other AML subtypes, the identification of APL conveys unique therapeutic and prognostic implications. In fact, (1) this leukemia is a medical emergency and up to 10% of early hemorrhagic deaths are currently recorded even in patients receiving modern state-of-the-art treatments19-27; (2) the optimized front-line approach (ATRA + chemotherapy) is different from that used in other AMLs, and is effective also in controlling the life-threatening coagulopathy.13-18 Although morphologic diagnosis is straightforward in the vast majority of hypergranular cases, it appears insufficient for the identification of each and every patient who would benefit from ATRA-containing treatments. In fact, ATRA is effective in leukemia cells expressing the PML/RARα protein, whereas cases with cytological features of APL but bearing variant translocations such as the t(11;17) expressing the PLZF/RARα fusion are ATRA-unresponsive.13-18,71 Vice versa, the APL microgranular variant, which expresses PML-RARα and is ATRA-sensitive,13-18 is, on morphological grounds, hardly distinguishable from AMLs with differentiation (M2) or from myelomonocytic leukemias (M4).72 The APL-specific genetic abnormality can be demonstrated at the chromosome, DNA, RNA, or protein level. We review here the main advantages and pitfalls of these methods, with special emphasis on RT-PCR of PML/RARα.
Cytogenetics and Southern blot.
Conventional karyotyping on banded metaphases enables documentation of the pathognomonic t(15;17) in the majority (up to 90%) of morphologically defined APL cases.13-18 A recent study of patients entered into the Medical Research Council (MRC) ATRA trial showed that the translocation could be cytogenetically documented in 81 of 93 (87%) cases with PCR-detectable PML/RARα transcript.42 Reasons for “false-negative” cytogenetics in APL include analysis of cells not belonging to the neoplastic clone (eg, erythroblasts), which may undergo mitosis in direct or short-term preparations,73 the existence of cryptic translocations or microinsertion,74,75 or difficulties in interpreting poor-quality preparations with few mitoses and/or fuzzy chromosomes.42 In most of these situations, FISH analysis using specific PML and RARα probes would successfully assist diagnosis and demonstrate the genetic rearrangement.74,75 Despite these limitations, conventional karyotype retains an important role and should always complement, and not be a substitute for, molecular diagnostics. In fact, chromosome partners of 17q other than 15q,76-78 complex translocations involving more than two chromosomes,79,80and additional abnormalities besides the t(15;17)81-83are only detectable by cytogenetic studies. Their identification provides potentially relevant clinical and biological information. Of considerable interest, in view of the different therapeutic requirements, is the identification of the rare t(11;17) APL variant.71
Southern blot is equally as specific as RT-PCR but laborious and time-consuming (5 to 10 days). In fact, at least two probes and several enzymatic digestions are required to identify breakpoints on the RARα second intron in all APL cases.84,85 Additional hybridizations with PML probes are needed to detect rearrangements on the 15q+ derivative, to determine the type of PML breakpoint, or to rule out a variant translocation.86 Because of such complexity, the use of Southern blot for routine diagnosis of APL has been almost abandoned in specialized laboratories.
Staining with anti-PML antibodies.
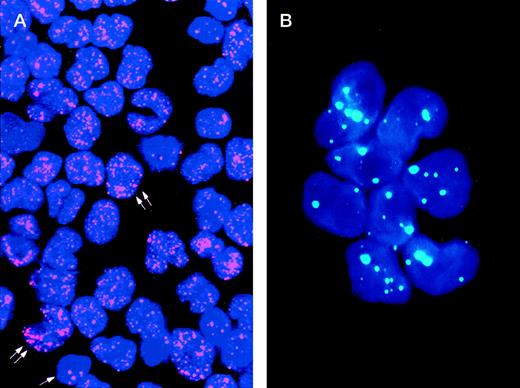
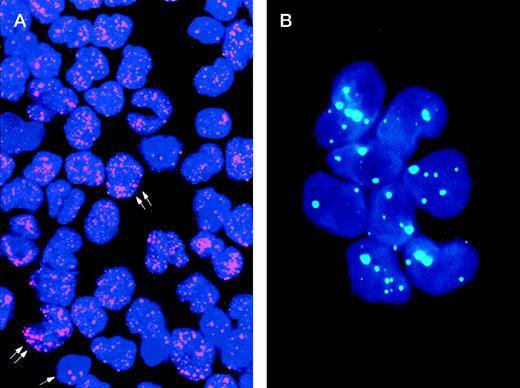
The study of the PML distribution pattern in leukemic cells provides a rapid, specific, low-cost, and relatively simple diagnostic approach.42-44 Different from the wild-type (speckled) staining, which corresponds to the localization of PML into 5 to 20 discrete nuclear particles (so-called “nuclear bodies”), APL cells show a characteristic and easily distinguishable nuclear PML positivity known as “microspeckled,” resulting from the disruption of the nuclear bodies and redistribution of the protein into greater than 50 small granules/per cell (Fig1).42-44 Either immunocytochemistry or immunofluorescence have been successfully used as detection systems. The monoclonal antibody (MoAb) PG-M3,87 directed against an amino-terminal PML epitope that is shared by all the known PML and PML-RARα isoforms, is particularly suitable for diagnostic use.44 In view of its reliability for rapid (2 hours required) and specific diagnosis, immunocytochemistry might be recommended as a valid alternative to molecular or karyotypic analysis.
Immunolabeling of a microgranular APL case (A) and of an acute nonlymphoid leukemia M4 subtype (B) with the PG-M3 MoAb. (A) Indirect immunofluorescence with rhodamine-labeled antibody and nuclear DAPI counterstain. Double arrows show a microspeckled distribution of the PML/RAR protein within the nuclei of APL cells. This pattern contrasts with the nuclear speckled positivity PML wild type of normal residual hematopoietic elements (single arrow). (B) Indirect immunofluorescence with fluoresceine-labeled antibody and nuclear DAPI counterstain. Blast cells show the nuclear speckled positivity.
Immunolabeling of a microgranular APL case (A) and of an acute nonlymphoid leukemia M4 subtype (B) with the PG-M3 MoAb. (A) Indirect immunofluorescence with rhodamine-labeled antibody and nuclear DAPI counterstain. Double arrows show a microspeckled distribution of the PML/RAR protein within the nuclei of APL cells. This pattern contrasts with the nuclear speckled positivity PML wild type of normal residual hematopoietic elements (single arrow). (B) Indirect immunofluorescence with fluoresceine-labeled antibody and nuclear DAPI counterstain. Blast cells show the nuclear speckled positivity.
RT-PCR of PML/RARα.
This is the only technique that defines the PML breakpoint type and that allows the definition of a correct strategy for subsequent MRD monitoring. The advantage of routinely using this assay at diagnosis to better address treatment was initially suggested by Miller et al,28 and subsequently validated in prospective multicenter trials published by the Gruppo Italiano Malattie Ematologiche Maligne dell’Adulto (GIMEMA),19 Medical Research Council (MRC),21 and Programa Español para el Tratamiento de Hemopatias Malignas (PETHEMA)22 groups. According to most investigators, high-quality RNA and efficient RT are the crucial determinants for successful RT-PCR of PML/RARα.88-90 Because of frequent leukopenia and the associated coagulopathy, the yield and quality of RNA from diagnostic samples are frequently poor. It is the experience of the GIMEMA study, now including almost 700 newly diagnosed APL patients, that local preparation of mononuclear cells (Ficoll-Hypaque [Nycomed Pharma AS, Oslo, Norway] isolation and storage in a guanidium-isothiocyanate [GTC] solution) and overnight delivery to a referral laboratory results in better RNA quality, compared with shipment of whole blood.19 For the purpose of rapid diagnosis, small volumes (0.5 mL) of GTC-suspended leukemic cells can be processed with bench microfuges, thereby avoiding the use of expensive equipment and reducing RNA extraction times to less than 1 hour. After RT, a hot-start PCR may be performed at diagnosis with a single amplification round. This procedure allows visualization of the results on the ethidium bromide–stained gel in 3 to 4 hours (including time required for RT).90 By avoiding or minimizing primer misannealing and dimerization, the hot-start method enables a more specific reaction that also results in enhanced sensitivity. Denaturation of RNA before RT and initial denaturation of first-round PCR products have also been recommended by some investigators for improved specificity and sensitivity of the assay.89 Blotting of PCR products and hybridization with specific probes is required if the amplification of nonspecific bands is suspected. This is particularly important in light of the frequent occurrence of PCR artifacts resulting from primer misannealing. As a control for RNA integrity and efficiency of the RT step, some investigators amplify one of the two normal genes involved in the translocation, whereas others prefer the analysis of an unrelated, ubiquitously and low expressed gene.91
In the clinical practice, patients with typical hypergranular APL are usually given specific ATRA-containing therapy before results of the genetic characterization are known. This attitude carries the sole risk of initiating ATRA in those rare cases with hypergranular morphology that lack the PML/RARα recombination. Given the possibility of rapidly identifying APL at the genetic level by anti-PML staining, an anti-PML specific antibody could be included in the characterization panel routinely used in all AMLs to minimize the risk of mistreating patients. This would also avoid the risk of missing those APLs with atypical morphology with no available cytogenetic and molecular characterization.
CLINICAL SIGNIFICANCE OF THE DISTINCT PML/RARα ISOFORMS
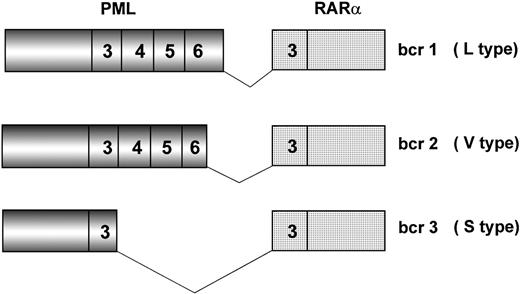
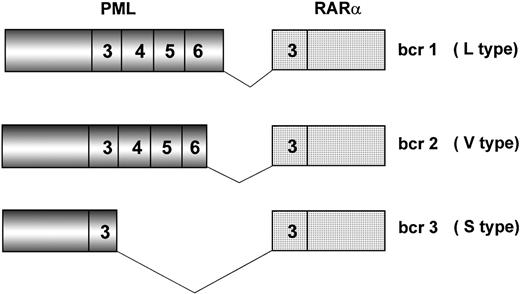
Because of chromosome 15 breakpoint variability (PML intron 6, bcr1; exon 6, bcr2; intron 3, bcr3), two transcripts of different length can be visualized by RT-PCR, referred to as long (L) and short (S). L includes bcr1 and bcr2 or variable (V) transcripts (Fig2), which can only be distinguished by DNA sequence or specific oligonucleotide hybridization. Together, bcr1 and bcr3 transcripts account for more than 90% of cases. Correlations between type of PML/RARα transcript and patients’ clinico-biologic features at diagnosis or response to therapy and outcome have been investigated in many studies.19,42,45,50,82 92-101 The majority of such studies, however, used methodologies that fail to distinguish the L from the V form of PML/RARα; therefore, the impact of including V form patients in the L form group is unknown.
Schematic representation of the three major PML/RAR fusion transcripts. Breakpoints on the RAR gene always occur within intron 2. Due to distinct breakpoints on the PML gene (intron 6, exon 6, and intron 3), different segments of this latter are fused to RAR exon 3, resulting in bcr1 (L type), bcr2 (V type), and bcr3 (S type) fusion transcripts, respectively. Numbers in boxes represent PML and RAR exons.
Schematic representation of the three major PML/RAR fusion transcripts. Breakpoints on the RAR gene always occur within intron 2. Due to distinct breakpoints on the PML gene (intron 6, exon 6, and intron 3), different segments of this latter are fused to RAR exon 3, resulting in bcr1 (L type), bcr2 (V type), and bcr3 (S type) fusion transcripts, respectively. Numbers in boxes represent PML and RAR exons.
With respect to presenting features, no correlations were found with sex, platelet number, presence of the coagulopathy, and ATRA syndrome.19,45,97,99 The bcr3 transcript was reported to correlate with higher white blood cell counts and M3v morphology,99 additional karyotype abnormalities,82 CD34 expression,92 and CD2 expression.92,93 Association with additional karyotype abnormalities and positive staining for CD2 was, however, not confirmed by others.42,94,95 The distribution of PML/RARα isoforms according to age showed a higher frequency of the bcr3 type in children with respect to adults in the Italian population,92 whereas Kane et al100 reported that the V form was the most frequent in a series of the US Pediatric Oncology Group.
The results of studies that analyzed response to treatment and clinical outcome according to the PML/RARα isoform need to be carefully evaluated, taking into consideration relevant differences in the therapeutic context. Shorter remission duration in patients with the bcr3 isoform were reported in two earlier studies in which ATRA alone was used as induction therapy.50,96 In line with these findings, in vitro studies have shown significant differences in retinoid binding and transcriptional activation properties of bcr1 and bcr3 PML/RARα isoforms.102 Subsequent surveys of patients receiving combined ATRA and chemotherapy (in most cases administered simultaneously) showed no statistically significant differences in complete remission and disease-free survival rates.19,99,101 However, it is worth noting that a trend toward less favorable prognosis is reported in all studies for patients with the bcr3 isoform.19,99,101 By analyzing molecular response to treatment in the GIMEMA protocol, we found no difference in breakpoint distribution between patients achieving RT-PCR negativity and those remaining positive after induction.19 Finally, a study of Gallagher et al98 reported a decreased response to ATRA in vitro in patients with a bcr2 (V) PML/RARα transcript.
Taken together, these data indicate that, although a bcr3 transcript type correlates with established adverse prognostic features in APL (ie, hyperleucocytosis, M3v), such association does not appear, at the present time, to translate into poorer outcome, at least in the context of modern ATRA + chemotherapy regimens. However, a long-term follow-up of ongoing clinical trials is awaited to better clarify this issue.
MOLECULAR RESPONSE TO TREATMENT AND MRD MONITORING
The presence of a specific and PCR-detectable tumor marker in leukemic cells allows investigators to molecularly assess response to therapy and MRD in every patient with APL. Many investigators have monitored MRD by RT-PCR during hematologic remission to better adjust treatment.18,103 Overall, there is general agreement that a positive PML/RARα test after consolidation is a strong predictor of subsequent hematologic relapse, whereas repeatedly negative results are associated with long-term survival in the majority of patients (Table1).28,45,52,88,104-113 However, these correlations are not absolute. In fact, either sporadic cases who remain PCR+ in long-term remission114 or, more frequently, patients who ultimately relapse after negative tests,45,52,88,103,107-111 have been reported. Several reasons may account for such exceptions. Most of initial studies were retrospective and heterogeneous in terms of patient selection, treatment, and sensitivity of the PCR assays used.28,45-50In addition, one major limitation of the assays used is their failure to precisely quantitate the amount of MRD, which in turn makes a comparison among the reported studies difficult. Recent prospective analyses on homogeneously treated patients, and with more rigorous methodologies, have further highlighted the prognostic value of RT-PCR monitoring in APL (Table 1).19,52,88,101 107-110 In this section, we will critically review the studies reported on this topic, with special emphasis on the findings of most recent large, prospective trials.
The following parameters are critical for the interpretation of MRD studies: (1) timing of sampling; (2) therapeutic context; (3) fusion gene (PML/RARα or RARα/PML reciprocal hybrid) and control gene of the PCR (actin, β2 microglobulin, PML, RARα, others); (4) sensitivity of the RT-PCR assay; (5) inter-laboratory standardization (particularly relevant in multicenter studies); (6) retrospective or prospective nature of the study; (7) number of tests per each patient. With concern to technical aspects, the recommendations previously discussed for diagnostic RT-PCR are valid and even more important in the setting of MRD studies. False-negative results are a major clinical concern and may result from poor RNA quality or limited sensitivity of the assays used. However, low RT-PCR sensitivity may also depend on properties of the PML/RARα transcript, such as low expression or relative instability.18,41,89 To increase sensitivity of the assay, a number of technical modifications have been suggested (Table 2).18,89,90 As internal PCR controls, dilutions of low-expressed genes (devoid of pseudogenes to rule out amplification of DNA) should be used to mimic the amounts of MRD searched for the specific translocation.91 Finally, continuous efforts should be done toward standardization of the PML/RARα RT-PCR assay to guarantee reproducibility of results among different laboratories. A certain degree of inter-laboratory discordance was documented by a recent external quality-control study involving 12 European institutions.115 The use of the newly developed real-time PCR technology may hold promise to provide adequate standardization at the quantitative level and more objective comparison of results.
With respect to timing of sampling, no differences in the 2-year disease-free survival were found in 139 patients of the GIMEMA trial who tested positive or negative postinduction.19 Similarly, the MRC reported no statistically significant differences in the 3-year relapse risk according to PCR status at the end of induction treatment.101 It is hoped that such early monitoring using quantitative techniques would result in more informative evaluations, thereby allowing investigators to better address further therapy in these patients.
As to the tests performed during consolidation, detection of residual disease after the third course (of four total given) correlated in the MRC study with an increased risk of relapse (62 v 29%,P < .01) and with poorer survival.101 RT-PCR tests during consolidation were performed in too few cases in our study,19 and no conclusion on their utility can be drawn at this time. In both the GIMEMA and MRC trials, patients were studied prospectively and with a PML/RARα RT-PCR assay showing a similar sensitivity threshold (1 positive cell in 104). In the MRC analysis, patients were additionally evaluated for the reciprocal RARα/PML hybrid amplification, which as also found by Tobal et al114 is reported to be more sensitive, with a detection limit of 1 in 105 or even greater. Although this approach did increase the detection rate of residual disease during consolidation, its use failed to identify all patients who ultimately relapsed.101 Furthermore, because RARα/PML is not expressed in a quarter of patients at diagnosis116 and because of occasional detection of residual RARα/PML transcripts in some long-term survivors, its role in molecular monitoring is uncertain at present.
All MRD studies conducted in patients receiving chemotherapy in addition to ATRA reported an exceedingly high proportion (>90%) of RT-PCR− cases at the end of consolidation,19,21,52,101 implying that MRD evaluation at this sampling time fails to identify the majority of patients that will eventually relapse (≈25% to 30%).19,21,52,101 Increased sensitivity of the PML/RARα assay might lead to better identification of this sizable fraction of patients who require additional therapy at the end of consolidation. However, improved sensitivity might carry the risk of detecting “indolent” transcript amounts, such as those occasionally reported in long-term survivors.18 114
Although assessment of the PCR status immediately after the end of consolidation has little prognostic role, subsequent monitoring during the early posttreatment period appears informative. We recently reported the results of a prospective study in which 163 patients of the GIMEMA trial were tested at regular preestablished time intervals after the end of treatment.52 Twenty of 21 patients who converted to PCR+ relapsed within a median time of 3 months, whereas the 3-year estimate of relapse risk for patients who tested negative more than two times postconsolidation was below 10%. In fact, only 8 of 142 who tested negative in at least two successive samples underwent relapse after a median follow-up of 18 months postconsolidation. Interestingly, 81% of the PCR+conversions were recorded within the first 6 months after the end of therapy.52 It is based on these findings that, in our trial, patients in clinical remission who convert to RT-PCR+, as confirmed in two successive BM samplings, are now administered early salvage treatment, before developing overt relapse.53
In summary, RT-PCR of PML/RARα appears less sensitive than the assays used to amplify other leukemia-associated hybrid genes; however, the assessment of remission status at the molecular level represents a significant clinical advance with respect to other poorly sensitive methods (morphology, karyotype). It is commonly accepted that the efficacy of present strategies or novel approaches for the treatment of this disease (arsenicals, liposomal ATRA, other retinoid derivatives, etc) need to be assessed taking into account the response at the RT-PCR level. However, repeated posttreatment sampling appears necessary to obtain prognostically relevant information. A status of persistent PCR-negativity confirmed in at least two successive marrow samples at the end of treatment might be regarded as our best presently available therapeutic goal. While further standardization of laboratory tests and automated quantitation (real-time PCR) will probably improve the current state-of-the-art and allow us to better identify distinct risk groups at the end of treatment, prospective monitoring during the first 6 to 12 months is recommended for early detection of relapse.
CONCLUSIONS AND FUTURE PERSPECTIVES
Elucidation of the genetic defect underlying the disease-specific t(15;17) has had a remarkable impact into the management of APL. However, while the relevance of diagnostic PML/RARα detection is out of question, further investigation is needed to better establish the clinical value of molecular monitoring. In particular, the following have to be clarified: (1) whether a greater sensitivity of the RT-PCR assay for PML/RARα will allow better identification of patients at risk of relapse at the end of consolidation; (2) if newer quantitative technologies may provide earlier monitoring that is clinically significant; and (3) whether anticipation of salvage therapy in patients treated for molecular recurrence is advantageous over the treatment of hematologic relapse. As to this latter issue, studies are underway,53 but it may be anticipated that therapy of molecular relapse should at least minimize the significant mortality rate observed during reinduction of overt disease.
The success of tailored treatment of APL encourages both basic and clinical investigators to search for more rational, and, hence, less toxic drugs that directly interfere with the function of the leukemia-associated fusion proteins (molecular treatment). Like PML/RARα and PLZF/RARα, AML1/ETO, the transforming protein associated with M2-AMLs, has been recently shown to function as a transcriptional repressor in a complex containing nuclear corepressors and histone-deacetylases.117,118 Also in this case, the nuclear complex formation is crucial to the biological activities of the fusion protein. Translocations involving the histone-deacetylases p300 and CBP have been recently described in rare cases of AML.119 Thus, formation of aberrant complexes with histone-modifying enzymes and alterations in chromatin structure might be a general mechanism of myeloid leukemogenesis. In vitro, drugs with inhibitory activity on histone-deacetylases have already been shown to interfere with the APL phenotype.38-40 It is tempting to speculate that the use in AMLs of drugs that modulate histone acetylation might, in the near future, represent another important example of molecular treatment.
ACKNOWLEDGMENT
We are grateful to Profs F. Mandelli and M.A. Sanz, and to Drs D. Grimwade, G. Avvisati, W. Arcese, and G. Cimino for helpful discussion and critical reading of the manuscript. We also thank Prof A.K. Burnett and Dr D. Grimwade for sharing their unpublished data. Finally, we are indebted to all the clinical and laboratory investigators of the GIMEMA and Associazione Italiana di Ematologia e Oncologia Pediatrica (AIEOP) groups for their invaluable contribution to the Italian cooperative study “AIDA” for the diagnosis and treatment of patients with acute promyelocytic leukemia.
Supported by AIL (Associazione Italiana contro le Leucemie), AIRC (Associazione Italiana per la Ricerca sul Cancro), CNR P.F. “Biotecnologie,” Italia-Usa Project on Therapy of tumors, and Fondazione Tettamanti.
REFERENCES
Author notes
Address reprint requests to Francesco Lo Coco, MD, Department of Cellular Biotechnology and Hematology, University La Sapienza of Roma, Via Benevento 6, 00161 Roma, Italy; e-mail:lococo@bce.med.uniroma1.it.