Abstract
The occurrence of protein C inhibitor (PCI) in human platelets and megakaryocytes was analyzed. As judged from enzyme-linked immunosorbent assays (ELISAs), PCI was present in platelets at a concentration of 160 ng/2 × 109 cells. Its specific activity was 5 times higher than that of plasma PCI. Consistently, mainly the 57-kD form (active PCI) and some high molecular weight (Mr) forms, but no bands corresponding to cleaved PCI, were detected when platelet lysates were immunoprecipitated with monoclonal anti-PCI-IgG and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. The localization of PCI in platelets was studied by immunofluorescence histochemistry and immunotransmission electron microscopy: PCI was detected in granules, in the open canalicular system, and on the plasma membrane. At these sites, colocalization with plasminogen activator inhibitor-1 was seen. Studies were performed to clarify whether platelet PCI is endogenously synthesized or taken up from plasma. Internalization of biotinylated-PCI was analyzed using platelets in suspension and gold-labeled streptavidin for visualization of incorporated biotin. Dose- and time-dependent uptake of PCI was found. PCI mRNA was detected in platelets by reverse transcriptase-polymerase chain reaction (RT-PCR) and Southern blotting, as well as in megakaryocytes by in situ hybridization of human bone marrow cryosections. We therefore conclude that platelets contain a functionally active PCI pool that is derived from both endogenous synthesis as well as internalization.
IT IS GENERALLY ACCEPTED that cell surfaces are essential for blood clotting reactions to take place. Platelets provide a surface that allows coagulation factors to align in a way to optimize their specific interactions (for reviews see Bloom et al1). Many of these interactions are performed by specific proteins present on the surface of platelets. Protein reactions that implicate the platelets in the process of hemostasis are those of adhesion to the cut end of a blood vessel, spreading of the adherent platelets on the exposed subendothelial surface, secretion of platelet constituents, formation of a mass of aggregated platelets, and acceleration of plasma coagulation, resulting in the formation of a fibrin clot. Subsequently, the formed fibrin clot retracts to a smaller volume, a process that is also platelet-dependent. Three principal effects result from platelet activation: secretion of the contents of intracellular granules, exposure of latent surface receptors for plasma proteins, and alterations in lipid structure of the platelet surface membrane, leading to acceleration of plasma coagulation. Platelets contain a variety of granules in which intracellular substances are selectively sequestered, including dense bodies, α granules, and lysosomes.1-3 It has been shown that not all of the proteins present in the α granules are derived from endogenous synthesis.4-6 Megakaryocytes, the precursors of platelets, and platelets themselves, incorporate proteins (eg, fibrinogen and albumin) from the surrounding medium and fuse them into α granules.2,5 7-10 Thus, platelets appear to contain a unique type of secretory granules, with contents originating both from endogenous synthesis in megakaryocytes and from endocytosis by megakaryocytes and platelets.
The protein C system is an important anticoagulant regulator of blood coagulation.11,12 The complex thrombin-thrombomodulin bound to the surface of the endothelial cells activates protein C by limited proteolysis, leading to the formation of activated protein C (APC), a serine protease. APC binds to protein S, which acts as receptor and cofactor for APC on the membrane of platelets and endothelial cells. The APC-protein S complex on the cell membrane inactivates coagulation cofactor proteins, factors Va and VIIIa, by limited proteolysis.1
The presence in plasma of an inhibitor of APC was first described by Marlar and Griffin in 1980.13 Subsequently, this inhibitor was purified from human plasma and characterized by Suzuki et al.14,15 Protein C inhibitor (PCI) is a single-chain polypeptide with a molecular weight (Mr) of 57,000. PCI is thought to be the primary regulator of the protein C system, although α2-macroglobulin and α1-antitrypsin also appear to play a role when large amounts of APC are generated in pathological settings.16-23Comparing the inhibition rate constants, PCI is a better inactivator of APC than α1-antitrypsin or α2-macroglobulin, but these 2 inhibitors are present in plasma at much higher concentrations than PCI. Furthermore, baboon models suggest that, in vivo, PCI is the preferred APC inhibitor until its concentration becomes limiting, and then α1-antitrypsin becomes the predominant inhibitor.24 However, there is no direct evidence for a role of PCI in APC regulation, because patients deficient in the protein have not been described. In addition to APC, PCI also inhibits thrombin and several other procoagulant and profibrinolytic proteinases, such as factor Xa, factor XIa, urokinase plasminogen activator (u-PA), tissue plasminogen activator (t-PA), and plasma kallikrein.25-27 Tissue kallikrein is also inhibited by PCI as well as acrosin and prostate-specific antigen.28-31 The inhibition of each enzyme is accompanied by formation of an enzyme-inhibitor complex and by the cleavage of the reactive site peptide bond of PCI. PCI is found in plasma at a concentration of 3.6 to 6.8 μg/mL in normal individuals.19 PCI is also present in urine25and in several other body fluids (eg, tears, saliva, milk, cerebrospinal fluid, synovial fluid, amniotic fluid, and Graaf follicular fluid).32 The highest concentrations have been described in seminal plasma (200 μg/mL).32 PCI synthesis has been shown in liver cells, in organs of the male and female reproductive tracts, in the pancreas, and in tubular cells of the kidney.32,33 PCI belongs to the subgroup of heparin-binding serpins, and heparin and other glycosaminoglycans can modulate the inhibitory activity of PCI.14,15,25,26 34
The presence in human platelets of an inhibitor of APC was described in 1989; this inhibitor, if released, interfered with the ability of phospholipid and washed platelet membranes to catalyze the anticoagulant effects of APC.35 It has also been shown that platelet releases significantly reduced the activated partial thromboplastin time (aPTT) in the presence of APC.36Furthermore, platelets contain substances capable of binding PCI (as, for example, fibrinogen),37 of being inhibited by PCI (as tPA or, after platelet activation, thrombin, APC, factor Xa, factor XIa, and plasma kallikrein), or of regulating the PCI activity.14,26,27 38 We therefore analyzed the possibility of whether PCI might be present in platelets. In this report, we can show that platelets in fact contain PCI, which is functionally active and which is derived from endogenous synthesis as well as from endocytotic uptake.
MATERIALS AND METHODS
Aprotinin (Trasylol) was obtained from Bayer-Austria (Vienna, Austria). Apyrase, benzamidine, prostaglandin E1, and heparin sodium salt from porcine intestinal mucosa were obtained from Sigma-Aldrich Chemicals (Vienna, Austria). Purified urinary PCI, rabbit-anti-PCI-IgG, peroxidase-conjugated rabbit-anti-PCI-IgG, and monoclonal-anti-PCI-IgG (4PCI) were prepared as described previously.30,31 39 The protein concentration of purified PCI was determined by amino acid analysis performed by Toplab (Munich, Germany). Biotinylated PCI was prepared using biotinyl-ε-amino caproic-N-hydroxysuccinimide ester from Boehringer Mannheim (Vienna, Austria), following the manufacturer’s instructions. The efficiency of the labeling as well as the inhibitory activity of the biotin-labeled PCI were controlled.
Preparation of platelet extracts.
Fresh venous blood from 5 healthy volunteers was collected on acid-citrate-dextrose solution (ACD; 130 mmol/L citric acid, 124 mmol/L trisodium citrate, 110 mmol/L dextrose; pH 6.5) at a ratio of 9 parts blood to 1 part anticoagulant. Platelet-rich plasma (PRP) was obtained by centrifugation at 120g for 10 minutes at 26°C. Seven parts PRP were diluted with 1 part calcium-free Tyrode’s buffer (137 mmol/L NaCl, 3 mmol/L KCl, 0.4 mmol/L NaH2PO4, 1 mmol/L MgCl2, 14 mmol/L NaHCO3, 5.5 mmol/L dextrose, 10 mmol/L HEPES; pH 7.4) containing 100 KIU/mL aprotinin, 2.5 U/mL apyrase, 10 mmol/L benzamidine, and 29 mg/L soybean-trypsin inhibitor and were further centrifuged at 1,060g for 10 minutes at 26°C to obtain a platelet pellet. Subsequently, platelets were washed 5 times and pelleted by centrifugation at 2,040g for 15 minutes at 26°C. The pellet containing washed human platelets was finally resuspended in modified calcium-free Tyrode’s buffer, and the platelet concentration was adjusted to 2 × 109cells/mL as determined in a standard hemacytometer. The isolation procedure was always completed within 2 hours.
To prepare platelet lysates, washed platelets (2 × 109 cells/mL) were centrifuged and resuspended in phosphate-buffered saline (PBS; 10 mmol/L phosphate buffer and 140 mmol/L NaCl; pH 7.4) containing 1% Triton X-100, 100 KIU/mL aprotinin, 10 mmol/L benzamidine, and 25 μg/mL soybean trypsin inhibitor. The sample was incubated in this buffer for 10 minutes with frequent vortexing and was then centrifuged at 2,000g for 15 minutes to remove big membrane fragments. The supernatant obtained was stored at −70°C until further use.
Quantification of total PCI antigen.
Total PCI antigen in platelet lysates and in a citrated plasma pool was determined by enzyme-linked immunosorbent assay (ELISA) as described previously,29 except that the microtiter plates were coated with 20 μg/mL 4PCI instead of 10 μg/mL.
Quantification of active PCI antigen.
A functional ELISA was developed to determine active PCI antigen based on its ability to bind u-PA. 4PCI, which does not interfere with PCI activity, was used as a catching antibody. It was acid treated as described,29 diluted in coating buffer (5.6 mmol/L Na2CO3, 35 mmol/L NaHCO3, 0.01% thimerosal, pH 9.6) to yield a concentration of 20 μg/mL, and incubated overnight at 4°C in wells of a microtiter plate. Remaining binding sites were blocked for 1 hour at 37°C with PBS-1% bovine serum albumin (BSA). After blocking and washing with PBS-0.5% Tween 20, dilutions of platelet lysates in PBS-1% BSA were pipetted into the wells and incubated for 150 minutes at 37°C. Dilutions of citrated pooled plasma consisting of equal volumes of plasma obtained from 20 healthy donors (10 men and 10 women; age range, 20 to 50 years) were used as standard. After washing again, u-PA (Technoclone, Vienna, Austria) was added at a concentration of 80 U/mL in PBS-1% BSA and incubated for 1 hour at 37°C. Thereafter, wells were washed and incubated with peroxidase-conjugated monoclonal anti-u-PA-IgG (Scu-PA-1; Technoclone) at a 1:200 dilution in PBS-1% BSA. After 1 hour at 37°C, the plates were washed and then incubated with ABTS substrate for 30 minutes at room temperature. The substrate reaction was stopped with 0.32% sodium fluoride, and absorbances were immediately measured at 405 nm/492 nm. The coefficients of variation for this functional PCI ELISA were calculated from duplicate determinations of 3 dilutions of 30 different plasma samples. The intra-assay coefficient of variation was 1.8%. The corresponding inter-assay coefficient of variation was 8.6%.
Quantification of total PAI-1 antigen.
PAI-1 antigen concentration in platelet extracts was determined using an ELISA kit from Technoclone. This ELISA measures free, complexed, and latent PAI-1 and is not affected by other plasminogen activator inhibitors.
Immunoprecipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blotting.
Immunoprecipitation of platelet lysates was performed in wells of a 96-well Nunc Immuno Maxisorb plate (Nunc, Roskilde, Denmark). The wells were coated overnight at 4°C with acid-treated 4PCI (20 μg/mL). The remaining binding sites were blocked and platelet lysates were added to the wells and incubated for 3 hours at 37°C. After washing (PBS-0.5% Tween 20), bound proteins were separated from the plate with 20 μL of 20% SDS/0.05% Bromphenol blue/glycerol (1:1). Samples were then removed and incubated for 5 minutes at 90°C. SDS-PAGE (10% acrylamide) was performed using a Mini-PROTEAN II electrophoresis cell from Bio-Rad (Vienna, Austria). Twenty microliters of immunoprecipitated samples was loaded to the wells and run at a constant current of 50 mA for 90 minutes with a cooling system. Two identical gels were run, and 1 of them was used as control. Purified PCI (2.5 μL) was also electrophoresed in each gel. Separated proteins were electrotransferred (75 minutes at 100 V) to Hybond–P polyvinylidene diflouride (PVDF) membranes (Amersham Life Sciences, Vienna, Austria) using a Mini-Trans-Blot Electrophoretic Transfer Cell from Bio-Rad. After protein transfer, the PVDF membranes were blocked with 1.5% nonfat dried milk diluted in blotting buffer (20 mmol/L Tris-HCl, pH 7.4, 500 mmol/L NaCl) and incubated overnight with or without (control) rabbit-anti-PCI-IgG (15 μg/mL) in blocking buffer. Thereafter, membranes were incubated for 1 hour with anti-rabbit-IgG biotinylated species-specific whole antibody from donkey (Amersham Life Sciences) at a 1:500 dilution in blocking buffer, followed by streptavidin-biotinylated horseradish peroxidase complex (Amersham Life Sciences) at a 1:500 dilution in blocking buffer. Finally, bound peroxidase was visualized with TMB substrate color reagent (3, 3′, 5, 5′,-tetramethylbenzimide).
Immunofluorescence microscopy.
Platelets were isolated from whole blood as previously described, except that the calcium-free-Tyrode’s buffer was enriched with apyrase and prostaglandin E1 but not with the other inhibitors. The platelets obtained were extensively washed and thereafter resuspended in a small volume of Tyrode’s buffer. Smears of the washed platelets were performed on poly-L-lysine-coated glass slides and allowed to dry for 3 hours. Afterwards, platelets were permeabilized in cold acetone for 1 minute. Unspecific reactive sites were blocked with PBS-5% BSA; subsequently, the sections were incubated overnight at 4°C with the first antibody diluted in PBS-1% BSA. Monoclonal anti-PCI-IgG (4PCI) at a concentration of 20 μg/mL was used as a first antibody. Staining specificity was assessed in negative control experiments performed by substitution of the first antibody with dilution buffer alone, by incubation with mouse-nonimmune-IgG, and by incubation with a monoclonal antibody against the nonsense protein heparin cofactor II. Staining specificity was also assessed in positive control experiments performed by incubation with a monoclonal antibody against PAI-1 (5PAI-12), a protein known to be present intracellularly in platelets. Additionally platelets were stained with a monoclonal antibody against factor D, which was found to be localized on the membrane of the platelets but not intracellularly.
Thereafter, the platelet smears were incubated with fluorescein-conjugated goat-antimouse-IgG (Vector Laboratories Inc, Burlingame, CA) diluted 1:100 in PBS-1% BSA for 1 hour at 37°C. After incubation and washing (PBS), a drop of Vectashield Mounting Medium (Vector Laboratories Inc) was placed onto the section and then coverslipped. Slides were viewed using a Leitz Dialux 22/22 EB microscope (Ernst Leitz Wetsler Gmbh, Lietzlow, Germany) equipped with a Nikon super high pressure mercury lamp (Nikon Corp, Tokyo, Japan) and a power supply model HB-10101 AF (Nikon Corp). Photographs were taken with a Leitz Vario-Orthomax 2 camera, at a magnification of 3,000 and an exposure time of 3 seconds.
Immunotransmission electron microscopy.
Platelets were isolated from whole blood as described above. After extensively washing of the platelet pellet obtained, the platelets were fixed with 4% paraformaldehyde/0.5% glutaraldehyde for 20 minutes at room temperature. In platelet stimulation experiments, platelets were incubated for 30 minutes at 37°C in a water bath and then activated with 1 μmol/L calcium ionophore A23187 under gentle agitation. After this activation, the platelets were fixed and further processed as described below. Human bone marrow cells obtained by iliac puncture from a healthy donor were sedimented by centrifugation at 2,000g for 10 minutes washed and fixed in 4% (wt/vol) paraformaldehyde for 20 minutes at room temperature.
Before embedding in LR White medium resin, washed platelets and bone marrow pellets were fixed again for 1 hour in 2.5% glutaraldehyde in 0.1 mol/L cacodylate buffer, pH 7.4, and then rinsed in 0.1 mol/L phosphate buffer, pH 7.4. Ultrathin serial sections of 150 nm were cut and mounted on gold grids. Nonspecific binding sites on the sections were blocked (PBS-5% BSA) and grids were incubated for 3 hours at 37°C with different concentrations (500, 250, 125, 75, 35, and 20 μg/mL) of 4PCI in PBS-1% BSA. In negative control experiments, the sections were incubated with nonimmune-mouse-IgG. After incubation with the first antibody and washing, platelet and bone marrow sections were incubated with 10 nm gold-labeled antimouse-IgG (Sigma-Aldrich) at a dilution of 1:50 in PBS-1% BSA. In double-labeling experiments, sections were incubated with a mixture of 75 μg/mL 4PCI and 75 μg/mL rabbit-anti-PAI-1-IgG (final concentrations) in PBS-1% BSA. In control experiments for double-labeling, incubations were performed in a mixture of 4PCI and rabbit-nonimmune-IgG or in rabbit-anti-PAI-1-IgG and mouse-nonimmune-IgG. After washing, grids were incubated in biotinylated-antimouse-IgG (final dilution, 1:200 in PBS-1% BSA) and goat-antirabbit-IgG coupled with 5 nm colloidal gold particles (final dilution, 1:100; Amersham Life Sciences). Further incubation was performed with streptavidin-gold 10 nm conjugate diluted 1:100 in PBS-1% BSA. The sections were then counterstained with 3% uranyl acetate and lead citrate and examined with a JEM 1200 EXII transmission electron microscope (JEOL Ltd, Tokyo, Japan). Photographs were taken on AGFA Scientia EM films (AGFA-Gevaert Ag, Leverkusen, Germany) at magnifications between 40,000 and 120,000.
Uptake of PCI by platelets.
Platelets were isolated as described above, with the only modification being that 1 μg/mL prostaglandin E1 was added to the washing buffer. The washed platelets were adjusted to a concentration of 1.6 × 109 cells/mL and incubated for 30 minutes at 37°C before starting this experiment. Afterwards, 200 μL of the platelet suspension was incubated at room temperature with 200 μL of biotinylated PCI (4.5 or 9 μg/mL, respectively). The uptake reaction was stopped after 2 different incubation times (7 and 15 minutes, respectively) by adding 5 parts of 4% paraformaldehyde/0.2% glutaraldehyde in calcium-free Tyrode’s buffer to 1 part of platelet/PCI solution. Cells were fixed for 20 minutes at room temperature, washed, and further processed for immuno-electron microscopy as described above. Sections of these platelets on gold grids were blocked and incubated for 3 hours at 37°C with 10 nm gold-labeled streptavidin diluted 1:25 in PBS-1% BSA. After this incubation, the grids were washed, counterstained, and examined with a transmission electron microscope.
Reverse transcriptase-polymerase chain reaction (RT-PCR) and Southern blotting.
Human blood was collected from healthy donors on ACD anticoagulant solution as described above. The PRP was diluted (7:1) with calcium-free-Tyrode’s buffer containing 2.5 U/mL apyrase, 100 KIU/mL aprotinin, and 10 mmol/L benzamidine and gel-filtered platelets were prepared by filtering this PRP through a 40-mL Sepharose CL-2B chromatography column (Pharmacia Biotech, Vienna, Austria). Fractions (1.5 mL) were collected and aliquots of each fraction were smeared on poly-L-lysine–coated slides and stained with Accutain (Sigma Diagnostics, Vienna, Austria). The slides were then checked by light microscopy. Fractions containing only platelets and no other blood cells were pooled, and platelets were washed 5 times and adjusted to 1 × 109 cells/mL.
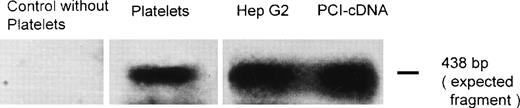
Total RNA from gel-filtered platelets and HepG2 hepatoma cells was isolated using RNeasy Mini Kit (QIAGEN, Vienna, Austria), and 5 μg each was reverse transcribed by using a sequence-specific primer and the first-strand cDNA Synthesis Kit for RT-PCR from Boehringer Mannheim. The RT-generated PCI cDNA fragments (1,157 bp) were amplified by PCR. As a control for this reaction, PCI-cDNA inserted into the plasmid vector pBluescript II KS (+/−) phagemid (Stratagene, La Jolla, CA) was also amplified. The following primers were used for amplification: 5′-primer (the binding site is located in exon III: from base 10727 to base 10748) TCAGTATCACTACCTCCTGGAC and 3′-primer (binding site in exon V: from base 12705 to base 12726) CTGTTGAACACTAGCCTCTGAG. The human PCI sequence data were obtained from EMBL Data Bank (accession nos. M64880through M64884). The design of both primers was performed in such a way that it allowed the differentiation between the product obtained from the reverse-transcribed platelet RNA (438 bp) and genomic DNA contaminations (1,978 bp). PCR amplification using 5 μL of the cDNA from the RT reaction was performed in 5 μL of 10× reaction buffer, 1.5 mmol/L MgCl2, 200 μmol/L deoxynucleotides, 10 pmol/L primers, and 2.5 U Taq polymerase in a final volume of 25 μL. The samples were subjected to amplification in a GeneAmp PCR System 2400 (Perkin Elmer Cetus Instruments, Emeryville, CA) to 94°C for 5 minutes; then 30 cycles of amplification were performed, each consisting of denaturation at 94°C for 1 minute, annealing at 61°C for 1 minute, and extension at 72°C for 1 minute, and then a final incubation at 72°C for 7 minutes was performed. Amplified products were separated in 1% agarose TBE gels and stained with ethidium bromide. After electrophoresis, the gel was transferred overnight to a Duralon-UV membrane (Stratagene). A PCI-cDNA fragment of 982 bp (from base 7994 to base 11344 in the above-described sequence) was 32P-labeled using Random Primed DNA Labeling Kit (Boehringer Mannheim) and used as a probe for the blotting. The nylon-membrane was equilibrated in Church’s buffer (0.5 mol/L Na2HPO4, pH 7.2, 7% SDS, 0.5 mol/L EDTA, pH 8) for 1 hour at 62°C and then incubated overnight at 62°C with 0.7 mL of denatured 32P-DNA probe in 6.3 mL of Church’s buffer. Thereafter, the membrane was extensively washed and subjected to autoradiography overnight at −70°C using a Kodak X-OMAT Scientific Imaging Film (Eastman Kodak, Rochester, NY).
In situ hybridization.
A PCI-cDNA fragment (593 bp; from base 7994 in exon II to base 10645 in exon III) was in vitro transcribed using the pCR II vector (Invitrogen, San Diego, CA) that contains the bacteriophage SP6 and T7 polymerase promoters. The ligation reaction for the PCI-cDNA fragment into the plasmid vector was performed by using the Rapid DNA Ligation Kit from Boehringer Mannheim. After transformation and cloning, the DNA template was linearized with HindIII (2.9 U) and in vitro transcribed with T7 RNA polymerase to obtain the sense RNA probe (655 bp). The antisense RNA probe (613 bp) was obtained by linearizing withEcoRI (14.15 U) and in vitro transcription with the SP6 RNA polymerase. The transcription procedure was performed by using the digoxigenin (DIG) RNA Labeling Kit obtained from Boehringer Mannheim. The human bone marrow samples to be used for in situ hybridization procedures were immediately embedded in Tissue Tek and snap-frozen in liquid nitrogen. Sections of these samples were allowed to settle on poly-L-lysine–coated glass slides and in situ hybridization was performed as described in the protocols for in situ hybridization to tissue sections, obtained from Boehringer Mannheim, which were based on published data.40 Cryosections were incubated with DIG-labeled antisense or sense PCI cRNA probes, which were further detected with anti-DIG-alkaline phosphatase Fab fragments, followed by NBT/BCIP substrate detection. Controls were always included to ensure the specificity of the detected signals. These controls were made in the target (hybridization with sense probe) as well as in the detection (omission of anti-DIG antibody).
RESULTS
Total PCI antigen and active PCI antigen in platelet lysates.
Platelet lysates and plasma samples were analyzed for total PCI antigen and active PCI antigen by ELISAs. The total PCI antigen concentration in platelet lysates (2 × 109 platelets/mL) was 160 ± 0.03 ng/mL (mean ± SD), as determined in 5 measurements performed in 2 independent experiments. The PCI plasma concentration measured with the same assay was 5.8 ± 0.02 μg/mL. Active PCI antigen in platelet lysates was 14% ± 2% (mean ± SD) of that measured in the citrated pooled plasma (100%) used as standard. This mean was calculated from duplicate measurements of 7 different dilutions. Therefore, the specific activity of PCI in platelet lysates was 5-fold higher as compared with the specific activity of PCI present in plasma. As a control, PAI-1 antigen was also determined in these platelet lysates, and the results obtained agree with published data41-43: 1,639 ± 23 ng/mL (mean ± SD) or 0.82 ng/106 platelets. We therefore assumed that the platelet lysates were in a condition to allow further reliable experiments.
Immunoprecipitation, SDS-PAGE, and Western blotting.
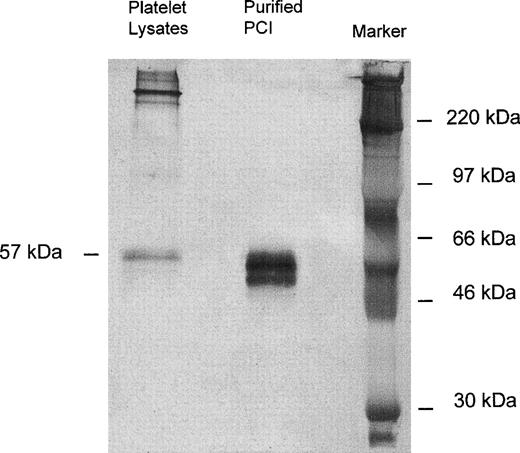
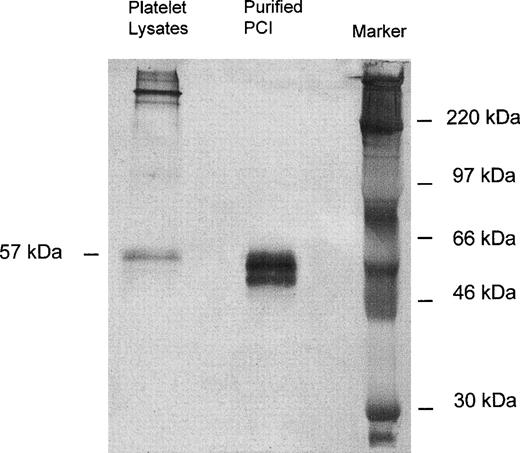
Immunoprecipitation of platelet lysates was performed with monoclonal-anti-PCI-IgG (4PCI), and immunoprecipitates were analyzed by SDS-PAGE and Western blotting with rabbit-anti-PCI-IgG. As can be seen from Fig 1, purified urinary PCI used as a control exhibited the typical pattern of closely spaced bands with molecular weights between 60 and 54 kD,14,15 44 with the most intense bands corresponding to active PCI (57 kD) and reactive site-cleaved PCI (54 kD), respectively. In platelet lysates there was also a prominent band at 57 kD, corresponding to the migration distance of active PCI (Fig 1). However, bands corresponding to cleaved PCI were hardly seen. In addition, platelet lysates contained several high molecular weight bands of PCI antigen (>220 kD).
Immunoprecipitation, SDS-PAGE, and Western blot of platelet lysates. Solubilized proteins contained in platelet lysates were subjected to immunoprecipitation with monoclonal anti-PCI-IgG as described in Materials and Methods. Precipitates were analyzed by SDS-PAGE (10% acrylamide) and immunoblotting using rabbit-anti-PCI-IgG. Bound antigen was detected with biotinylated-antirabbit-IgG followed by streptavidin-peroxidase and with TMB color substrate reagent. Purified urinary PCI was used as a control. In platelet lysates, only the 57-kD band corresponding to the active PCI was observed; bands corresponding to cleaved PCI were hardly seen.
Immunoprecipitation, SDS-PAGE, and Western blot of platelet lysates. Solubilized proteins contained in platelet lysates were subjected to immunoprecipitation with monoclonal anti-PCI-IgG as described in Materials and Methods. Precipitates were analyzed by SDS-PAGE (10% acrylamide) and immunoblotting using rabbit-anti-PCI-IgG. Bound antigen was detected with biotinylated-antirabbit-IgG followed by streptavidin-peroxidase and with TMB color substrate reagent. Purified urinary PCI was used as a control. In platelet lysates, only the 57-kD band corresponding to the active PCI was observed; bands corresponding to cleaved PCI were hardly seen.
Immunofluorescence microscopy.
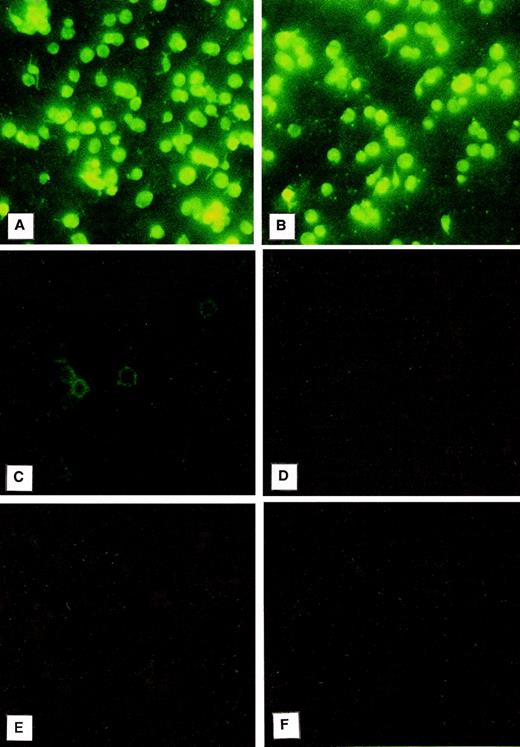
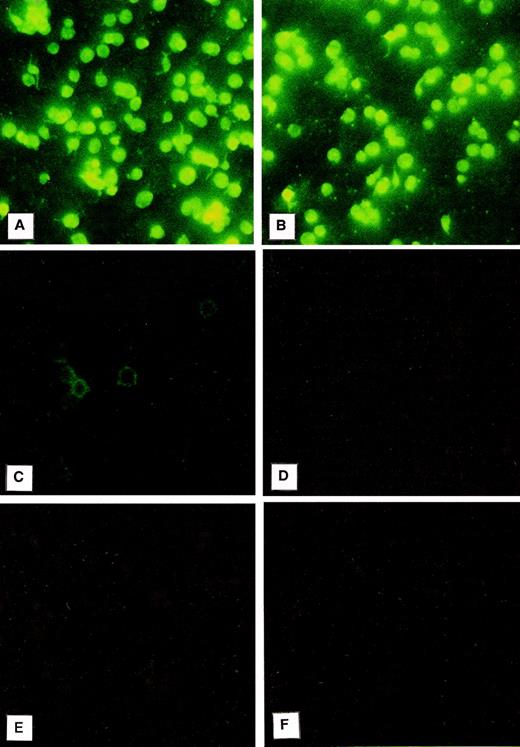
Immunofluorescence was used to evaluate the localization of PCI in platelets (Fig 2). For this purpose, the platelet smears were incubated with a monoclonal antibody to PCI (4PCI). The PCI staining pattern observed (Fig 2A) looked very similar to that of PAI-1 (Fig 2B), which is known to be present in the α granules and which was therefore chosen as an intracellular positive control for this experiment.41-43 The platelets incubated with 4PCI exhibited a fluorescent homogeneous pattern as well as a spotted staining pattern consistent with intracellular localization of the protein (Fig 2A). This pattern was different from that obtained with a monoclonal antibody to factor D, which showed a rim pattern of staining, characteristic for exclusive surface and not for intracellular localization (Fig 2C). No staining was seen in control experiments using antibodies against heparin cofactor II (Fig 2D) or nonimmune-mouse-IgG (Fig 2F) or when the first antibody was replaced by dilution buffer alone (Fig 2E).
Immunofluorescence microscopy: PCI in washed platelets. Smears of washed platelets isolated from whole blood were incubated with a monoclonal antibody against PCI (A), PAI-1 (B), factor D (C), or heparin cofactor II (D); in other negative control experiments with dilution buffer alone (E); or with a nonimmune mouse IgG (F), followed by fluorescein-conjugated goat-antimouse-IgG. PCI showed fluorescence intracellularly as well as PAI-1, which was used as a positive control (A and B, respectively), whereas a rim pattern of staining consistent with an exclusively surface localization of the protein, which was observed with the antibody to factor D (C), was not observed in case of PCI. Negative control experiments performed with an antibody to heparin cofactor II (D), with mouse-nonimmune IgG (F), or by incubation with dilution buffer alone (E) did not show any fluorescence. (Original magnification × 3,000; exposure time: 3 seconds.)
Immunofluorescence microscopy: PCI in washed platelets. Smears of washed platelets isolated from whole blood were incubated with a monoclonal antibody against PCI (A), PAI-1 (B), factor D (C), or heparin cofactor II (D); in other negative control experiments with dilution buffer alone (E); or with a nonimmune mouse IgG (F), followed by fluorescein-conjugated goat-antimouse-IgG. PCI showed fluorescence intracellularly as well as PAI-1, which was used as a positive control (A and B, respectively), whereas a rim pattern of staining consistent with an exclusively surface localization of the protein, which was observed with the antibody to factor D (C), was not observed in case of PCI. Negative control experiments performed with an antibody to heparin cofactor II (D), with mouse-nonimmune IgG (F), or by incubation with dilution buffer alone (E) did not show any fluorescence. (Original magnification × 3,000; exposure time: 3 seconds.)
Immunogold-transmission electron microscopy.
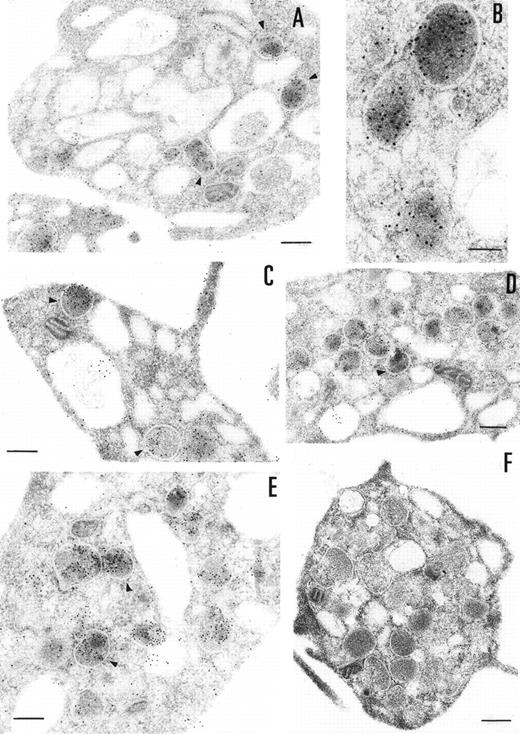
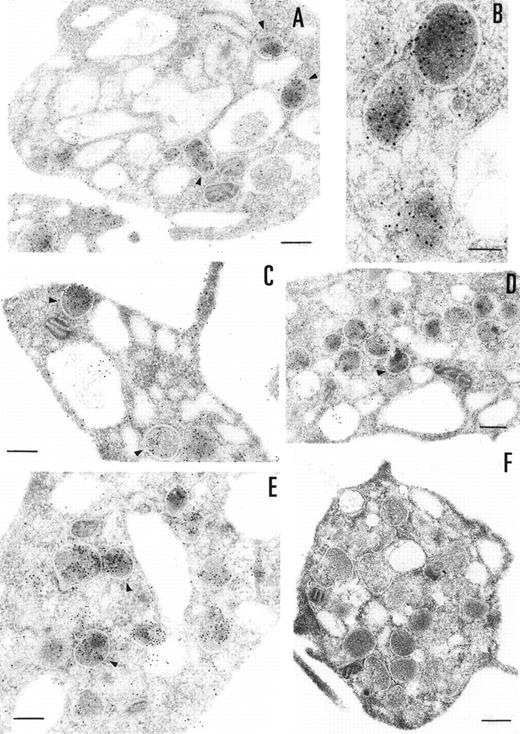
The ultrastructural localization of PCI was analyzed by immuno-electron microscopy using postembedding immunogold labeling. In these sections, platelets appeared unstimulated. The different structures were well preserved, with the only exception of some regions of the membranes (plasma membrane, membranes of the granula, and membranes of the open canalicular system). The negative control experiments performed by substituting the first specific antibody with nonimmune-mouse-IgG (Fig 3F) or omitting the first antibody and incubating with dilution buffer alone (not shown) did not show any labeling. The staining with the 4PCI antibody (Fig 3A through E), localized PCI in the α granules (Fig 3A through E, arrowheads) in the open canalicular system (OCS; bound predominantly to the membranes or bound to some precipitates in the lumen of these structures) as well as on the plasma membrane. The platelet cytoplasmic matrix reflected minimal background levels of labeling as did other organelles such as mitochondria. As far as α granules are concerned (Fig 3A through E, arrowheads), gold particles were evenly distributed over all α granules. The labeling in these granules was predominantly in the electron dense zone or nucleoid and also sometimes bound to the membrane of the granules. The labeling of the plasma membrane (Fig 3A through E) was observed either at the internal or at the external surface of the trilaminar structure. The distribution of PCI labeling after platelet activation with calcium ionophore A23187 was also analyzed (not shown). Differences in the distribution of staining between stimulated and unstimulated platelets were quantitatively evaluated by locating more than 500 gold particles in each case. Activation of the platelets showed a considerable decrease in staining in the α granules.
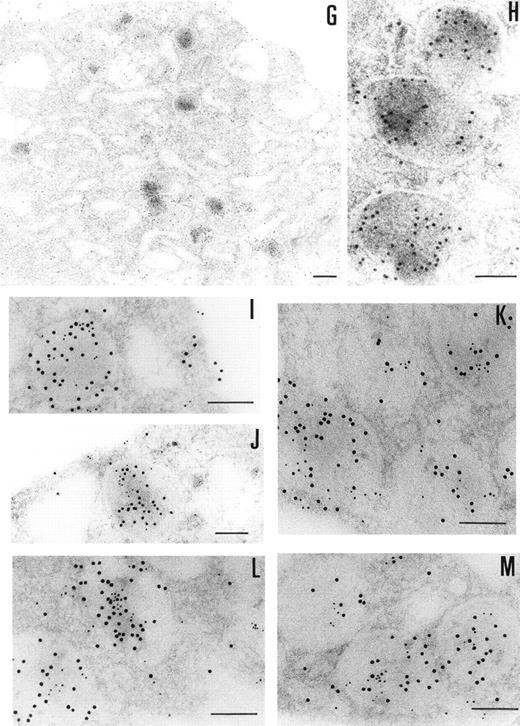
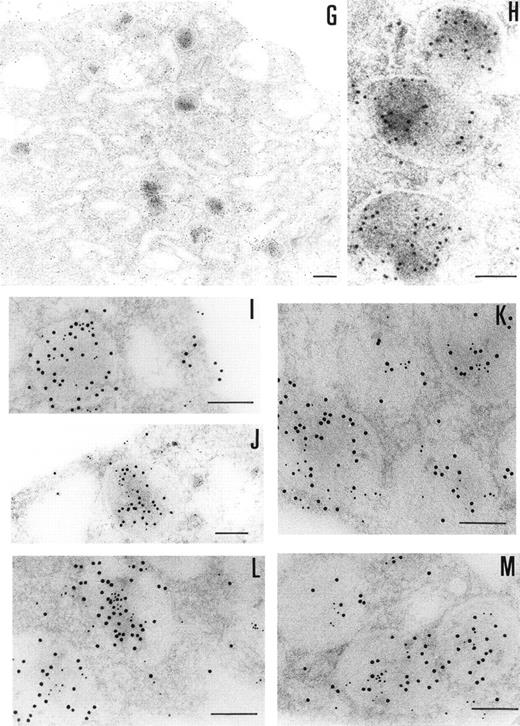
Immunoelectronmicroscopy of platelet and megakaryocytes sections. Washed platelets and megakaryocytes were proceeded for transmission electron microscopy as described in Materials and Methods. Sections incubated with 75 μg/mL of 4PCI (A through E) showed positive staining in the OCS, in the granules (arrowheads), and on the plasma membrane. Sections incubated with nonimmune-IgG (negative control, F) were devoid of gold staining. PCI labeling in the granules (A through E, arrowheads) was distributed over all granules and predominantly in the electron dense zone or nucleoid, although sometimes they were found to bind to the granule membrane (A through E, arrowheads). The PCI staining pattern in megakaryocytes (G and H) was very similar to that seen in platelets. Gold particles often appeared in granules identifiable as granules, as well as on the plasma membrane, and in the open canalicular system. Experiments of colocalization of PCI with PAI-1 (I through M) were performed as described in Materials and Methods. The gold particles of 10 and 5 nm indicate the presence of PCI and PAI-1, respectively. Colocalization demonstrates that the granules observed were indeed granules. (A and C through G) Bars = 200 nm; (B and H through M) bars = 100 nm.
Immunoelectronmicroscopy of platelet and megakaryocytes sections. Washed platelets and megakaryocytes were proceeded for transmission electron microscopy as described in Materials and Methods. Sections incubated with 75 μg/mL of 4PCI (A through E) showed positive staining in the OCS, in the granules (arrowheads), and on the plasma membrane. Sections incubated with nonimmune-IgG (negative control, F) were devoid of gold staining. PCI labeling in the granules (A through E, arrowheads) was distributed over all granules and predominantly in the electron dense zone or nucleoid, although sometimes they were found to bind to the granule membrane (A through E, arrowheads). The PCI staining pattern in megakaryocytes (G and H) was very similar to that seen in platelets. Gold particles often appeared in granules identifiable as granules, as well as on the plasma membrane, and in the open canalicular system. Experiments of colocalization of PCI with PAI-1 (I through M) were performed as described in Materials and Methods. The gold particles of 10 and 5 nm indicate the presence of PCI and PAI-1, respectively. Colocalization demonstrates that the granules observed were indeed granules. (A and C through G) Bars = 200 nm; (B and H through M) bars = 100 nm.
The presence of PCI in megakaryocytes was also investigated by the same immunogold staining procedures (Fig 3G and H). The megakaryocytes seen in the bone marrow sections displayed a smooth surface, with a nucleus presenting several rounded lobes with abundant euchromatin (not shown). Mature megakaryocytes displaying morphologic evidence of platelet production were also found (not shown). The pattern of PCI labeling in these cells was very similar to that seen in platelets. PCI was localized in granules identified as α granules, as judged from the size, form, and electron density (Fig 3G and H). The cisternae of the OCS or similar structures were also positive for PCI (Fig 3G). Gold particles appeared also bound to the plasma membrane (Fig 3G). One interesting finding in this experiment was the presence of PCI in the nucleus of megakaryocytes (data not shown). Granulocytes present in the same bone marrow sections showed also positive PCI labeling at different organelles as well as in the nucleus (not shown). Control experiments performed by substituting the first antibody (4PCI) with nonimmune-mouse-IgG did not show any staining either in megakaryocytes or in granulocytes (data not shown). To confirm the localization of PCI within the α granules, additional experiments were performed using PAI-1 as an α granule marker (Fig 3I through M). Gold particles of 2 different sizes were used to identify the 2 proteins of interest: PCI (10 nm) and PAI-1 (5 nm). Colocalization of PCI with PAI-1 confirmed that PCI-containing granules were indeed α granules (Fig 3I through M). In addition to the α granules, colocalization of PCI and PAI-1 was also seen in the OCS and on the plasma membrane (data not shown).
Uptake of biotinylated-PCI by platelets.
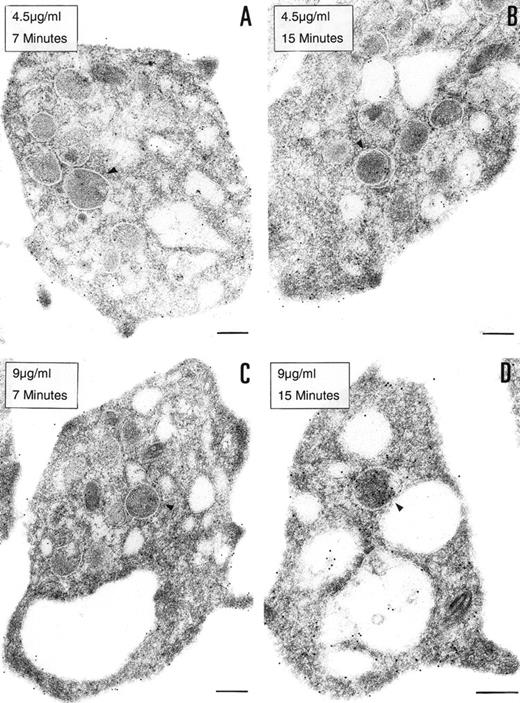
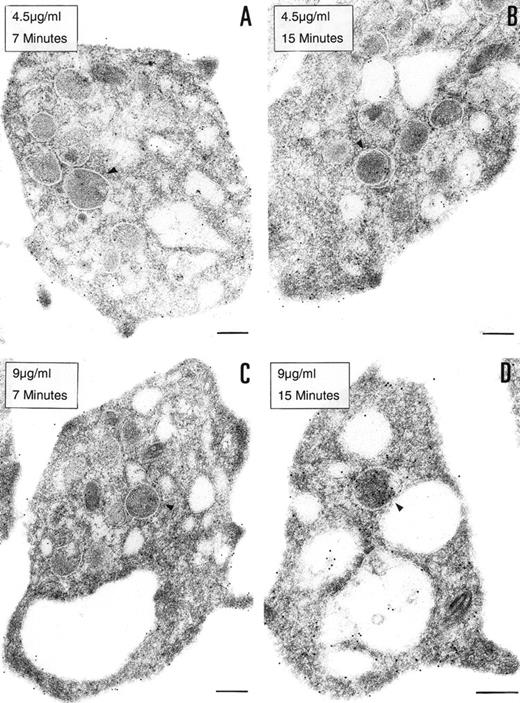
To determine if the PCI found in platelets is derived from an exogenous origin or from endogenous synthesis, platelets in suspension were incubated with 2 different concentrations of biotinylated PCI (4.5 or 9 μg/mL) for 7 or 15 minutes, respectively (Fig 4). Biotinylated PCI was chosen for the uptake experiments, based on published data that describe a possible internalization of solid-phase ligands, as colloidal gold-protein conjugates, and on postligand binding events induced by this kind of conjugates,45 as well as based on other data that demonstrate that platelets incubated with free biotin remained unlabeled, not showing unspecific uptake of biotin.46 The detected labeling was sparse, with only a few gold particles. A dose- and time-dependent uptake of PCI from the extracellular medium was observed (Fig 4A through D). The morphological study demonstrates that PCI was bound to platelets sequestered in the OCS and that some of these molecules were internalized, appearing in the cytoplasm without being surrounded by a membrane or in the protein-storing structures, the α granules (arrowheads). The proportion of labeling in the α granules was found to increase with the time and concentration (Fig 4A through D). No evidence for an endocytotic process mediated by receptors or other ligands was thus observed. Labels were not seen in coated pits or coated vesicles.
Uptake of biotinylated PCI by platelets. Washed platelets in suspension were incubated with 2 different concentrations of biotinylated PCI for 2 different periods of time, namely, 4.5 μg/mL for 7 minutes (A) and 15 minutes (B) or 9 μg/mL for 7 minutes (C) and 15 minutes (D). Uptake was stopped by addition of fixative and platelets were then processed for transmission electron microscopy. Biotinylated PCI was detected by incubating platelets sections with 10 nm gold-labeled streptavidin. Dose- and time-dependent internalization of biotinylated PCI was observed, which conduced the biotinylated PCI molecules to the granules (arrowheads). Bars = 200 nm.
Uptake of biotinylated PCI by platelets. Washed platelets in suspension were incubated with 2 different concentrations of biotinylated PCI for 2 different periods of time, namely, 4.5 μg/mL for 7 minutes (A) and 15 minutes (B) or 9 μg/mL for 7 minutes (C) and 15 minutes (D). Uptake was stopped by addition of fixative and platelets were then processed for transmission electron microscopy. Biotinylated PCI was detected by incubating platelets sections with 10 nm gold-labeled streptavidin. Dose- and time-dependent internalization of biotinylated PCI was observed, which conduced the biotinylated PCI molecules to the granules (arrowheads). Bars = 200 nm.
RT-PCR and Southern blotting.
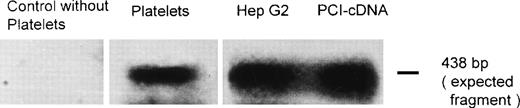
RT-PCR and Southern blot analysis were performed to provide evidence as to whether platelet PCI might also be derived from endogenous synthesis. After blotting and autoradiography, a band corresponding in size to the expected fragment (438 bp) was observed using total platelet RNA (Fig 5). A band of the same size was seen with total RNA from HepG2 cells used as a positive control (Fig 5). Both bands had the same size as the fragment amplified from PCI-cDNA (Fig 5). No genomic DNA contaminations were detected, as judged from the absence of a 1,978-bp band containing introns 3 and 4 of PCI (not shown). No band was observed in the control without platelets (Fig 5). These results suggest that platelet PCI is also derived from endogenous synthesis. Therefore, PCI present in the α granules seems to be derived from 2 sources: endogenous synthesis and uptake from the surrounding medium.
RT-PCR and Southern blotting. Total RNA obtained from platelets and Hep G2 cells was subjected to RT-PCR and Southern blotting using primers specific for a 438-bp fragment of PCI-cDNA. A PCI-cDNA fragment inserted into the plasmid vector pBluescript II KS (+/−) phagemid was also amplified by PCR with the same primers. Additionally, a negative control was performed by amplifiying a sample without platelets. After blotting, the membrane was incubated with a32P-labeled PCI-cDNA fragment and subjected to autoradiography as described in Materials and Methods.
RT-PCR and Southern blotting. Total RNA obtained from platelets and Hep G2 cells was subjected to RT-PCR and Southern blotting using primers specific for a 438-bp fragment of PCI-cDNA. A PCI-cDNA fragment inserted into the plasmid vector pBluescript II KS (+/−) phagemid was also amplified by PCR with the same primers. Additionally, a negative control was performed by amplifiying a sample without platelets. After blotting, the membrane was incubated with a32P-labeled PCI-cDNA fragment and subjected to autoradiography as described in Materials and Methods.
In situ hybridization of bone marrow cryosections.
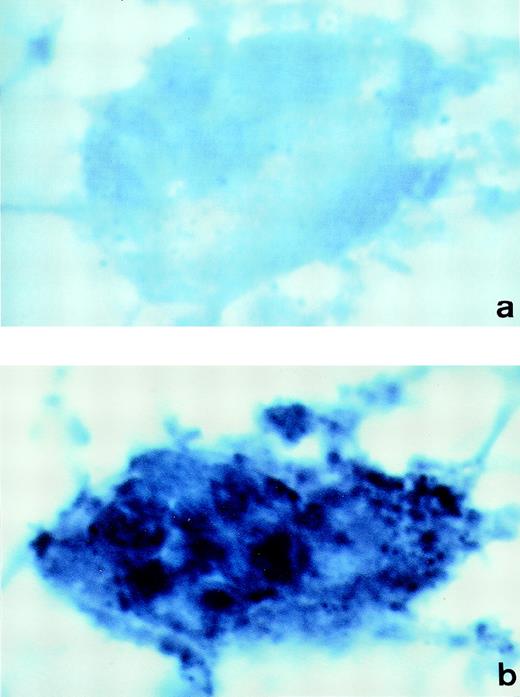
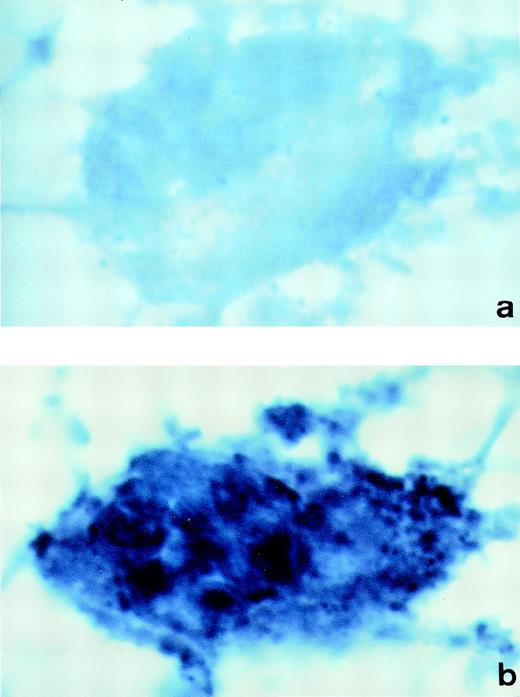
To demonstrate synthesis of PCI in megakaryocytes, human bone marrow cryosections were analyzed for the presence of PCI-mRNA (Fig 6). In situ hybridization was performed using DIG-labeled antisense or sense PCI cRNA probes, which were further detected with anti-DIG-alkaline phosphatase Fab fragments, followed by NBT/BCIP substrate detection. The megakaryocytes were easily recognized in these sections at lower magnifications because of their large size as compared with other cells present in the bone marrow and because of their typical morphology. The analysis of labeling with antisense PCI cRNA probe was found to be positive (Fig6b): cells showed dark blue precipitates, identified as positive hybridization signals. In addition to megakaryocytes, other nucleated cells in bone marrow also contained PCI-mRNA (data not shown). As judged from their morphology, these cells resembled granulocytes, which were also positive for PCI antigen as judged from immuno-electron microscopy. However, these cells were not further examined in the present study. Sections incubated with sense PCI cRNA probes (Fig 6a) or with antisense probe but without incubation with the anti-digoxigenin antibody (data not shown) yielded negative results. In conclusion, PCI mRNA was detected in megakaryocytes obtained from bone marrow preparations, which reflects again that not all of the PCI molecules present in platelets are derived from an endocytotic process and that some of them are sequestered into the storage granules after endogenous synthesis in megakaryocytes.
In situ hybridization of human bone marrow cryosections. Sections of human bone marrow were incubated with DIG-labeled-cRNA sense (a) or antisense probe (b). Bound cRNA probes were detected with anti-digoxigenin-alkaline phosphatase Fab fragments, followed by incubation with NBT/BCIP color solution. The megakaryocytes observed in sections incubated with the PCI-cRNA antisense probe showed dark blue precipitates (b). Control sections incubated with the sense probe yielded negative results (a). (Original magnification × 1,000.)
In situ hybridization of human bone marrow cryosections. Sections of human bone marrow were incubated with DIG-labeled-cRNA sense (a) or antisense probe (b). Bound cRNA probes were detected with anti-digoxigenin-alkaline phosphatase Fab fragments, followed by incubation with NBT/BCIP color solution. The megakaryocytes observed in sections incubated with the PCI-cRNA antisense probe showed dark blue precipitates (b). Control sections incubated with the sense probe yielded negative results (a). (Original magnification × 1,000.)
DISCUSSION
It has been suggested previously that platelets might contain an inhibitor of APC.35,36 We therefore analyzed in this study the presence of PCI in washed human platelets. As judged from an ELISA that recognizes all forms of PCI antigen (active, complexed, and cleaved), PCI is present in platelets at a concentration of 160 ng/2 × 109 platelets. Using an ELISA specific for functional PCI antigen and pooled citrated plasma as a reference, we have shown that this PCI in the platelets is active and, furthermore, that the specific activity of platelet PCI is approximately 5 times higher than that of plasma PCI. This could be caused by binding of platelet PCI to substances stimulating its activity, as shown previously for proteoglycans on epithelial kidney cells.47To analyze in a semiquantitative way the different forms of PCI antigen in platelets, platelet lysates immunoprecipitated with monoclonal anti-PCI-IgG were analyzed in Western blots. The main PCI antigen bands detected exhibited Mrs of 57,000 (corresponding in size to active PCI)44 and greater than 220,000. Bands corresponding to cleaved, inactive PCI (54 kD or less) were practically absent. The identity of the high Mr bands is still unknown and will be analyzed separately.
Having shown the presence of PCI in platelet lysates, we analyzed the localization of PCI in resting platelets. Using immunofluorescence microscopy, a relatively homogenous pattern of distribution of PCI in resting platelets was observed, together with a spotted pattern in many domains, indicating intracellular localization of PCI and not only external binding to the plasma membrane. A similar pattern of fluorescence has been described for other proteins localized in subcellular structures of platelets, eg, in the α granules, in the dense granules, or in the OCS.7,8,48 Immunotransmission electron microscopy showed localization of PCI in the α granules, in the OCS, and on the surface of the platelets associated to the plasma membrane. A possible redistribution of PCI in platelets after activation was investigated by activating the platelets with calcium ionophore. The results were consistent with the hypothesis that, after activation of the platelets, the PCI-pool stored in the α granules is secreted, presumably, to the extracellular space. Soluble proteins have been identified that are stored in the platelet granules and are secreted upon platelet activation,2,3,42,49,50 and some of them were found to play a role in the arrest of bleeding (physiological hemostasis) or in the formation of vaso-occlusive thrombi (pathological thrombosis).43
Many proteins stored in α granules are endogenously synthesized and packed within the platelet precursor, the megakaryocyte, such as platelet factor 4, thrombospondin, and von Willebrand factor.2 Other proteins in the α granules, such as fibrinogen and albumin, are accumulated via a different mechanism involving endocytosis and pinocytosis from the surrounding extracellular medium at both the megakaryocyte and circulating platelet levels.5,7,9,10 A slight uptake into the platelets analyzed was observed. This uptake was time- and dose-dependent, and biotinylated-PCI was detected on the plasma membrane, in the OCS, and in the α granules. It can therefore be assumed that PCI is bound to the platelets and sequestered in the open canalicular system and that some of these molecules are internalized, appearing in the protein-storing structures, the α granules. No evidence for an endocytotic process mediated by receptors or other ligands was observed; labels were not seen in coated pits or vesicles. In vivo, due to the high levels of PCI in plasma as compared with platelets, the process of internalization of PCI could occur by fluid-phase internalization or pinocytosis, as observed for other proteins (eg, albumin).51
We were also able to show by RT-PCR that platelets contain PCI mRNA, suggesting endogenous PCI synthesis in megakaryocytes. To confirm this hypothesis and because we cannot completely rule out contamination of the gel filtered platelets with white blood cells, we performed in situ hybridization of bone marrow cryosections. Positive staining for PCI mRNA was seen in megakaryocytes. Therefore, endogenous synthesis of PCI occurs in the precursors of the platelets, in which we were also able to localize PCI protein. The localization of PCI antigen within megakaryocytes looked very similar to that of platelets: it was seen in small granules, which might be identified as α granules, in the channels of the OCS, and on the surface of the cell, at the plasma membrane. Interestingly, megakaryocytes were not the only cells in bone marrow that were positive for PCI. Other cell types morphologically indistinguishable from granulocytes showed also positive staining in their granules as well as in structures similar to vesicles dispersed along the cytoplasm. An unexpected observation was furthermore the presence of PCI labeling in the nucleus of megakaryocytes and granulocytes. Nuclear staining was not seen with nonimmune-IgG and therefore seems to be specific. Analysis of these data is too preliminary at the moment and could be a subject for further investigations.
In conclusion, the results presented in this work demonstrate the presence in platelets of 2 PCI pools with different origins: from megakaryocytic synthesis and from plasma uptake. Furthermore, they indicate that this PCI is stored in platelets in a more active form than that found in plasma and that it is secreted after platelet activation. Although extrapolation to physiologic and pathophysiologic conditions is difficult, platelet activation in vivo may be responsible for local increase in PCI activity, which could play a role at sites of platelet accumulation, ie, at sites of clot formation. We have shown previously that PCI plasma levels are elevated in survivors of myocardial infarction,52 suggesting a role of PCI in the development of thrombotic processes. Our present findings suggest that platelets might contribute to the local elevation of PCI. These locally increased PCI levels could not only inhibit the activity of the anticoagulant protein C pathway,11 but also the activation of fibrinolysis,25 resulting in a tendency of clots to persist, thereby being involved in the development of thrombotic diseases such as myocardial infarction or deep venous thrombosis.
ACKNOWLEDGMENT
The authors are grateful to Dr F. Keil (Department of Internal Medicine I, University of Vienna, Vienna, Austria) for his help obtaining bone marrow samples. We also thank Prof E. Koller and Dr I. Volf (Department of Medical Physiology, University of Vienna) for their helpful suggestions.
Supported in part by Grants No. P 10823-Med and P 12308-Gen from the Austrian Science Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Margarethe Geiger, MD, Department of Vascular Biology and Thrombosis Research, University of Vienna, Schwarzspanierstrasse 17, A-1090 Vienna, Austria; e-mail:margarethe.geiger@univie.ac.at.