Abstract
Stem cell factor (SCF) is expressed as an integral membrane growth factor that may be differentially processed to produce predominantly soluble (S) (SCF248) or membrane-associated (MA) (SCF220) protein. A critical role for membrane presentation of SCF in the hematopoietic microenvironment (HM) has been suggested from the phenotype of the Steel-dickie(Sld) mice, which lack MA SCF, and by studies performed in our laboratory (and by others) using long-term bone marrow cultures and transgenic mice expressing different SCF isoforms.Steel17H (Sl17H) is an SCF mutant that demonstrates melanocyte defects and sterility in males but not in females. The Sl17H allele contains a intronic mutation resulting in the substitution of 36 amino acids (aa’s) in the SCF cytoplasmic domain with 28 novel aa’s. This mutation, which affects virtually the entire cytoplasmic domain of SCF, could be expected to alter membrane SCF presentation. To investigate this possibility, we examined the biochemical and biologic properties of the Sl17H-encoded protein and its impact in vivo and in vitro on hematopoiesis and on c-Kit signaling. We demonstrate that compound heterozygous Sl/Sl17H mice manifest multiple hematopoietic abnormalities in vivo, including red blood cell deficiency, bone marrow hypoplasia, and defective thymopoiesis. In vitro, both S and MA Sl17H isoforms of SCF exhibit reduced cell surface expression on stromal cells and diminished biological activity in comparison to wild-type (wt) SCF isoforms. These alterations in presentation and biological activity are associated with a significant reduction in the proliferation of an SCF-responsive erythroid progenitor cell line and in the activation of phosphatidylinositol 3-Kinase/Akt and mitogen-activated protein-Kinase signaling pathways. In vivo, transgene expression of the membrane-restricted (MR) (SCFX9/D3) SCF in Sl/Sl17H mutants results in a significant improvement in peripheral red blood cell counts in comparison toSl/Sl17H mice.
STEM CELL FACTOR (SCF; also known as Kit-ligand [KL], mast cell growth factor [MGF], and Steel[Sl] factor) is the ligand for the receptor tyrosine kinase encoded by the c-kit proto-oncogene.1,2 The c-Kit receptor is a membrane spanning protein that is a member of the platelet-derived growth factor (PDGF) receptor family and possesses intrinsic kinase activity.3 Mutations of c-Kit are associated with the dominant White Spotting (W) phenotype in mice, whereas mutations of the c-Kit ligand are associated with the Sl mutant phenotype.2,4-8 Mutations at either locus produce similar abnormalities in mice, with many mutations resulting in embryonic lethality due to severe anemia.9 Viable mice homozygous for mutations at either locus typically are deficient in erythrocytes, mast cells, progenitors for multiple hematopoietic lineages, and melanocytes (resulting in black-eyed white mice) and these mice are sterile.9
Several cytokines and/or growth factors, such as colony-stimulating factor-1 (CSF-1), interleukin-1 (IL-1), and tumor necrosis factor (TNF), have been described that function as both membrane and soluble proteins.10,11 The majority of these transmembrane growth factors are proteolytically processed to release soluble protein.12,13 The membrane-anchored counterparts of these proteins also possess biological activity and can promote several distinct biological functions.13-15 SCF can induce proliferation, survival, adhesion/migration, and differentiation in c-Kit–expressing cells of hematopoietic and nonhematopoietic lineages. Both soluble (S) (SCF248) and membrane-associated (MA) (SCF220) SCF can mediate direct cell-cell contact with c-Kit–positive cells.16-19 Our laboratory has demonstrated that the generation of soluble SCF is dependent on 3 distinct proteolytic cleavage sites. The primary site in exon 6 is preferentially used, in vitro, in stromal cells expressing SCF. A secondary site located in exon 7 is used only in the absence of the primary site.15 Chymase digestion of a third site (encoded in exon 6) results in a protein product of 158 amino acids (aa’s).20 This site is sensitive to chymase digestion by mast cells but not by stromal cells.20 Proteolytic cleavage at all 3 sites results in biologically active SCF protein.14,15,20 Additionally, we have shown that mutagenesis of the primary cleavage site in exon 6 and the secondary site in exon 7 in the SCF cDNA leads to the generation of membrane-restricted (MR) and biologically active form of SCF (SCFX9/D3), when expressed in murine stromal cells in vitro and in vivo.15 21
Some viable Sl alleles (eg, Steel-panda [Slpan] and Steel-contrasted[Slcon]) exhibit alterations in the enhancer regions of the Sl gene that affect the levels of SCF mRNA in various tissues.22 A small number of Sl mutations disrupt the coding sequence of SCF, which changes the structure of the expressed SCF protein. Steel-dickie (Sld) is the result of an intragenic 4-kb deletion that removes sequences encoding transmembrane and cytoplasmic domains.23,Sld mice produce only a soluble mutant form of the SCF protein.16,23The fact that Sld mice are viable, but display multiple hematopoietic cell deficiencies, are sterile, and lack coat color suggests that the Sld SCF protein retains some biological activity and that the MA isoform must mediate proliferation, survival, or migration of several hematopoietic and nonhematopoietic lineages in vivo. In this regard, we have recently shown that transgene expression of the MR SCF but not soluble SCF inSld mutants significantly improves the hematologic abnormalities associated with these mice.20 Also, exclusive expression in vivo of the MA SCF (SCF220) results in mast cell deficiency, but no other hematopoietic or nonhematopietic abnormalities.24 These observations suggest that sequences located in the cytoplasmic domain of SCF or that membrane presentation or both is pivotal for c-Kit activation.
Another viable Sl allele, Steel17H(Sl17H), contains a mutation in the polypyrimidine tract of intron 7, leading to abnormal splicing and the absence of exon 8 encoded sequences that comprise the cytoplasmic domain of SCF. This results in the substitution of 28 novel aa’s with only the first cytoplasmic aa (lysine) read in the correct frame.25 The first 3 aa’s of the cytoplasmic domain of native SCF are lysines. Positively charged aa’s within the cytoplasmic domain located immediately adjacent to the transmembrane region have been shown to be important for anchoring transmembrane proteins in the membrane.26,27 Mutations that reduce the net positive charge in this region can lead to instability of protein in the cell membrane.26 In addition, mutations in the cytoplasmic tail of membrane growth factors, particularly the removal of the COOH-terminal valine (which is also present in SCF), lead to reduced cell surface expression of these growth factors.28Therefore, we hypothesized that the mutation in theSl17H would affect stromal cell membrane presentation of SCF. In this study, we cloned both the soluble and MA isoforms of Sl17H and studied the presentation, biological activity, and in vivo consequences of expression of this mutant in blood cell development.
MATERIALS AND METHODS
Generation of Sl/Sl17H transgenic mice.
Experiments involving mice described here were reviewed and approved by Animal Use Committee of Indiana University School of Medicine (Indianapolis, IN). C3H–Sl17H/+ mice were a kind gift of the ABL-Basic Research Program, National Cancer Institute (Frederick, MD). These mice were maintained by breeding into C3H background. C3H/HeJ mice were obtained from Jackson Laboratories (Bar Harbor, ME). Transgenic mice were generated by microinjecting into the pronuclei of fertilized C3H/HeJ eggs a 1.3-kbNde I/Kpn I fragment comprising either the human phosphoglycerate kinase promoter (hPGK) and cDNA encoding the soluble (hPGK-SCF248) or the MR (hPGK-SCFX9/D3) form of SCF, as described previously.21 Microinjected eggs were transferred to the oviducts of pseudo-pregnant outbred Swiss-Webster females. Offspring were tested for the presence of transgene by analyzing tail DNA. Briefly, tail DNA from mice was digested overnight in digestion buffer (100 mmol/L NaCl, 10 mmol/L Tris base, pH 8, 25 mmol/L EDTA, pH 8, 1% sodium dodecyl sulfate [SDS], and 150 μg/mL proteinase K) at 50°C and extracted the next day with phenol and chloroform. The high molecular weight DNA was subsequently digested with EcoRI, electrophoresed, transferred to filters, and probed using a full-length 32P-labeled cDNA murine SCF probe. To examine the in vivo role of the 2 isoforms of SCF on hematopoietic lineages in Sl17H mutants, we first crossed transgenic mice overexpressing either the soluble or the MR form of SCF to WC/ReJ-Sl/+ mice to obtain Sl/+ mice that overexpress either isoform of SCF. These mice were identified based on their phenotype (ie, forehead blaze and diluted belly) and Southern blot analysis. We have previously shown that these mice express equivalent amounts of SCF transgene.21 Transgene positive Sl/+ male mice were further crossed to C3H-Sl17H/+ mice to obtainSl/Sl17H transgene-positive or -negative mice.
Peripheral blood analysis.
Total peripheral red blood cell (RBC) and white blood cell (WBC) counts were analyzed on tail vein bleeds with a hemocytometer and Coulter Model ZM electronic particle counter (Coulter Electronics, Hialeah, FL). For WBC counts, RBCs were lysed using Zapoglobin (Coulter Electronics) according to the manufacturer’s recommendations. Peripheral blood hematocrits were performed by spinning capillary tubes for 5 minutes in a model MB micro-capillary centrifuge (IEC, Boston, MA).
Cell preparations, antibodies, and flow cytometric analysis.
Mice were killed by cervical dislocation, and thymus glands and bone marrow (BM) were surgically excised from adult wild-type (wt) andSl17H transgenic and nontransgenic mice. Thymus suspensions were prepared and filtered through nylon-mesh to remove debris. BM was harvested and cellularity was determined using a Coulter Model ZM. Phycoerythrin (PE)-conjugated monoclonal antibodies (MoAbs) were directed against CD4, CD8, and CD69. All the PE- and fluorescein isothiocyanate (FITC)-conjugated MoAbs, including the isotype control antibodies, were purchased from Pharmingen (San Diego, CA). Cells (1 × 106) were incubated at 4°C for 30 minutes with 1 μg of the first MoAb. Cells were washed 3 times with phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA). Cells were then incubated with 1 μg of the second MoAb for 30 minutes at 4°C, subsequently washed 3 times with PBS containing 0.1% BSA, and analyzed by fluorescence-activated cell sorter (FACS; Becton Dickinson, San Jose, CA). To determine the cell surface expression of SCF on Sl/Sl4 stromal cells transfected with cDNAs encoding either the soluble or the MA isoforms of wt and Sl17H protein, flow cytometric analysis with an anti-SCF antibody was performed. Briefly, 1 × 106 stromal cells were stained separately with either 1 μg/mL of primary anti-SCF antibody (Genzyme, Cambridge, MA) or a control antibody for 30 minutes at 4°C. Afterwards, the cells were washed twice with PBS/0.1% BSA and subsequently stained with 1 μg of secondary PE-conjugated anti-IgG (Santa Cruz Biotechnology, Santa Cruz, CA) under identical conditions and analyzed by FACS.
Cloning of the Sl17H soluble (Sl17H-SCF248) and MA (Sl17H-SCF220) cDNAs.
Murine wt-SCF248 andSl17H-SCF248 cDNAs were blunt-end subcloned into the HincII site of Bluescript (Stratagene, La Jolla, CA). Subsequently, BamHI/Xho I fragment containing the entire coding sequence of SCF was cloned into the PSG5 expression vector (Stratagene). wt-SCF220 andSl17H-SCF220 cDNAs were cloned into the V19.8 expression vector.29 Briefly, polymerase chain reaction (PCR) was performed on reverse-transcribed (RT) total RNA derived from testes of a Sl/Sl17H mouse. Amplification and cDNA synthesis was performed according to the manufacturer’s instructions (Perkin-Elmer, Branchburg, NJ). PCR fragments were purified by agarose gel electrophoresis before cloning. The upstream SCF primer was flanked at its 5′ end by an artificial BamHI site, and the downstream primer was flanked by a Pst I site. This allows cDNAs amplified by PCR to be cloned between the BamHI and Pst I sites of bluescript and subsequently into the expression vector V19.8. Nucleotide sequencing was performed to determine the absence of exon 8 encoding sequences on double-stranded DNA encoding bothSl17H-SCF248 andSl17H-SCF220.
Generation of stable transfectants of Sl/Sl4 stromal cells expressing wt and Sl17H-SCF cDNAs.
Transfection of plasmid DNA into Sl/Sl4 stromal cells devoid of endogenous SCF mRNA and protein was performed using DOTAP (Boehringer Mannheim, Indianapolis, IN) according to the manufacturer’s recommendations. Sl/Sl4 cells were grown to subconfluence in Dulbecco’s modified Eagle’s medium (DMEM) with 10% calf serum (CS). Plasmid mixtures containing cDNAs of wt or Sl17H SCF and a hygromycin-resistance gene in p48 (10:1 ratio) were incubated with DOTAP at room temperature for 10 minutes and then diluted with fresh medium.30 The medium was then removed from the cells and replaced with the transfection mixture, and the cells were allowed to grow overnight. The following day, fresh medium containing hygromycin (300 U/mL; Boehringer Mannheim) was added to the cells. Well-isolated hygromycin-resistant colonies arising in 10 to 14 days were transferred to 24-well plates and expanded. Total cellular RNA was prepared from each clone using Tri-reagent (Molecular Research Center, Cincinnati, OH) and clones were screened for SCF expression by RT-PCR.
Expression of wt-SCF248, wt-SCF220,Sl17H-SCF248, andSl17H-SCF220 cDNAs inSl/Sl4 stromal cells.
Total cellular RNA from at least 3 stromal cell transfectants expressing either wt or Sl17H SCF was purified using Tri Reagent (Molecular Research Center) according to the manufacturer’s instructions and was used as template. Semiquantitative RT-PCR was performed on at least 2 clones of each stromal cell transfectants expressing wt-SCF248, wt-SCF220,Sl17H-SCF248, andSl17H-SCF220 cDNAs using actin as an internal control. Briefly, RNA was used to synthesize single-stranded cDNA by RT and random hexamers (Perkin-Elmer). cDNA was amplified in a 100 μL reaction mixture by PCR using ampliTaq DNA polymerase in 35 cycles of 1 minute of denaturation at 94°C, 2 minutes of annealing at 55°C, and 3 minutes of synthesis at 72°C using a 5′-GGAGATCTGCGGGAATCC-3′ sense primer and 5′-GTCCACAATTACACCTCTTG-3′ antisense primer based on published sequences.16 RT-PCR products were examined on a 2.5% agarose gel. As a control for integrity of total RNA, primer specific for actin (5′-TGGTGGGAATGGGTCAGAAGGACTC-3′ sense primer and 5′-TTGGCATAGAGGTCTTTACGGATGT-3′ antisense primer) were used to amplify cDNA using the described conditions.31 The predicted amplification product of these primers is 732 bp.
Metabolic labeling and immunoprecipitation.
For protein analysis, subconfluent cells were starved for 30 minutes in methionine-free DMEM, 10% dialyzed CS. Afterwards, the cells were labeled with 0.5 mCi/mL of [35S]Met for 30 minutes and chased with cold media for various time points. Conditioned medium was collected from each cell line after 1, 2, and 3 hours, filtered through a 0.45-μm filter, and stored at −80°C until used. Labeled cells were washed with PBS and lysed in lysis buffer (0.5% sodium-deoxycholate, 0.5% Nondiet P-40, 50 mmol/L NaCl, 25 mmol/L Tris-Cl [pH 8.0], and 1 mmol/L phenylmethylsulfonyl fluoride). Immunoprecipitation was performed using a rabbit polyclonal antibody raised against rat-SCF (kindly supplied by Dr Larry Bennett, Amgen, Thousand Oaks, CA). Two milliliters of polyclonal antisera was conjugated to 1 mL protein A-agarose using the Affinica purification kit (Schleicher and Schuell, Keene, NH) according to the manufacturer’s instructions. The antibody-conjugated protein A-agarose was resuspended in 3 mL of PBS with 0.05% sodium azide and stored at 4°C. For immunoprecipitation, labeled cells were thawed overnight at 4°C concentrated 2× for 30 minutes using a Centriprep-10 (Amicon, Beverly, MA) device according to the manufacturer’s instructions and transferred to 1.5-mL microfuge tubes. Twenty-five microliters of conjugated antibody was added to each sample and allowed to incubate overnight at 4°C. The samples were centrifuged for 3 minutes at 4°C, the supernatant was discarded, and the immunoprecipitates were washed 3 times in wash buffer (0.5 mol/L NaCl, 20 mmol/L Tris-HCl [pH 7.5], and 1% Triton) and once with 20 mmol/L Tris-Cl (pH 7.5). The protein was released from the beads by boiling for 5 minutes. An equal volume of 2× sample loading buffer (100 mmol/L Tris-HCl [pH 6.8], 200 mmol/L dithiothreitol, 4% SDS, 0.02% bromophenol blue, and 20% glycerol) was added to each sample and boiled for 5 minutes, and the proteins were analyzed on 12% or 15% SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Biological activity of transfected cell lines.
The effect of Sl17H-SCF248 andSl17H-SCF220 protein on proliferation of an SCF-dependent erythrocytic progenitor cell line, G1E-ER2,32 was assayed using thymidine incorporation. The presence of soluble and MA SCF activity in stable stromal cell transfectants was analyzed using a coculture assay. On the day before assay, stromal cells were treated with 5 μg/mL mitomycin C, then washed 3 times in PBS, counted, and seeded at 3 × 104cells/well in 0.1% gelatin-coated 96-well plates. These cultures were incubated in DMEM, 10% CS at 37°C. After 24 hours, 5 × 104 G1E-ER2 cells were added to the stromal cells and cultured for 24 to 48 hours. Subsequently, 1.0 μCi of [3H] thymidine was added to each well for 6 to 8 hours at 37°C. Cells were then harvested using an automated cell harvester (96-well harvester; Brandel, Gaithersburg, MD), and thymidine incorporation was determined in a scintillation counter.
MAP-Kinase and Akt activation by stromal cells expressing either the wt-SCF220 or theSl17H-SCF220 isoform of SCF.
Activation of mitogen-activated protein (MAP)-Kinase (Erk-1 and Erk-2) was determined by utilizing a phospho-specific MAP-Kinase antibody (Thr202/Tyr204; New England Biolabs, Beverly, MA). The antibody detects Erk-1 and Erk-2 MAP-Kinase only when they are catalytically activated by phosphorylation at Thr202 and Tyr204. Activation of the phosphatidylinositol 3 (PI-3) Kinase/Akt pathway was determined by using a phospho-specific Akt (Ser 473) antibody (New England Biolabs). Akt is a downstream signaling molecule from PI-3Kinase.33-35 Briefly, Sl/Sl4 cells expressing either the wt-SCF220 or theSl17H-SCF220 were treated as described above. These cells were washed and plated on 6-well gelatin-coated plates (1 × 106/well) and cultured for 36 to 48 hours. A c-Kit+, SCF-responsive erythroid progenitor cell line, G1E-ER2, was factor-starved for 6 to 8 hours in medium containing 1 mg/mL BSA and Iscove’s modified Dulbecco’s medium (IMDM). Subsequently, 6 to 8 × 106cells were loaded onto stromal cells expressing either the wt-SCF220 or theSl17H-SCF220 SCF and were further cocultured for various time points at 37°C. Thereafter, cells were harvested and lysed in lysis buffer (10 mmol/L K2HPO4, 1 mmol/L EDTA, 5 mmol/L EGTA, 10 mmol/L MgCl2, 1 mmol/L Na2VO4, 50 mmol/L β-glycerol-phosphate, 10 μg/mL leupeptin, 1 μg/mL pepstatin, and 10 μg/mL aprotinin) at 4°C for 30 minutes. Cell lysates were clarified by centrifuging for 30 minutes at 10,000g at 4°C. Western blot analyses were performed according to the manufacturer’s instructions (New England Biolabs).
RESULTS
Impaired hematopoiesis in Sl17H mutant mice.
Previous work (Brannan et al25) has shown that theSl17Hallele results in impaired melanocyte and germ cell development. At the molecular level, this result is due to a splicing defect caused by a mutation in the 3′ splice acceptor site of intron 7, specifically, a T→A transversion within the polypyrimidine tract in the 3′ splice acceptor site of intron 7 (Fig 1A).25 This point mutation impedes splicing; as a consequence, exon 8 is skipped and exon 7 is spliced directly to exon 9 (Fig 1B). A comparison of the wt andSl17H sequences showed that exon 8 is completely missing from Sl17H mRNA (Fig 1).25 Exon 8 begins 1 aa carboxy-terminal to the transmembrane domain and encodes 23 of 36 aa’s of the SCF cytoplasmic domain. The result of this deletion is a frameshift; only the first aa (lysine) of theSl17H cytoplasmic domain is read in frame; the next 27 aa’s are read in an alternative reading frame before a stop codon is encountered (Fig 1B).25
Schematic representation of the Sl17Hgenomic mutation. (A) Sl17H contains a T→A transversion in the 3′ splice acceptor site of intron 7. The mutation is marked with an asterisk (*). Lowercase letters indicate intron 7 sequences that are flanked on the 5′ end with exon 7 (▨) and at the 3′ end with exon 8 (). The short dotted line indicates abnormal splicing, which skips exon 8 sequences inSl17H. (B) Schematic representation of the wt andSl17H SCF protein. N, amino terminus;EC, extracellular domain; TM, transmembrane domain (▪▨); CD, cytoplasmic domain (▪); C, carboxy terminus. Boundaries for exons 7, 8, and 9 are indicated by arrows (↑). The gap (∨) in the Sl17H protein due to the absence of exon 8 sequences is indicated below the wt SCF protein. The last 36 and 28 aa’s encoded by the wt and theSl17H cDNAs, respectively, are shown below each of the schematic diagrams.
Schematic representation of the Sl17Hgenomic mutation. (A) Sl17H contains a T→A transversion in the 3′ splice acceptor site of intron 7. The mutation is marked with an asterisk (*). Lowercase letters indicate intron 7 sequences that are flanked on the 5′ end with exon 7 (▨) and at the 3′ end with exon 8 (). The short dotted line indicates abnormal splicing, which skips exon 8 sequences inSl17H. (B) Schematic representation of the wt andSl17H SCF protein. N, amino terminus;EC, extracellular domain; TM, transmembrane domain (▪▨); CD, cytoplasmic domain (▪); C, carboxy terminus. Boundaries for exons 7, 8, and 9 are indicated by arrows (↑). The gap (∨) in the Sl17H protein due to the absence of exon 8 sequences is indicated below the wt SCF protein. The last 36 and 28 aa’s encoded by the wt and theSl17H cDNAs, respectively, are shown below each of the schematic diagrams.
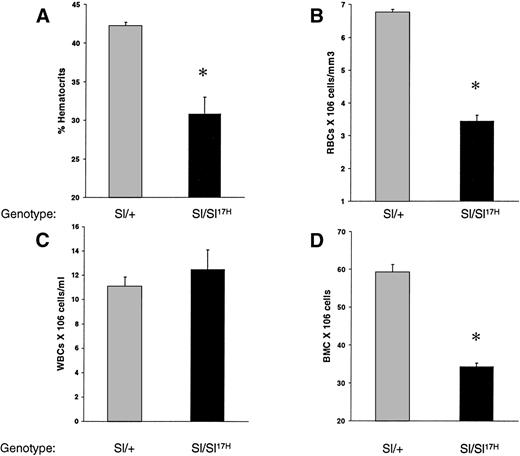
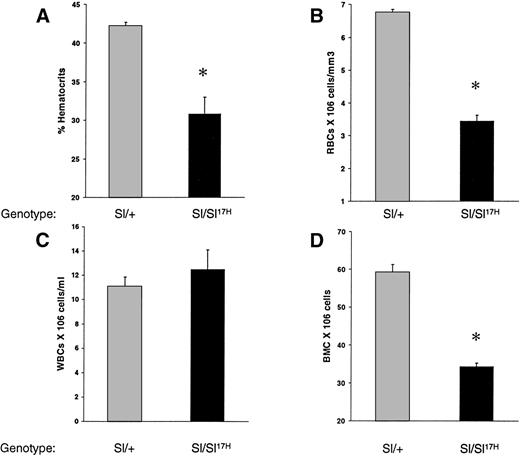
To examine the in vivo consequence(s) of the absence of normal cytoplasmic domain SCF aa’s and the presence of 28 novel aa’s in the cytoplasmic domain of the Sl17H-encoded SCF protein on hematopoiesis, we compared peripheral blood and BM in Sl/+ versus Sl/Sl17H compound heterozygotes. As shown in Fig 2, the Sl17H allele has a deleterious effect on hematopoiesis. Sl/Sl17Hmice demonstrate significantly lower hematocrit levels (Fig 2A), peripheral RBCs (Fig 2B), and BM cellularity (Fig 2D) compared withSl/+ mice. In contrast, peripheral WBC counts were comparable (Fig 2C).
A comparison of hematologic values between 12-to 14-week-old Sl/Sl17H and control Sl/+mice. (A) Hematocrit levels, (B) peripheral RBC counts, (C) peripheral WBC counts, and (D) BM cellularity. A minimum of 3 mice were examined in each group. Data shown are the mean ± SEM. *P < .05.
A comparison of hematologic values between 12-to 14-week-old Sl/Sl17H and control Sl/+mice. (A) Hematocrit levels, (B) peripheral RBC counts, (C) peripheral WBC counts, and (D) BM cellularity. A minimum of 3 mice were examined in each group. Data shown are the mean ± SEM. *P < .05.
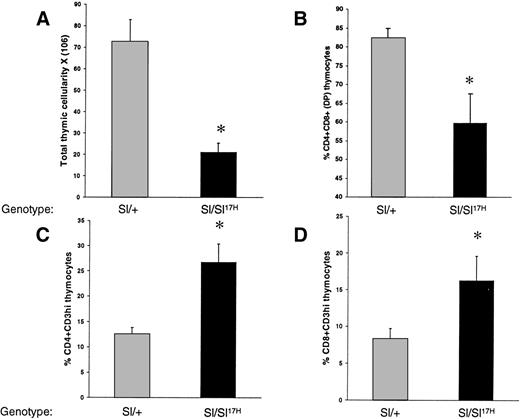
Recent studies have shown that c-Kit is expressed on thymocytes and that SCF/c-Kit interactions play an important role in T-cell development but not in mature T cells.9,36-38 In addition, our laboratory has shown that membrane presentation of SCF is critical for normal T-cell development.38 To determine if theSl17H-encoded protein affects thymopoiesis, we compared total thymic cellularity and thymocyte subset distribution inSl/+ and Sl/Sl17H mice. As seen in Fig 3A, Sl/Sl17H mice demonstrate a 70% decrease in total thymic cellularity in comparison to Sl/+ mice. Thymocytes from mutant mice were examined for maturation using the expression of CD4, CD8, and CD3. In comparison to control mice, mutant mice exhibit a significant reduction in the percentage of immature CD4+CD8+ double-positive (DP) thymocytes (Fig 3B). In addition, a small but significant increase in the percentage of CD4−CD8−double-negative (DN) thymocytes is also seen in these mice (data not shown). The percentage of CD4+ single-positive (SP) and CD8+ SP thymocytes is also significantly increased in these mice (data not shown). A reduction in the CD4+CD8+ T-cell subset and an increase in the SP thymocytes is consistent with a more mature phenotype (ie, CD4+CD3hi or CD8+CD3hi) of thymocytes in the mutant mice. To further examine T-cell maturation, we performed 2-color flow cytometric analysis on thymocytes using MoAbs directed against CD4 and CD3 or CD8 and CD3. As shown in Fig 3C and D, in comparison to controls, thymocytes from mutant mice demonstrate a significantly elevated percentage of cells of a mature CD4+CD3hi or CD8+CD3hiphenotype. These data suggest that the expression ofSl17H-SCF results in abnormal T-cell development.
A comparison of thymic cellularity and T-cell subset distribution between Sl/Sl17H and controlSl/+ mice. (A) Thymocytes were harvested and counted as described in Materials and Methods. At least 5 mice from each group were examined. Data shown are the mean ± SEM. *P < .05. Thymocytes from mutant and control mice were harvested and analyzed by flow cytometry for the expression of (B) CD4 and CD8, (C) CD4 and CD3, and (D) CD8 and CD3 antigens. The percentage of various T-cell subsets are indicated.
A comparison of thymic cellularity and T-cell subset distribution between Sl/Sl17H and controlSl/+ mice. (A) Thymocytes were harvested and counted as described in Materials and Methods. At least 5 mice from each group were examined. Data shown are the mean ± SEM. *P < .05. Thymocytes from mutant and control mice were harvested and analyzed by flow cytometry for the expression of (B) CD4 and CD8, (C) CD4 and CD3, and (D) CD8 and CD3 antigens. The percentage of various T-cell subsets are indicated.
Expression of Sl17H-SCF248 andSl17H-SCF220 cDNAs in stromal cells: Comparison with wt SCF cDNAs.
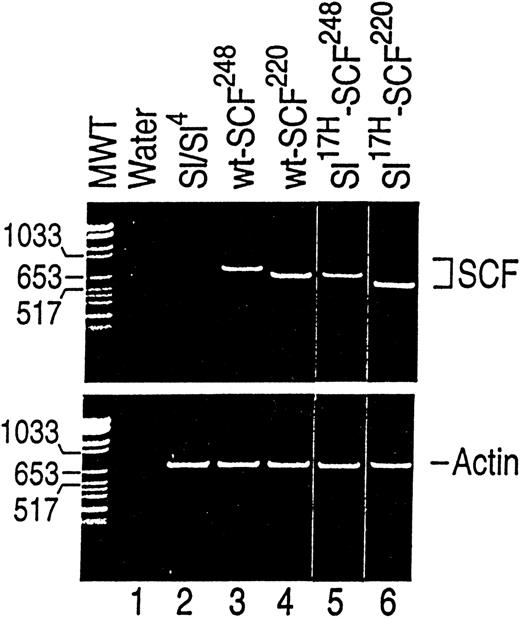
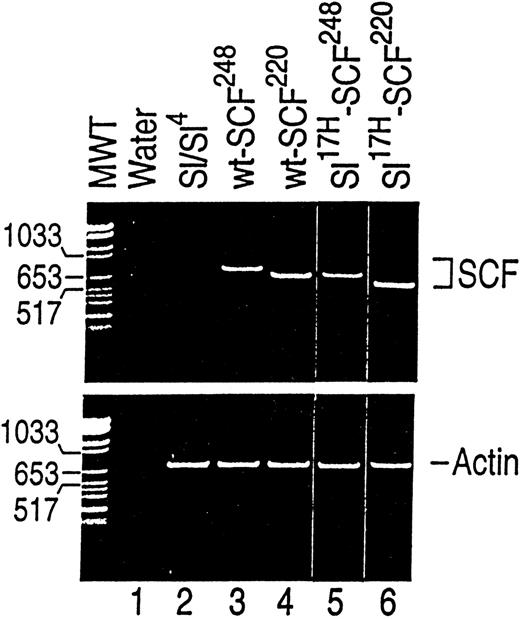
To further examine the biochemical, biological, and cellular abnormalities associated with the Sl17H-encoded protein, we isolated and cloned cDNAs representing theSl17H-SCF248 (soluble) and theSl17H-SCF220 (MA) SCF as described in Materials and Methods. wt and Sl17H cDNAs were cotransfected into Sl/Sl4 cell line with the hygromycin expression plasmid, p48. Hygromycin-resistant colonies were selected and analyzed for the expression of the introduced cDNAs using RT-PCR. The presence of RT-PCR transcripts of the predicted size was documented in at least 2 clones of each genotype. As shown in Fig 4 (and as expected), no band is present in Sl/Sl4 stromal cells derived from homozygous mice deleted of SCF coding sequence (Sl/Sl). Bands of 733 bp and 623 bp representing wt-SCF248 and wt-SCF220 can be seen in Fig 4 (lanes 3 and 4). Representative clones expressing shorter PCR products encoding Sl17H-SCF248 (665 bp; lane 5) and Sl17H-SCF220 (555 bp; lane 6) can also be seen in Fig 4. The bottom panel demonstrates RT-PCR products amplified using actin primers for RNA loading and integrity.
Analysis of wt and Sl17H mutant cDNA expression in stably transfected in stromal cells.Sl/Sl4 stromal cells were transfected with cDNAs encoding the wt-SCF248 (soluble) and wt-SCF220(MA) or the Sl17H-SCF248 (soluble) andSl17H-SCF220 (MA) mutant isoforms of SCF. SCF-specific primers (upper panel) were used that amplify both the wt and the Sl17H mutant forms of SCF. For semiquantitative analysis and RNA integrity, actin-specific primers (lower panel) were used. RT-PCR products were examined on a 2.5% ethidium bromide-containing agarose gel. Upper panel, SCF-specific primers. Lane 1, water control; lane 2, parentalSl/Sl4 cell line; lane 3, wt-SCF248(733 bp); lane 4, wt-SCF220 (623 bp; exon 6); lane 5,Sl17H-SCF248 (665 bp; exon 8); and lane 6, Sl17H-SCF220 (555 bp; exons 6 and 8). Lower panel, actin-specific primers (732 bp). The molecular weight (MW) marker is shown on the left.
Analysis of wt and Sl17H mutant cDNA expression in stably transfected in stromal cells.Sl/Sl4 stromal cells were transfected with cDNAs encoding the wt-SCF248 (soluble) and wt-SCF220(MA) or the Sl17H-SCF248 (soluble) andSl17H-SCF220 (MA) mutant isoforms of SCF. SCF-specific primers (upper panel) were used that amplify both the wt and the Sl17H mutant forms of SCF. For semiquantitative analysis and RNA integrity, actin-specific primers (lower panel) were used. RT-PCR products were examined on a 2.5% ethidium bromide-containing agarose gel. Upper panel, SCF-specific primers. Lane 1, water control; lane 2, parentalSl/Sl4 cell line; lane 3, wt-SCF248(733 bp); lane 4, wt-SCF220 (623 bp; exon 6); lane 5,Sl17H-SCF248 (665 bp; exon 8); and lane 6, Sl17H-SCF220 (555 bp; exons 6 and 8). Lower panel, actin-specific primers (732 bp). The molecular weight (MW) marker is shown on the left.
To examine if the Sl17H-encoded protein is secreted from stromal cells, we compared the biosynthesis of the wt-SCF248 and Sl17H-SCF248proteins. Sl/Sl4 stable stromal cell transfectants expressing similar levels of RNA (shown in Fig 4) were pulsed with [35S]-Met and chased for various time points. Subsequently, conditioned medium consisting of secreted SCF and cell lysates were harvested for immunoprecipitation. The immunoprecipitated proteins were analyzed on SDS-PAGE gels. Figure 5 compares the time course of protein synthesis of secreted (left panel) and cell-associated (right panel) SCF from stromal cells expressing either the wt or theSl17H protein. As seen in the left panel, a 30-kD glycosylated SCF product is secreted from cells expressing both the wt and the Sl17H SCF. As previously reported and as shown in Fig 5 (right panel), the cell-associated primary SCF translation products of both wt and Sl17HcDNAs are progressively processed primarily by carbohydrate modification as they are transported through the endoplasmic reticulum and the Golgi compartments.14 In addition to small molecular mass proteins representing the unglycosylated forms of SCF, with time, a 43-kD mature cell-associated SCF product is observed in both the wt and the Sl17H stromal cell transfectants, as seen in the right hand panel (Fig 5). These data suggest that theSl17H–SCF protein is proteolytically processed and secreted by stromal cells in a fashion similar to wt protein, whereas the cell-associated form of Sl17 -encoded protein is clearly made and contained in stromal cells.
Comparison of the biosynthesis of wt-SCF248(soluble) and Sl17H-SCF248 (soluble) isoforms of SCF. Stable stromal cell transfectants were starved of methionine-free medium, labeled with 0.5 mCi/mL of [35S]-methionine, and chased for 0, 1, 2, and 3 hours with cold medium. Conditioned medium was collected from each cell line. Immunoprecipitation was performed using a rabbit polyclonal antibody. Synthesis of soluble (left panel) and cell-associated (right panel) fractions of SCF from stromal cells transfected with cDNAs listed on the left are shown. A 30-kD glycosylated SCF product is secreted from cells expressing the two cDNAs (left panel). In addition to small molecular mass proteins representing the unglycosylated forms of SCF, with time, a 43-kD mature cell-associated SCF product is observed in both the wt and the Sl17H stromal cell transfectants, as seen in the right-hand panel.
Comparison of the biosynthesis of wt-SCF248(soluble) and Sl17H-SCF248 (soluble) isoforms of SCF. Stable stromal cell transfectants were starved of methionine-free medium, labeled with 0.5 mCi/mL of [35S]-methionine, and chased for 0, 1, 2, and 3 hours with cold medium. Conditioned medium was collected from each cell line. Immunoprecipitation was performed using a rabbit polyclonal antibody. Synthesis of soluble (left panel) and cell-associated (right panel) fractions of SCF from stromal cells transfected with cDNAs listed on the left are shown. A 30-kD glycosylated SCF product is secreted from cells expressing the two cDNAs (left panel). In addition to small molecular mass proteins representing the unglycosylated forms of SCF, with time, a 43-kD mature cell-associated SCF product is observed in both the wt and the Sl17H stromal cell transfectants, as seen in the right-hand panel.
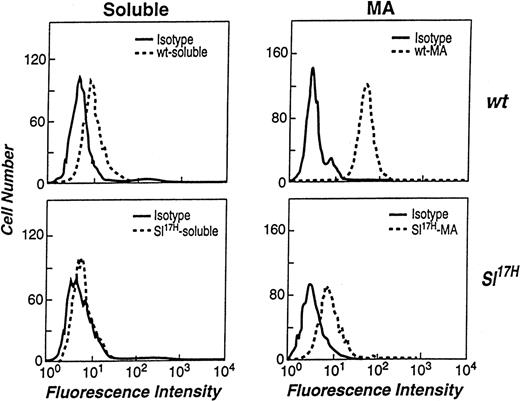
We and others have previously demonstrated that both soluble and MA isoforms of SCF can be detected on the cell surface by flow cytometry.14,21,39 However, the levels of cell surface expression of the 2 isoforms differ significantly, due to rapid proteolytic processing of the soluble SCF in comparison to the MA isoform.14,15 39 To examine if the presence of novel aa in the cytoplasmic domain of the Sl17H protein affects the cell surface expression of SCF, we compared wt andSl17H stromal cell transfectants by flow cytometry. At least 2 clones of stromal cell lines expressing either the wt or theSl17H SCF isoforms were stained with an anti-SCF antibody and analyzed. The left panels in Fig 6 demonstrate that the cell surface expression of soluble Sl17H-SCF248(lower panel) is undetectable in comparison to soluble wt-SCF248 (upper panel) in representative clones of each genotype. Because we have previously demonstrated a critical role for MA SCF in normal erythropoiesis, we next compared the cell surface expression of the MA wt-SCF220 andSl17H-SCF220 isoforms. Compared with wt-SCF220 expression (upper panel), a significant reduction in the surface expression ofSl17H-SCF220 (lower panel) is seen inSl/Sl4 stromal cell transfectants. Shown are representative clones of each genotype. These data suggest that the surface presentation of Sl17H is defective, despite apparently normal biosynthesis and half-life.
Flow cytometric analysis of the stromal cell surface expression of wt-SCF248 and wt-SCF220 orSl17H-SCF248 andSl17H-SCF220 isoforms of SCF. (Top left panel) wt-SCF248 (soluble), (bottom left panel)Sl17H-SCF248 (soluble), (top right panel) wt-SCF220 (MA), and (bottom right panel)Sl17H-SCF220 (MA) stromal cell lines were stained with either isotype control (solid lines) or antimouse SCF (dotted lines) antibody. Shown is 1 of 2 clones for each genotype showing similar results.
Flow cytometric analysis of the stromal cell surface expression of wt-SCF248 and wt-SCF220 orSl17H-SCF248 andSl17H-SCF220 isoforms of SCF. (Top left panel) wt-SCF248 (soluble), (bottom left panel)Sl17H-SCF248 (soluble), (top right panel) wt-SCF220 (MA), and (bottom right panel)Sl17H-SCF220 (MA) stromal cell lines were stained with either isotype control (solid lines) or antimouse SCF (dotted lines) antibody. Shown is 1 of 2 clones for each genotype showing similar results.
Impaired biological activity of the Sl17H protein expressed in hematopoietic microenvironment derived stromal cells.
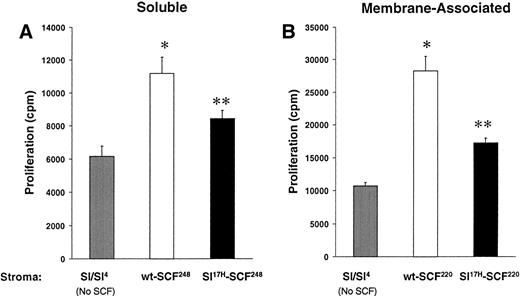
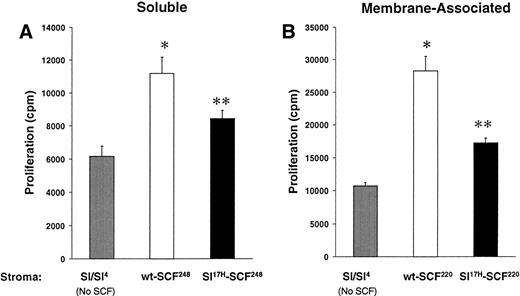
These data suggest that impaired erythropoiesis in vivo inSl17H mice may be due to reduced membrane SCF presentation. To examine the biological consequences of expression of the Sl17H protein in more detail, we compared the proliferation of an SCF-dependent erythrocytic progenitor cell line, G1E-ER2, in response to stromal cell presentation of wt andSl17H SCF isoforms. Analysis of the SCF-induced proliferation was performed using a thymidine incorporation assay. G1E-ER2 cells were cocultivated with mitomycin C-treated stromal cell transfectants expressing either the wt or the Sl17Hsoluble or MA protein. Figure 7A and B demonstrate increased proliferation of G1E-ER2 cells in response to stromal cell transfectants expressing either wt orSl17H isoforms compared withSl/Sl4 stromal cells. However, expression of either the soluble or the MA Sl17H protein in stromal cells results in significantly less proliferation of G1E-ER2 cells in comparison to wt SCF isoforms (Fig 7A and B). These data suggest that presentation of the Sl17H-encoded protein by stromal cells is associated with diminished proliferation due to either defective SCF/c-Kit interactions, reduced stromal cell presentation, or both.
A comparison of the biological activity of wt-SCF248 and wt-SCF220 withSl17H-SCF248 andSl17H-SCF220 isoforms of SCF. G1E-ER2 cells were cocultured for 24 to 48 hours with parentalSl/Sl4 stromal cells or (A) stable stromal cell transfectants expressing wt-SCF248 orSl17H-SCF248 SCF or (B) stable stromal cell transfectants expressing wt-SCF220 orSl17H-SCF220 SCF. Proliferation was measured by thymidine incorporation assay. Results show the mean ± SEM of a representative experiment performed at least twice with more than 1 clone in replicates of 6. *P < .05Sl/Sl4 v wt (SCF248 and SCF220); **P < .05 Sl/Sl4 v Sl17H (SCF248 and SCF220); and P < .05 wt (SCF248 and SCF220) v Sl17H(SCF248 and SCF220). Note difference in scale between (A) and (B).
A comparison of the biological activity of wt-SCF248 and wt-SCF220 withSl17H-SCF248 andSl17H-SCF220 isoforms of SCF. G1E-ER2 cells were cocultured for 24 to 48 hours with parentalSl/Sl4 stromal cells or (A) stable stromal cell transfectants expressing wt-SCF248 orSl17H-SCF248 SCF or (B) stable stromal cell transfectants expressing wt-SCF220 orSl17H-SCF220 SCF. Proliferation was measured by thymidine incorporation assay. Results show the mean ± SEM of a representative experiment performed at least twice with more than 1 clone in replicates of 6. *P < .05Sl/Sl4 v wt (SCF248 and SCF220); **P < .05 Sl/Sl4 v Sl17H (SCF248 and SCF220); and P < .05 wt (SCF248 and SCF220) v Sl17H(SCF248 and SCF220). Note difference in scale between (A) and (B).
Impaired MAP-Kinase and PI-3Kinase activation of c-Kit+ erythroid progenitors by MA isoform ofSl17H.
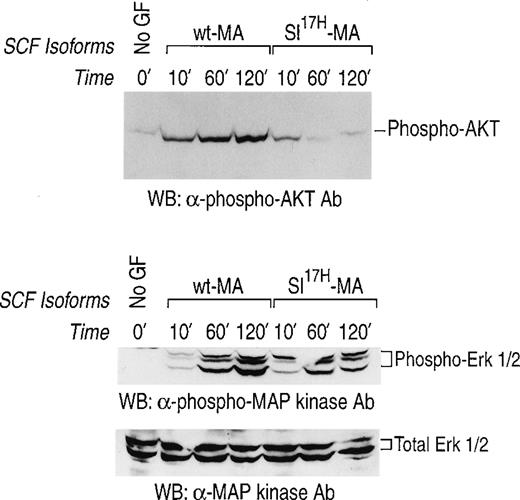
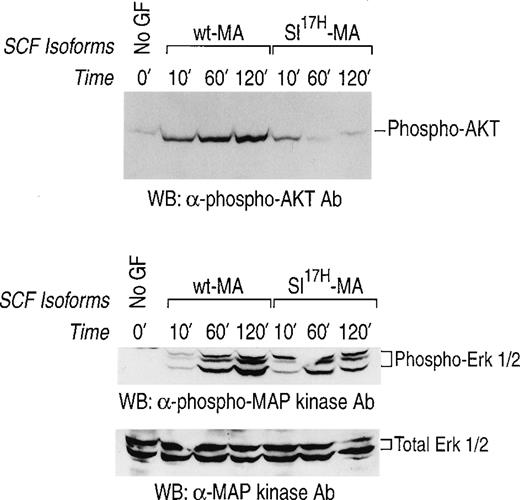
We have previously demonstrated prolonged activation of c-Kit in response to stromal cell presentation of MA SCF.21,40 To examine the consequence(s) of impaired membrane presentation of theSl17H protein by stromal cells on c-Kit signaling, we examined the activation of the downstream signaling pathways, MAP-Kinase and PI-3Kinase/Akt, in G1E-ER2 cells. Activation of the MAP-kinase pathway was determined by examining the phosphorylation of Erk-1 and Erk-2. Activation of PI-3Kinase was determined by examining the phosphorylation of Akt, a downstream effector of PI-3Kinase.33-35 The G1E-ER2 cells were cocultured on stromal cells expressing either the wt-MA or theSl17H-MA SCF for various time points, and cellular lysates were analyzed. As shown in Fig 8, coculture of erythroid progenitors on wt-MA results in enhanced and sustained activation of Akt (upper panel) and of Erk-1 and Erk-2 (middle panel) over a period of 120 minutes. Coculturing G1E-ER2 cells on stromal cells expressing the Sl17H-MA results in significant reduction of Akt activation (upper panel) and Erk-1 and Erk-2 (middle panel) activation in comparison to wt SCF. The bottom panel represents total protein in each lane. These experiments were performed at least 3 times using 2 different clones of each genotype with similar results.
Activation of MAP-Kinase and PI-3Kinase/Akt in erythroid progenitors by stromal cells expressing either the wt-SCF220 (MA) orSl17H-SCF220 (MA) isoform of SCF. Factor-starved erythroid progenitors were cocultured with mitomycin C-treated stromal cells expressing either the wt or theSl17H MA SCF for various time points. Subsequently, at various times, cell lysates were collected and subjected to Western blot analysis with a rabbit anti-phospho Akt antibody and a rabbit anti-phospho MAP-Kinase antibody and the enhanced chemiluminescence detection system. The phosphorylated form of Akt (upper panel) and MAP-Kinase (Erk-1 and Erk-2) (middle panel) are indicated. In both the upper and middle panels, lane 1 corresponds to unstimulated starved erythroid progenitor cells and lanes 2, 3, and 4 correspond to stromal cells expressing the wt-SCF220 (MA) form of SCF cocultured with erythroid progenitor cells for 10, 60, and 120 minutes, respectively. Lanes 5, 6, and 7 correspond to stromal cells expressing the Sl17H-SCF220 (MA) form of SCF cocultured with erythroid progenitor cells for 10, 60, and 120 minutes, respectively. The bottom panel demonstrates the loading control for total protein in each lane. These experiments were performed at least 3 times with 2 different clones of each genotype.
Activation of MAP-Kinase and PI-3Kinase/Akt in erythroid progenitors by stromal cells expressing either the wt-SCF220 (MA) orSl17H-SCF220 (MA) isoform of SCF. Factor-starved erythroid progenitors were cocultured with mitomycin C-treated stromal cells expressing either the wt or theSl17H MA SCF for various time points. Subsequently, at various times, cell lysates were collected and subjected to Western blot analysis with a rabbit anti-phospho Akt antibody and a rabbit anti-phospho MAP-Kinase antibody and the enhanced chemiluminescence detection system. The phosphorylated form of Akt (upper panel) and MAP-Kinase (Erk-1 and Erk-2) (middle panel) are indicated. In both the upper and middle panels, lane 1 corresponds to unstimulated starved erythroid progenitor cells and lanes 2, 3, and 4 correspond to stromal cells expressing the wt-SCF220 (MA) form of SCF cocultured with erythroid progenitor cells for 10, 60, and 120 minutes, respectively. Lanes 5, 6, and 7 correspond to stromal cells expressing the Sl17H-SCF220 (MA) form of SCF cocultured with erythroid progenitor cells for 10, 60, and 120 minutes, respectively. The bottom panel demonstrates the loading control for total protein in each lane. These experiments were performed at least 3 times with 2 different clones of each genotype.
Expression of membrane-restricted SCF improves erythroid deficiency in Sl/Sl17H mice.
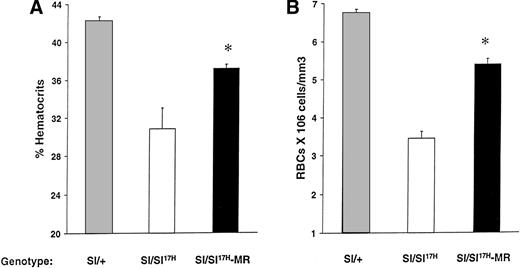
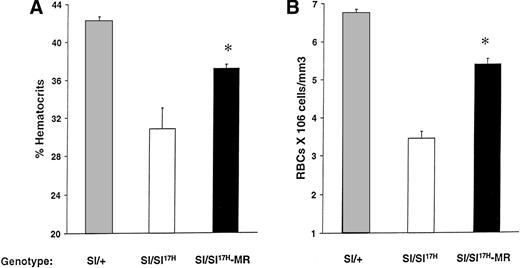
Because the above-mentioned studies demonstrate impaired cell surface presentation of the Sl17H-SCF, we hypothesized that the expression of the MR form of SCF (SCFX9/D3) should improve the phenotypic abnormalities in these mice. To test this hypothesis, we performed genetic crosses of transgenic mice expressing SCFX9/D3 cDNA into Sl/Sl17H mutants to generate Sl/Sl17H-MR mice. We have previously used this approach to examine the effects of SCFX9/D3 transgene is Sl/Sld mutants.21 As shown in Fig 9, expression of the MR-SCF transgene in Sl/Sl17H mice significantly improves their hematocrits (Fig 9A) and peripheral RBC counts (Fig 9B) in comparison to Sl/Sl17H mice. However, in comparison toSl/+ mice, the expression of SCFX9/D3 transgene did not completely correct these deficiencies, a result that may relate to the level or timing of transgene expression during development. No effects were seen on the coat color, a result similar to our findings when wt SCF isoforms were expressed in Sldmice.21
Rescue of erythroid lineage deficiencies by transgene expression of SCFX9/D3 (MR) in Sl/Sl17Hmice. A comparison of (A) hematocrit levels and (B) peripheral RBC counts between Sl/+, Sl/Sl17H, andSl/Sl17H-MR mice. A minimum of 5 Sl/+, 3Sl/Sl17H, and 6 Sl/Sl17H-MR mice were examined in each group. Results show the mean ± SEM. *P < .05.
Rescue of erythroid lineage deficiencies by transgene expression of SCFX9/D3 (MR) in Sl/Sl17Hmice. A comparison of (A) hematocrit levels and (B) peripheral RBC counts between Sl/+, Sl/Sl17H, andSl/Sl17H-MR mice. A minimum of 5 Sl/+, 3Sl/Sl17H, and 6 Sl/Sl17H-MR mice were examined in each group. Results show the mean ± SEM. *P < .05.
DISCUSSION
Despite significant molecular and biochemical characterization of SCF, little is known regarding the physiological role(s) of soluble and MA SCF isoforms. Sld, a viable Sl mutant, arose as result of an an intragenic deletion removing the exons encoding the transmembrane and cytoplasmic domains of SCF.23 The Sld allele appears to be capable of producing biologically active secreted protein, although the truncated protein lacks both exon 7- and exon 8-encoded membrane-proximal aa’s and is therefore not proteolytically processed in the same fashion as the wt isoform of soluble SCF.16 The severe hematologic deficiencies in compound heterozygousSl/Sld mice have suggested that the MA isoform of SCF is critical in c-Kit function, and the cytoplasmic domain of SCF is potentially important in mediating juxtracrine signaling.
We have used the naturally occurring mutation Sl17Hto analyze the biological importance of the SCF cytoplasmic domain as it relates to cell surface presentation, biosynthetic processing, bioactivity, and juxtracrine signaling by using in vitro and in vivo genetic approaches. The major conclusions of our study are (1) the cytoplasmic domain sequences of SCF are important for cell surface presentation and is abnormal in Sl17H mice; (2) membrane presentation is important for c-Kit/SCF–mediated juxtacrine signaling, which is impaired in Sl17H mice; (3) membrane presentation of SCF is critical for the proliferation/survival of c-Kit–expressing hematopoietic cells and is also defective inSl17H mice; and (4) Sl/Sl17Hmice are deficient in RBC production, suggesting a critical role for membrane presentation in normal erythroid blood cell development.
Our findings that the wt sequences in the cytoplasmic domain of SCF are required for expression of SCF on the cell surface are in agreement with reports showing several soluble and membrane-anchored proteins in which cytoplasmic domain mutations interfere with cell surface expression. A single point mutation in the cytoplasmic domain of both the membrane-anchored P-glycoprotein and the c-Kit receptor causes these proteins to be inefficiently processed.41,42 In addition, mutations of the COOH-terminal cytoplasmic residues of the α-proteinase inhibitor or the thyroxine-binding globin also causes these proteins to be inefficiently processed.43 Proteins with cytoplasmic domain mutations may not interact properly with molecules involved in transport from the endoplasmic reticulum (ER) to the cell surface. Also, incorrect folding as a consequence of mutations in the cytoplasmic domain may be responsible for the observed defects on maturation.44 45
We find soluble SCF in the supernatant of HM-derived stable stromal cell transfectants expressing the Sl17H mutant SCF by IP at levels comparable to that of wt SCF. However, our in vitro biological data suggest that stromal cells making soluble SCF are less capable of stimulating the proliferation/survival of c-Kit+cells. These data suggest that the altered cytoplasmic domain sequences of the mutant protein do not eliminate the secretion of soluble SCF, although they do impair the biological activity of the mutant protein. In this regard, it is possible that the presence of novel aa’s in the cytoplasmic tail of the mutant protein prevents SCF monomers from efficiently forming dimers. Some evidence suggests that bivalent binding of ligands is key for inducing receptor dimerization and subsequently receptor activation.46 Recent studies using radiolabeled SCF added to serum have shown that 49% to 72% of the circulating SCF exists in monomeric form.47 Furthermore, a detailed examination of dimerization-defective variants of SCF show substantially reduced mitogenic activity, whereas the activity of disulfide-linked SCF dimer was 10-fold higher than that of wt SCF.47 These results suggest a correlation between dimerization capacity and biological activity.
Based on these findings, one would predict that SCF and other cytokines that are synthesized as membrane-anchored proteins are expressed on the cell surface as dimers to efficiently activate their cognate receptors. Indeed, recent biochemical studies have shown that most of the membrane SCF exists as a dimeric protein, and the presence of novel aa’s in theSl17Hmutant may result in inefficient dimerization and reduced adhesion of mast cells to COS cells, in comparison to wt SCF COS cell transfectants.48 However, the interpretation of these data may not be straightforward. These experiments were performed on membrane fractions derived from COS cell transfectants that already expressed 50% less cell surface SCF, making it difficult to conclude if the diminished Sl17H dimerization in COS cell transfectants is due to a reduction in total cell surface protein or due to an actual impairment in dimerization as a result of the presence of 28 novel aa’s in the cytoplasmic domain.
An important determinant for anchoring proteins in the membrane is the presence of positively charged aa’s located immediately adjacent to the transmembrane region within the cytoplasmic domain.26,27 Mutations that reduce the net positive charge in this region can lead to membrane protein instability.27In the case of SCF, the first 3 aa’s of the cytoplasmic domain are Lys-Lys-Lys (+3). In the Sl17H protein, these aa’s are Lys-Tyr-Ala (+1), a net reduction in positive charge. This reduction in positive charge could significantly reduce the stability of SCF within the cell membrane and, consequently, its ability to interact with c-Kit. Our flow cytometric data demonstrating reduced cell surface expression and biological data showing reduced activity by stromal cell transfectants expressing the soluble and MASl17H SCF would support this notion. In this regard, our results are consistent with previously published studies in which up to 50% reduction in the amount of cell surfaceSl17H mutant SCF in COS cells and in primary BM-derived stromal cells was reported.48 49 Our data, along with the observations made by others, would suggest that a reduction in the function of cell surface expressed Sl17H mutant SCF is due to reduced SCF membrane presentation. Importantly, the apparent decrease in PI-3Kinase/Akt activation and MAP-Kinase activation in erythroid progenitor cell line by the mutant MA SCF further supports the notion that appropriate presentation of SCF is necessary for full activation of 2 important signaling pathways downstream from Kit involved in cell proliferation.
The Sl17H mutation has previously been shown to affect melanogenesis and gametogenesis.25 In addition,Sl17H homozygous mutants (Sl17H/Sl17H) also demonstrate reduced mast cells and impaired homing of stem/progenitor cells to the spleen.48 In this report, we demonstrate that BM cellularity, peripheral RBC counts, hematocrit levels, and T-cell development were appreciably affected in Sl/Sl17Hcompound heterozygous mice. The reduced cell surface presentation we observed likely contributes to these hematopoietic deficiencies. We and others have previously shown that Sld mice also suffer from impaired erythropoiesis. It has been hypothesized that this deficiency is due to the lack of MA SCF in Sldmice. In this regard, we have recently shown thatSld mice expressing MR SCF as a transgene produce significantly more RBCs in comparison to Sld mice or Sld mice expressing the soluble form of SCF.21 Recent work by Tajima et al24 have confirmed that soluble SCF is not required for RBC production. We observed reduced cell surface presentation of theSl17H-encoded protein, and a significant decrease in the proliferation and activation of downstream signaling molecules, in an erythrocytic progenitor cell line by Sl17H MA SCF. These data, along with the correction in RBC deficiency seen inSl/Sl17H mice expressing the MR form of SCF, suggest a critical role for the cell-surface presentation of SCF in normal erythropoiesis.
R.K. was initially supported by a postdoctoral fellowship from the Leukemia Society of America and is currently being supported by a National Institutes of Health (NIH) postdoctoral fellowship (NRSA F32 DK09752). Supported by grants from the NIH to D.A.W. (RO1DK48605).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Reuben Kapur, PhD, Herman B Wells Center for Pediatric Research, Cancer Research Building, 1044 W Walnut, Room 402C, Indianapolis, IN 46202-5225; e-mail: rkapur@iupui.edu.



![Fig. 5. Comparison of the biosynthesis of wt-SCF248(soluble) and Sl17H-SCF248 (soluble) isoforms of SCF. Stable stromal cell transfectants were starved of methionine-free medium, labeled with 0.5 mCi/mL of [35S]-methionine, and chased for 0, 1, 2, and 3 hours with cold medium. Conditioned medium was collected from each cell line. Immunoprecipitation was performed using a rabbit polyclonal antibody. Synthesis of soluble (left panel) and cell-associated (right panel) fractions of SCF from stromal cells transfected with cDNAs listed on the left are shown. A 30-kD glycosylated SCF product is secreted from cells expressing the two cDNAs (left panel). In addition to small molecular mass proteins representing the unglycosylated forms of SCF, with time, a 43-kD mature cell-associated SCF product is observed in both the wt and the Sl17H stromal cell transfectants, as seen in the right-hand panel.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1915/5/m_blod41811005x.jpeg?Expires=1763732397&Signature=3PXRMgjIzwNNbVxFBMjvVxCGDtf3QZZTcennvNZUcKm9SVI7731C5W8hYvp7bXm1kRCCB52u3Acc8mRrKALM5opW17gQt4VOxxmmp9GU84BVkBqWUQjR6BlKAm3CKPGz4Kp5tqGLHwxNah8r9rqDU3DgKaocq32~yFwSD2fIi2uz2bpjYCW~It8zlsCZk~RtqrlENn3DG5lN31VdSMIIqLT~77XZ-d13Tidu4hofab1t-Ty9h0VTXoOnaEfL2pVBkhaep2otQUH0Dqf5g90vWDoc30PvIOWv-2bRXg4OU9wXePSBWD46JTeZs2~oM~c7CQi3wNpDq4q~F5bMVNg1tA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Fig. 5. Comparison of the biosynthesis of wt-SCF248(soluble) and Sl17H-SCF248 (soluble) isoforms of SCF. Stable stromal cell transfectants were starved of methionine-free medium, labeled with 0.5 mCi/mL of [35S]-methionine, and chased for 0, 1, 2, and 3 hours with cold medium. Conditioned medium was collected from each cell line. Immunoprecipitation was performed using a rabbit polyclonal antibody. Synthesis of soluble (left panel) and cell-associated (right panel) fractions of SCF from stromal cells transfected with cDNAs listed on the left are shown. A 30-kD glycosylated SCF product is secreted from cells expressing the two cDNAs (left panel). In addition to small molecular mass proteins representing the unglycosylated forms of SCF, with time, a 43-kD mature cell-associated SCF product is observed in both the wt and the Sl17H stromal cell transfectants, as seen in the right-hand panel.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1915/5/m_blod41811005x.jpeg?Expires=1763732398&Signature=Lb67RlpU40hxeL0TxEMjMnwV9DMChaa4bGrAUkZJCrnYTO1TeIqyvwwpAv9XQxRuBZu1fZdUmd7oIKpFveMD7h6Dy4eWRCHL72VfzSyZEmgbIsFkbpawNehI~DJ50jwwDHfmeRPhM35rmK0aIGWh8fGoX9KuPdU5GGcUd9QJvmekSIcUTtmMFzjVtkWumq3QiRnMNzf2vJoqV0f2kRMpy1VJjyFGfhXzlLjwJNDczL5dAdmQ9yeBP6gnVvlYmxjNI-FUi3Sx9kDWH-FS~14pgz2VR8pcwDRTdwtDgtljW4EvNjkmxO71NqdnRJJEdHWOwWGhxaHI1pdEpsZAxg~aTQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)