M-Ras, a recently identified homologue of p21 Ras, is widely expressed, with levels of the 29-kD protein in spleen, thymus, and NIH 3T3 fibroblasts equaling or exceeding those of p21 Ras. A G22V mutant of M-Ras was constitutively active and its expression in an interleukin-3 (IL-3)–dependent mast cell/megakaryocyte cell line resulted in increased survival in the absence of IL-3, increased growth in IL-4, and, at high expression levels, in factor-independent growth. Expression of M-Ras G22V, however, had a negative effect on growth in the presence of IL-3, suggesting that M-Ras has both positive and negative effects on growth. Expression of M-Ras G22V in NIH-3T3 fibroblasts resulted in morphological transformation and growth to higher cell densities. M-Ras G22V induced activation of thec-fos promoter, and bound weakly to the Ras-binding domains of Raf-1 and RalGDS. Expression of a mutant of M-Ras G22V that was no longer membrane-bound partially inhibited (40%) activation of thec-fos promoter by N-Ras Q61K, suggesting that M-Ras shared some, but not all, of the effectors of N-Ras. An S27N mutant of M-Ras, like the analogous H-Ras S17N mutant, was a dominant inhibitor of activation of the c-fos promoter by constitutively active Src Y527F, suggesting that M-Ras and p21 Ras shared guanine nucleotide exchange factors and are likely to be activated in parallel. Moreover, M-Ras was recognized by the monoclonal anti-Ras antibody Y13-259, commonly used to study the function and activity of p21 Ras. Mammalian M-Ras and a Caenorhabditis elegans orthologue exhibit conserved structural features, and these are likely to mediate activation of distinctive signaling paths that function in parallel to those downstream of p21 Ras.
THE H-, N-, AND K-Ras proteins, collectively termed here p21 Ras proteins, have been implicated in a wide variety of cellular responses, ranging from proliferation and differentiation1,2 to cell death.3 They are encoded by 3 genes: Harvey (H), N, and Kirsten (K), which is alternatively spliced and generates 2 proteins: K-RasA and K-RasB. The p21 Ras proteins are attached to the inner leaflet of the plasma membrane by carboxy-terminal lipid modifications. They function as biological switches and, upon hydrolysis of bound guanosine triphosphate (GTP) to guanosine diphosphate (GDP), undergo allosteric changes in 2 adjacent, highly conserved regions of the molecule, termed “switch I” (residues 32-38 in H-Ras) and “switch II” (residues 60-76 in H-Ras).4,5 In their active, GTP-bound state, the p21 Ras proteins bind a series of effector molecules that include serine/threonine kinases such as Raf-1, A-Raf, B-Raf,6,7 and MEKK18; the p110 catalytic subunit of PI-3 kinase9; and Ral-GDS.10 Interaction of p21 Ras-GTP with all of these effectors involves its “effector loop” that extends from residues 32-40 and encompasses switch I, although genetic and x-ray crystallographic analyses indicate that the mechanisms through which these different effectors interact with p21 Ras are structurally distinct.11 12
Activating mutations of p21 Ras occur in some 20% to 30% of acute myeloid leukemias (AML) and myelodysplastic syndromes,13and can reach frequencies as high as 90% in pancreatic carcinomas.14 However, in contrast to the results of experiments in NIH-3T3 fibroblasts, where expression of activated mutants of p21 Ras results in transformation and reduced dependence on growth factors, expression of activated mutants of p21 Ras in cells of hematopoietic origin, for example in AML blasts, does not in general confer factor-independence.13 Ablation of functional K-Ras genes in mice results in anemia and intrauterine death, indicating that p21 K-Ras proteins have a critical role in the development of the hematopoietic system.15
Clues to the function of p21 Ras proteins in normal hematopoietic cells have come from experiments suggesting that p21 Ras proteins are activated by ligation of receptors for growth factors,16,17antigen,18,19 or other ligands, and that inhibition of the activation of p21 Ras can block the growth20 or development of lymphohematopoietic cells.21,22 This evidence has largely depended on two research tools: the monoclonal anti-Ras antibody Y13-259, used to measure activation of p21 Ras and to inhibit its function, and dominant-negative mutants such as S17N Ras, used to inhibit activation of p21 Ras. Y13-259 binds to p21 Ras isoforms via an epitope in switch II,23 thus blocking their interaction with Raf-1,24 and inhibits growth when microinjected into cells.25 The standard assay for assessing activation of p21 Ras is based on immunoprecipitation with Y13-259 and measurement of the ratio of GTP:GDP in the precipitate, and is the basis of reports that have concluded that p21 Ras is activated in cells of hematopoietic origin by hematopoietic growth factors16,17 or by ligation of antigen receptors.18,19 Dominant negative mutants such as p21 Ras S17N have an increased affinity for p21 Ras GDP26 and are thought to sequestrate guanine nucleotide exchange factors (GEFs) and, thus, block activation of endogenous p21 Ras. Expression of p21 Ras S17N has been used to implicate p21 Ras in the development of B and T lymphocytes21,22 as well as in signaling downstream of oncogenes, growth factors, cytokines, and other ligands.20 26-28
The p21 Ras proteins are members of a larger family that also includes R-Ras and R-Ras2/TC21.29,30 Two recent reports describe a new member of this family termed M-Ras,31 referring to its cloning from muscle, or R-Ras3,32 on the basis of homology with R-Ras and R-Ras2. Mutants with decreased GTPase activity induced the formation of microspikes in Swiss 3T3 cells,31 focus formation and anchorage-independent growth in NIH 3T3 cells, and moderate activation of Erk (extracellular signal-regulated kinase).32 Both groups reported that the pattern of expression was restricted, to brain and heart,32 or to brain and skeletal muscle.31
Here we show that in contrast to these reports, expression of M-Ras is not restricted to brain or muscle and occurs in thymus, spleen, and cell lines of hematopoietic origin, as well as in epithelial cells. Expression of constitutively active M-Ras in an IL-3–dependent hematopoietic cell line resulted in factor-independent growth. Moreoever, M-Ras could not be distinguished from p21 Ras in functional tests based on the monoclonal antibody Y13-259 or the dominant negative M-Ras mutant S27N. Analysis of the protein sequences of M-Ras and other members of the Ras-family showed that M-Ras displays distinctive motifs that differ from those found in R-Ras and R-Ras2 on the one hand, and p21 Ras on the other. Our data suggest that M-Ras is likely to be activated by the same stimuli that activate p21 Ras, but that it activates parallel pathways that regulate cellular growth.
MATERIALS AND METHODS
Materials.
Plasmids encoding the GST-fusion proteins GST-Raf-1-RBD and GST-RalGDS-RBD were kindly provided by Dr D. Shalloway (Cornell University, New York, NY) and Dr J. Bos (Utrecht University, Utrecht, The Netherlands), respectively, Src Y527F by Dr J. Cooper (Fred Hutchinson Center, Seattle, WA), and N-Ras Q61K by Dr R. Kay (The Terry Fox Laboratory, Vancouver, Canada). Biotinylated or native antibodies against the HA-epitope (12CA5) were purchased from Boehringer Mannheim (Boehringer Mannheim, Laval, Quebec, Canada) and Cy Chrome5-conjugated streptavidin from Pharminogen Canada (Mississauga, Ontario, Canada).
Generation of antisera.
Rabbits were injected with peptides corresponding to residues 187-204 (KKKTKWRGDRATGTHKLQ, pep I) and 142-159 (KEMATKYNIPYIETSAKD, pep II) of M-Ras, respectively. For immunizations both peptides were fused to the C-terminus of the tetanus toxoid epitope QYIKANSKFIGITEL.33For affinity purification of the polyclonal antisera, the peptides were covalently bound to NHS-activated Sepharose (Pharmacia Biotech, Uppsala, Sweden). Antibodies bound to the affinity-purification column were washed with 10 column volumes of phosphate-buffered saline (PBS), eluted with 3 mL of 100 mmol/L glycine (pH 2.5), and dialyzed against 2 L of PBS overnight.
Cell culture, transfections, and retroviral infections.
All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 2 mmol/L L-glutamine, 100 U penicillin/50 U streptomycin (Stem Cell Technologies, Vancouver, Canada), and 10% fetal calf serum R6-X cells (293-cells and BOSC23) or 10% calf serum (NIH3T3). R6-X, an interleukin-3 (IL-3)–dependent bipotential, mast cell/megakaryocyte line,34 was passaged in the presence of medium conditioned by the cell line WEHI-3B (2% of 10-fold concentrated medium, W3) as a source of IL-3. NIH 3T3 cells were kindly provided by Dr R. Kay. For retroviral infections, R6-X cells or NIH3T3 cells were incubated for 10 hours with retroviruses produced as described,35 in the presence of 10 μg/mL polybrene and, in the case of R6-X cells, 2% W3 and 2% of a 10-fold concentrate of medium conditioned by X063 cells secreting IL-4.36 After 10 hours, the R6-X cells were washed and resuspended in medium supplemented with IL-4 alone. In the case of 3T3 cells and, in some experiments, R6-X cells, infected clones were selected in puromycin at 1 μg/mL (Sigma, Mississauga, Ontario, Canada), and in the case of R6-X in the presence of 2% W3 as a source of IL-3. Analysis of growth and survival of polyclonal or clonal population of virally infected cells R6-X cells involved cell counting, assessment of uptake of 3,4,5-dimethyltiazole-2,5-diphenyltetrazolium bromide (MTT), or clonal growth in agar. For assessment of growth or survival using MTT, cells were plated at varying densities as indicated, in 96-well, flat-bottomed plates, with titrated amounts of growth factors as indicated. MTT uptake was assessed 3 to 5 days later. Colony assays were performed by plating cells in medium and indicated growth factors in a final concentration of 0.3% agar (Difco, Detroit, MI).34 R6-X cells were cloned by picking colonies from agar. In some cases, populations were enriched in enhanced green fluorescent protein (EGFP)-expressing cells by fluorescence-activated cell sorting using a FACSCalibur instrument (Becton Dickinson, San Jose, CA). Experiments were performed on retrovirally infected R6-X cells derived from 2 independent infections with consistent results.
Generation of mutations by site-directed mutagenesis.
The M-Ras cDNA was subcloned into pBluescript for site-directed mutagenesis and sequencing. Constructs were cloned with N-terminal myc- or HA-tags or C-terminal myc-tags into the eukaryotic expression vectors pEF-BOS or pcDNA3.1, or, for retroviral infections, into pMX-PIE (a gift of Dr Alice Mui, DNAX, San Francisco, CA). This vector drives expression of the EGFP from an internal ribosomal entry site (IRES) downstream of the Ras cDNA. Identities of all constructs were verified by DNA-sequencing.
Northern blot analysis and reverse transcription-polymerase chain reaction (RT-PCR).
Northern blots were performed according to the manufacturer’s instructions (CLONTECH, Palo Alto, CA) with a probe corresponding to the 120 carboxy-terminal amino acids of M-Ras. RT-PCR was performed on 2 μg of total RNA from the indicated cell lines. Oligonucleotides used were GTGAGTGCCGGTGGCCCTGTCTCCCTCGC (RT-reaction), TTTGGTTGCCATTTCTTTTCCTTGGTCCC (PCR antisense), ACCAGCGCTGTTCCAAGTGAAAAC- CTTCC (PCR sense), CACATATAAACTAGTAGTGGTGGGAGATGG (nested PCR sense), CTTAGGTGCATCAGATCCACCTTGTTG (nested PCR antisense). As negative control, the RT-reaction was performed on 2 μg of t-RNA. The identities of the amplified fragments were verified by a restriction digest.
Reporter gene assays.
293-cells were cotransfected with 0.25 to 0.5 μg of the luciferase reporter gene under control of the full-length c-fos promoter and the indicated plasmids. Cells were harvested 48 hours after transfection and lysed in 200 μL of lysis buffer (Promega, Madison, WI). Luciferase activity was determined with a luminometer (Dynex, Chantilly, VA) and the values normalized to total protein using BCA Protein Reagent A (Pierce, Rockford, IL).
Immunoblotting, immunoprecipitation, and affinity precipitation.
Transiently transfected 293-cells were washed twice in PBS and lysed in 500 μL of 50 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1% NP-40, 5 mmol/L EDTA, in the presence of 10 μg/mL each of leupeptin, aprotinin, soybean trypsin inhibitor, 0.7 μg/mL pepstatin, and 40 μg/mL phenylmethyl sulfonyl fluoride (PMSF). Denatured proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with ‘anti-HA’ antibody 12CA5 at 0.2 μg/mL or anti-Ras antibody Y13-259 at 15 μg/mL, as described.16 To decrease the Ig-light chain signal, we used secondary antibodies recognizing specifically the Ig-heavy chain. Affinity purifications to assay binding of M-Ras to the Ras-binding domains of Raf-1 and Ral-GDS were performed as described.37Briefly, GST-fusion proteins were affinity-purified with glutathione beads from lysates of Escherichia coli. Fifteen microliters of packed bead volume was mixed for 40 minutes with lysates of 293-cells expressing the indicated proteins in 25 mmol/L HEPES (pH 7.5), 150 mmol/L NaCl, 10% glycerol, 0.25% Na-deoxycholate, 1% NP-40, 25 mmol/L NaF, 10 mmol/L MgCl2, 1 mmol/L EDTA, 1 mmol/L Na vanadate, 10 μg/mL leupeptin, and 10 μg/mL aprotinin. The beads were washed with lysis buffer, boiled, and the eluted proteins resolved on 15% denaturing polyacrylamide gels. Cell fractionations were performed by suspending cells in homogenization-buffer (10 mmol/L TRIS-HCl [pH 7.5], 1 mmol/L EDTA, 10 μg/mL each of leupeptin, aprotinin, and soybean trypsin inhibitor, 0.7 μg/mL pepstatin, and 40 μg/mL PMSF). Cells were broken by brief sonication, the nuclei were pelleted, and the supernatant subjected to ultra-centrifugation for 20 minutes at 150,000g. The resulting supernatant was designated as cytosolic fraction (c) and the pelleted fraction as membrane enriched fraction (m).
RESULTS
Uniquely conserved characteristics of the C-terminus of M-Ras in mammals and Caenorhabditis elegans.
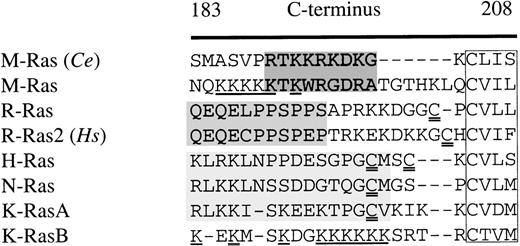
We identified a novel member of the Ras-family in the murine expressed sequence tag (EST) database. The cDNA encoded a small GTPase that was also described as M-Ras31 and R-Ras3.32 We also identified an orthologous protein in the C elegans database. M-Ras exhibits a high degree of homology with other members of the Ras-family, with 47% identity with H-Ras and 52% with R-Ras2. Although it is identical in the switch I region (residues 42-48), it displays only 3 conserved exchanges in the switch II region (residues 70-86). Significantly, none of the residues in switch II shown to be involved in binding of Y13-259 to p21 Ras23 differed between H-Ras and M-Ras. Comparison of the carboxy-terminal sequences of the murine and C elegans M-Ras orthologues with those of p21 Ras, R-Ras, and R-Ras2 indicated that M-Ras had distinctive characteristics and suggested that M-Ras was a discrete branch of this phylogenetic tree. Figure 1 shows alignments of the translated protein sequences of the C-termini of M-Ras and its C elegans orthologue with those of other members of the family of small GTPases. In contrast to R-Ras and R-Ras2, M-Ras and its C elegans orthologue lacked the cysteine-residues (double-underlined in Fig 1) close to the C-terminus that are necessary for palmitoylation. Moreover, murine and C elegans M-Ras lacked a proline-rich motif characteristic of the C-termini of R-Ras and R-Ras2, which we term the R-Ras box (shaded medium gray in Fig 1). Instead, M-Ras and its C elegansorthologue exhibited a distinctive motif, rich in basic residues (shaded dark gray in Fig 1). It included a threonine residue at position 190 of murine M-Ras which, as noted by Matsumoto et al,31 might serve as a target for phosphorylation. The C-terminus of murine M-Ras displayed a stretch of lysine residues (underlined in Fig 1) which resembles that occurring at the C-termini of K-RasB and also Rap-1. Other residues conserved between M-Ras and its C elegans orthologue included a tryptophan at position 60 of M-Ras (a threonine in p21 Ras), a hydrophobic residue at position 51 (an arginine in p21 Ras), and, at position 159, a D/EPP motif, which is likely to alter tertiary structure.
Comparison of the amino acid sequence of the C-terminus (residues 183-208) of M-Ras and a C elegans orthologue of M-Ras (Ce) with R-Ras, R-Ras2, H-, N-, and K-RasA/B. All sequences are murine with the exception of R-Ras2 (human). Gaps are indicated by a hyphen (-). The CAAX-motifs are boxed. Shaded areas indicate regions of homology between M-Ras and its C elegans orthologue (dark gray), the proline-rich R-Ras box (medium gray), and conserved regions in H-Ras, N-Ras, and K-RasA (light gray). Poly-lysine motifs in M-Ras and K-RasB are underlined. Cysteine residues that might be palmitoylated are double underlined. C eleganssequence accession no. is CE00958.
Comparison of the amino acid sequence of the C-terminus (residues 183-208) of M-Ras and a C elegans orthologue of M-Ras (Ce) with R-Ras, R-Ras2, H-, N-, and K-RasA/B. All sequences are murine with the exception of R-Ras2 (human). Gaps are indicated by a hyphen (-). The CAAX-motifs are boxed. Shaded areas indicate regions of homology between M-Ras and its C elegans orthologue (dark gray), the proline-rich R-Ras box (medium gray), and conserved regions in H-Ras, N-Ras, and K-RasA (light gray). Poly-lysine motifs in M-Ras and K-RasB are underlined. Cysteine residues that might be palmitoylated are double underlined. C eleganssequence accession no. is CE00958.
M-Ras is widely expressed.
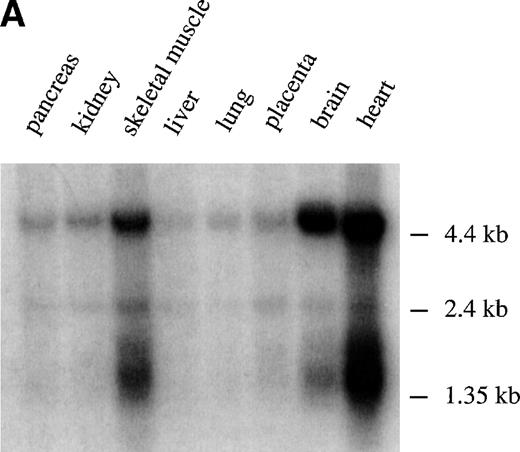
To assess the pattern of expression of M-Ras mRNA, RT-PCR and Northern blot analysis was performed. Using RT-PCR, we observed expression of M-Ras mRNA in all cell lines examined, including hematopoietic cell lines, fibroblasts, and epithelial cells derived from breast, skin, and kidney (Table 1). Northern Blot analysis showed 2 major transcripts of approximately 1.5 kb and 4.6 kb that were expressed highly in brain and heart tissues with lesser but significant levels in liver, lung, pancreas, placenta, and kidney (Fig 2A). Thus, in contrast with the conclusions of other reports,31 32 M-Ras was expressed widely.
Tissue distribution of M-Ras. (A) Northern blots with poly (A)+ RNAs from the indicated human tissues were probed with the C-terminal portion of human M-Ras cDNA. (B) Lysates of 293-cells transiently expressing HA-tagged M-Ras or N-Ras were resolved on SDS-PAGE and immunoblotted with anti-HA antibody 12CA5. The same blot was stripped and immunoblotted with Y13-259, the M-Ras specific pep I antibodies, and the cross-reactive pep II antibodies. (C) Lysates of the indicated tissues and of NIH 3T3 fibroblasts were subjected to immunoprecipitation with 15 μg Y13-259. The immunoprecipitates were resolved on SDS-PAGE and immunoblotted with M-Ras–specific antibodies ( pep I, top panel) or antibodies that cross-reacted between M-Ras and p21 Ras ( pep II, bottom panel).
Tissue distribution of M-Ras. (A) Northern blots with poly (A)+ RNAs from the indicated human tissues were probed with the C-terminal portion of human M-Ras cDNA. (B) Lysates of 293-cells transiently expressing HA-tagged M-Ras or N-Ras were resolved on SDS-PAGE and immunoblotted with anti-HA antibody 12CA5. The same blot was stripped and immunoblotted with Y13-259, the M-Ras specific pep I antibodies, and the cross-reactive pep II antibodies. (C) Lysates of the indicated tissues and of NIH 3T3 fibroblasts were subjected to immunoprecipitation with 15 μg Y13-259. The immunoprecipitates were resolved on SDS-PAGE and immunoblotted with M-Ras–specific antibodies ( pep I, top panel) or antibodies that cross-reacted between M-Ras and p21 Ras ( pep II, bottom panel).
Relative levels of p21 and M-Ras proteins in spleen and thymus.
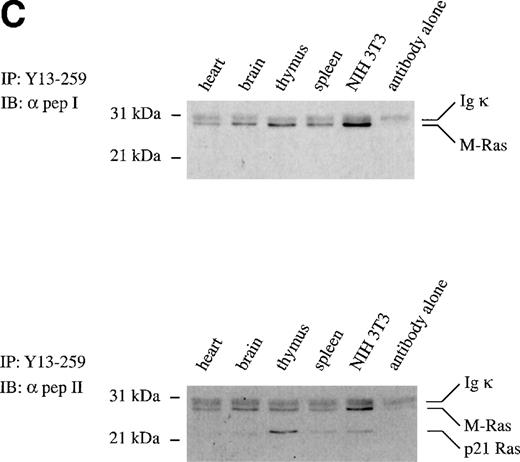
To examine the levels of expression of the endogenous M-Ras protein, we generated preparations of polyclonal antibodies that were specific for M-Ras (α pep I, Fig 2B) or that recognized both M-Ras and, with approximately 3-fold less efficiency, p21 Ras (α pep II, Fig 2B). Immunoprecipitations were performed on whole-cell lysates of spleen, thymus, and various other tissues and cell lines using Y13-259. The immunoprecipitates were resolved on SDS-PAGE and immunoblotted either with the M-Ras–specific antibodies (α pep I) or the antibodies recognizing both M-Ras and p21 Ras (α pep II). M-Ras was detected as a discrete band of an apparent molecular weight of 29 kD in all tissues and cell lines examined (Fig 2C, top panel). Use of the cross-reactive α pep II antibody permitted comparisons of relative levels of expression of M-Ras and p21 Ras. Immunoblotting of these immunoprecipitates showed levels of expression of M-Ras in the spleen and thymus that respectively exceeded or equalled those of p21 Ras. M-Ras protein was also detected in heart, brain, and NIH 3T3 fibroblasts (Fig 2C, bottom panel).
M-Ras is recognized by the monoclonal anti-Ras antibody Y13-259.
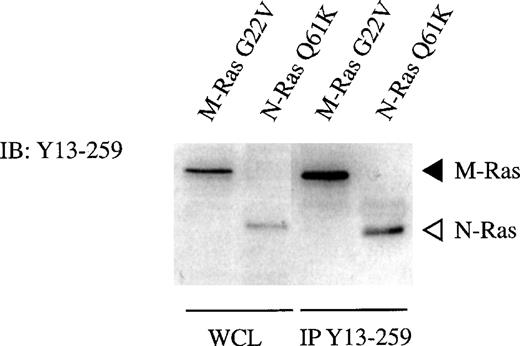
The monoclonal anti-p21 Ras antibody Y13-259 has been used extensively to monitor activation or function of p21 Ras. On the basis of the similarity of the switch II regions of M-Ras and p21 Ras, we next investigated the reactivity of M-Ras with the monoclonal anti-p21 Ras antibody Y13-259. Y13-259 recognized M-Ras and N-Ras with equivalent efficiency in immunoblotting of lysates of 293-cells expressing HA-tagged M-Ras or N-Ras (Figs 2C and 3). Importantly, for the interpretation of assays of the activation of p21 Ras, Y13-259 immunoprecipitated M-Ras and N-Ras from those cell lysates with equivalent efficiency (Fig 3).
M-Ras is recognized by the monoclonal anti-Ras antibody Y13-259. Lysates of 293-cells transiently expressing myc-tagged M-Ras or HA-tagged N-Ras were subjected to immunoprecipitation with 10 μg of Y13-259. The immunoprecipitates, in parallel with samples of the same lysates (WCL), were resolved on SDS-PAGE and immunoblotted with Y13-259 as described.
M-Ras is recognized by the monoclonal anti-Ras antibody Y13-259. Lysates of 293-cells transiently expressing myc-tagged M-Ras or HA-tagged N-Ras were subjected to immunoprecipitation with 10 μg of Y13-259. The immunoprecipitates, in parallel with samples of the same lysates (WCL), were resolved on SDS-PAGE and immunoblotted with Y13-259 as described.
Polyclonal populations of an IL-3–dependent cell line expressing a G22V mutant of M-Ras exhibit increased survival and growth in IL-4.
To investigate the oncogenic potential of M-Ras, we constructed a G22V mutant of M-Ras (M-Ras G22V) that, by analogy with mutants of other small GTPases, was predicted to be constitutively active. We used a retroviral vector to express M-Ras G22V in a murine IL-3–dependent cell-line, R6-X. The vector encoded a puromycin-resistance gene driven by an SV40 promoter and an EGFP gene that was expressed downstream of an IRES site, from the same mRNA as M-Ras G22V. Polyclonal populations of R6-X cells infected with either the M-Ras G22V retrovirus or a control virus encoding EGFP alone were derived by either selection in the presence of IL-3 and puromycin or by fluorescence-activated cell sorting (termed “sorted” populations). M-Ras expression was verified by immunoblotting and 2-color flow cytometry using a monoclonal antibody specific for the HA tag present on M-Ras G22V. The expression of M-Ras G22V varied widely within the polyclonal populations and on a cell-for-cell basis was correlated with expression of EGFP.
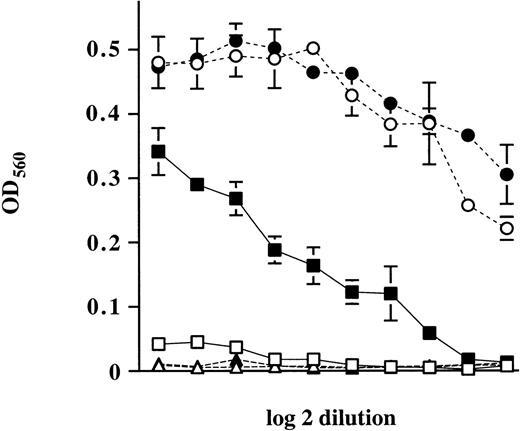
We observed that expression of M-Ras G22V resulted in marked changes in survival. When cultured in the absence of IL-3, polyclonal populations of R6-X cells expressing M-Ras G22V (R6-X M-Ras G22V) exhibited increased survival relative to the parental cells or R6-X cells expressing only EGFP (data not shown). This ability of R6-X cells expressing M-Ras G22V to survive in the absence of IL-3 was confirmed in an experiment in which parental R6-X or a polyclonal population of R6-X expressing M-Ras G22V were plated in agar in the absence of IL-3 for 4 days, after which IL-3 was added. The delayed addition of IL-3 to cultures of parental cells did not result in any colony growth. In contrast, R6-X cells expressing M-Ras G22V showed increased resistance to deprivation of IL-3, and many colonies grew in response to the delayed addition of IL-3 (data not shown). R6-X cells expressing M-Ras G22V also differed from parental cells in their ability to grow in IL-4. Parental R6-X cells failed to grow continuously in IL-4 alone, consistent with evidence that IL-4 is not a true growth factor.38 In contrast, polyclonal populations of R6-X cells expressing M-Ras G22V exhibited enhanced survival and growth in the presence of IL-4 (Fig 4).
Enhancement of growth of polyclonal populations of R6-X M-Ras G22V by IL-4. Polyclonal populations of R6-X M-Ras G22V (closed symbols) or of parental cells (open symbols) were plated at 500 cells per well in 96-well plates in medium alone (▴, ▵) or medium supplemented with decreasing amounts of synthetic IL-3 starting at 2 μg/mL (•, ○) or recombinant IL-4 (10 ng/mL) (▪, □). Cell numbers were assessed at day 6 by uptake of MTT. Results are shown as optical densities (OD) of triplicate cultures ± SEM.
Enhancement of growth of polyclonal populations of R6-X M-Ras G22V by IL-4. Polyclonal populations of R6-X M-Ras G22V (closed symbols) or of parental cells (open symbols) were plated at 500 cells per well in 96-well plates in medium alone (▴, ▵) or medium supplemented with decreasing amounts of synthetic IL-3 starting at 2 μg/mL (•, ○) or recombinant IL-4 (10 ng/mL) (▪, □). Cell numbers were assessed at day 6 by uptake of MTT. Results are shown as optical densities (OD) of triplicate cultures ± SEM.
High levels of expression of active M-Ras result in factor-independent growth but reduced growth in the presence of IL-3.
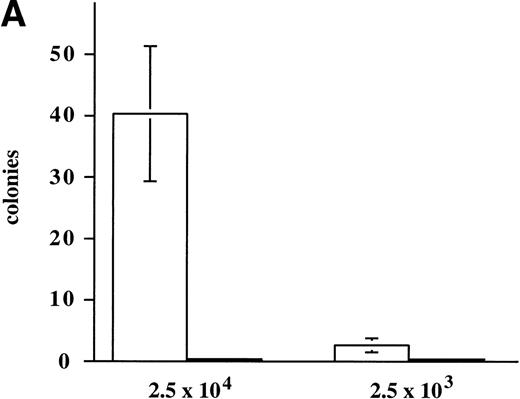
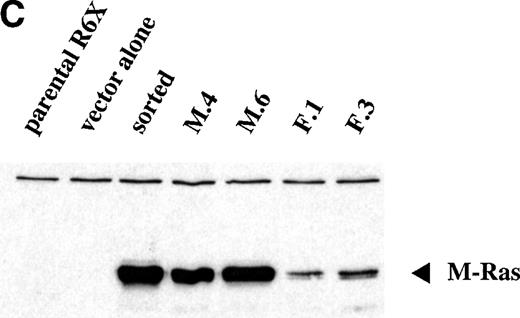
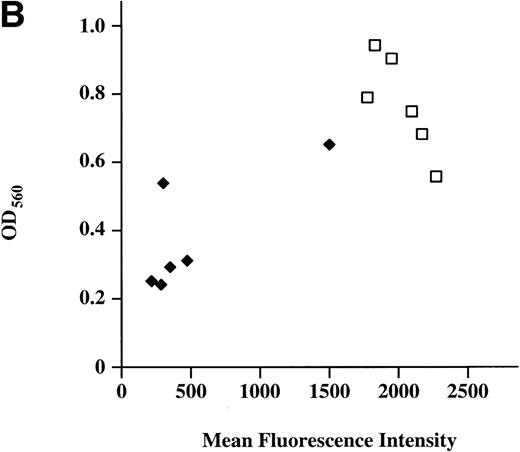
Continued observation of polyclonal populations of cells expressing M-Ras G22V cultured in the absence of IL-3 showed a subpopulation of cells that failed to die and slowly increased in number. Plating of polyclonal populations of R6-X M-Ras G22V in agar in the absence of IL-3 for 7 days resulted in the growth of small colonies at a low frequency (<0.2%) of the cells plated (Fig 5A). This low frequency of cells capable of forming colonies in the absence of IL-3 could have reflected functional heterogeneity of the polyclonal population, or, alternatively, an intrinsically low cloning efficiency of R6-X M-Ras G22V cells in the absence of IL-3. To address this question, we cloned cells from randomly selected colonies that had grown in medium alone, and we cloned cells from randomly selected colonies that had developed at high frequency (35% of plated cells) in the presence of IL-3. Two series of clones of R6-X M-Ras G22V were expanded in IL-3, 1 derived from colonies that had developed in medium alone (termed the M series), and 1 derived from colonies that had developed in IL-3 (termed the F series). These 2 series of clones were then washed and their survival in the absence of IL-3 compared. Cells of the M series of clones, derived from colonies grown in the absence of IL-3, failed to die when cultured in medium alone. In contrast, with a few notable exceptions, most of the clones derived from the F series of colonies grown in IL-3 exhibited significant cell death when deprived of IL-3. Inspection by fluorescent microscopy of cultures of the 2 series of clones growing optimally in IL-3 showed that cells of all of the M series clones, derived from colonies grown in the absence of IL-3, were bright green, whereas the F series clones, derived from the randomly chosen colonies grown in IL-3, varied in brightness, with most exhibiting only a low EGFP signal. There was a strong correlation between survival in the absence of IL-3 and brightness (Fig 5B). Similarly, when these clones were pated in agar in the absence of IL-3, there was a clear correlation between the brightness of the EFP signal and the size and frequency of colonies (data not shown). These results suggested that the ability to survive and grow in the absence of IL-3 correlated with levels of expression of M-Ras G22V. This was confirmed directly by assessing levels of M-Ras G22V by immunoblotting of cell lysates with anti-HA antibodies (Fig 5C).
Expression of constitutively active M-Ras G22V in an IL-3–dependent cell line results in factor-independent growth. (A) Polyclonal, puromycin-resistant R6-X M-Ras G22V cells (□) or parental R6-X cells (▪) were plated in agar in triplicate in the absence of IL-3 at 2.5 × 104 per mL or 2.5 × 103/mL (as indicated). Colonies were counted at day 7 using a colony-microscope. (B) Correlation of levels of expression of M-Ras G22V with factor-independent growth. Clones of R6-X M-Ras G22V cells derived from colonies grown in medium alone (M series; ⧫) or in the presence of IL-3 (F series; □) were expanded in IL-3, washed, and incubated in medium alone for days, when survival and growth were assessed by uptake of MTT as described above. The uptake of MTT (expressed as OD units) was plotted against the mean intensity of fluorescence (MFI) of EGFP assessed by flow cytometry of the same clones grown in the presence of IL-3. (C) Correlated expression of EGFP and M-Ras G22V. Twenty micrograms of whole-cell lysates of 4 of the clones shown in (B) above, 2 exhibiting high EGFP expression (M.4, MFI 1775, and M.6, MFI 2271) and 2 with low EGFP expression (F.1, MFI 216, and F.3, MFI 474), were immunoblotted using a monoclonal antibody specific for the HA-tag present on the M-Ras G22V. Also shown are lysates of parental R6-X cells and of a “sorted,” polyclonal population of M-Ras G22V-expressing R6-X cells, selected by fluorescence-activated cell sorting (FACS) and culture in IL-4 (sorted), and of a population of R6-X cells infected with empty vector (vector alone).
Expression of constitutively active M-Ras G22V in an IL-3–dependent cell line results in factor-independent growth. (A) Polyclonal, puromycin-resistant R6-X M-Ras G22V cells (□) or parental R6-X cells (▪) were plated in agar in triplicate in the absence of IL-3 at 2.5 × 104 per mL or 2.5 × 103/mL (as indicated). Colonies were counted at day 7 using a colony-microscope. (B) Correlation of levels of expression of M-Ras G22V with factor-independent growth. Clones of R6-X M-Ras G22V cells derived from colonies grown in medium alone (M series; ⧫) or in the presence of IL-3 (F series; □) were expanded in IL-3, washed, and incubated in medium alone for days, when survival and growth were assessed by uptake of MTT as described above. The uptake of MTT (expressed as OD units) was plotted against the mean intensity of fluorescence (MFI) of EGFP assessed by flow cytometry of the same clones grown in the presence of IL-3. (C) Correlated expression of EGFP and M-Ras G22V. Twenty micrograms of whole-cell lysates of 4 of the clones shown in (B) above, 2 exhibiting high EGFP expression (M.4, MFI 1775, and M.6, MFI 2271) and 2 with low EGFP expression (F.1, MFI 216, and F.3, MFI 474), were immunoblotted using a monoclonal antibody specific for the HA-tag present on the M-Ras G22V. Also shown are lysates of parental R6-X cells and of a “sorted,” polyclonal population of M-Ras G22V-expressing R6-X cells, selected by fluorescence-activated cell sorting (FACS) and culture in IL-4 (sorted), and of a population of R6-X cells infected with empty vector (vector alone).
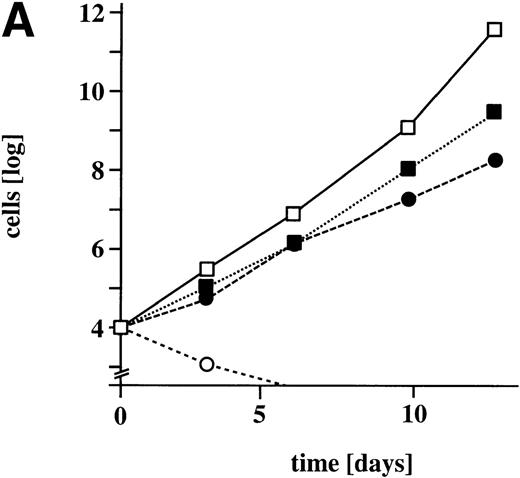
Although there was a clear positive correlation between the levels of expression of M-Ras G22V and the ability to grow in the absence of IL-3, the reverse was the case for growth in the presence of IL-3. A comparison of the growth in the presence and absence of IL-3, of parental R6-X cells and a population of R6-X cells selected for high levels of expression of M-Ras G22V (Fig 6), showed that although over 10 days the M-Ras G22V expressing cells expanded 6,000-fold in the absence of IL-3, they expanded 10-fold less in the presence of IL-3 than did the parental cells.
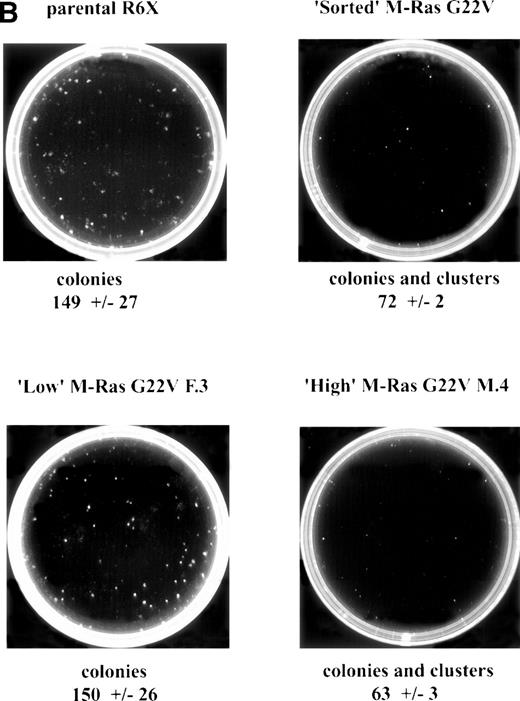
Expression of M-Ras G22V in an IL-3–dependent line results in factor-independent growth in the absence of IL-3, but decreased growth in its presence. (A) Parental R6-X (open symbols) or an FACS-sorted population expressing M-Ras G22V (closed symbols) were cultured in triplicate at 104/mL in medium alone (circles •, ○) or in the presence of IL-3 (2% W3) (squares ▪, □). Cells were counted at the indicated intervals and counts are shown as the mean ± SEM. (B) Parental R6-X cells or clones expressing high (clone F.3; MFI 474) or low (clone M.4; MFI 1775) levels of M-Ras G22V, or the polyclonal “sorted” population expressing high levels of M-Ras G22V cells (sorted), were plated in agar for 7 days in 2% W3 at 3 × 102 per mL. The mean ± SEM of numbers of colonies or in the case of M.4 and St, colonies and clusters, in triplicate cultures is shown under the images of representative plates of each triplicate. To visualize colonies, plates were stained with acridine orange (1 μg/mL) and the images were captured using a Canberra Packard photodocumentation system (Alpha Innotech Corp, San Leandro, CA).
Expression of M-Ras G22V in an IL-3–dependent line results in factor-independent growth in the absence of IL-3, but decreased growth in its presence. (A) Parental R6-X (open symbols) or an FACS-sorted population expressing M-Ras G22V (closed symbols) were cultured in triplicate at 104/mL in medium alone (circles •, ○) or in the presence of IL-3 (2% W3) (squares ▪, □). Cells were counted at the indicated intervals and counts are shown as the mean ± SEM. (B) Parental R6-X cells or clones expressing high (clone F.3; MFI 474) or low (clone M.4; MFI 1775) levels of M-Ras G22V, or the polyclonal “sorted” population expressing high levels of M-Ras G22V cells (sorted), were plated in agar for 7 days in 2% W3 at 3 × 102 per mL. The mean ± SEM of numbers of colonies or in the case of M.4 and St, colonies and clusters, in triplicate cultures is shown under the images of representative plates of each triplicate. To visualize colonies, plates were stained with acridine orange (1 μg/mL) and the images were captured using a Canberra Packard photodocumentation system (Alpha Innotech Corp, San Leandro, CA).
When clones expressing different levels of M-Ras G22V were plated in agar in the presence of IL-3, those expressing low levels of M-Ras G22V developed colonies with the same size, morphology, and frequency as parental cells (Fig 6B). In contrast, colonies developed from clones expressing the highest levels of M-Ras G22V were much smaller and less dispersed than those of the parental cells and occurred at lower frequency, with many of the clonogenic cells forming clusters of less than 30 cells (Fig 6B). This antagonistic effect of activated M-Ras on IL-3–stimulated growth also depended on the concentration of IL-3. Thus, when polyclonal populations of R6-X cells, most of which expressed lower or medium levels of M-Ras G22V, were plated in agar at supraoptimal concentrations of IL-3, the colonies that grew were half as frequent and smaller and less dispersed than those grown from parental cells. However, at 10-fold lower optimal concentrations of IL-3, the number and size of colonies derived from parental cells and the polyclonal population of M-Ras G22V expressing cells were similar (data not shown).
M-Ras G22V transforms NIH 3T3 fibroblasts.
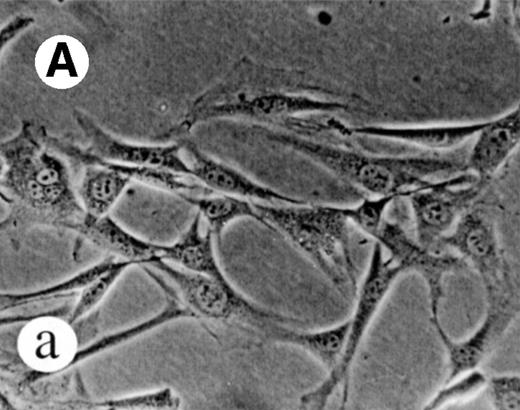
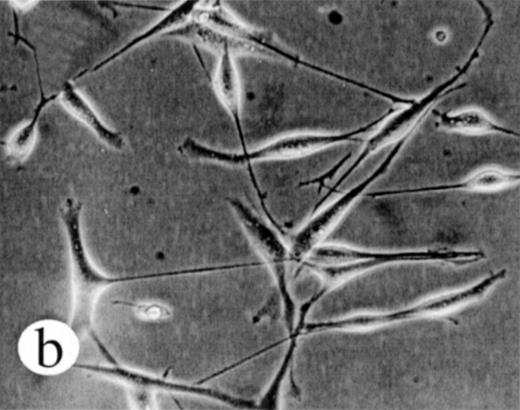
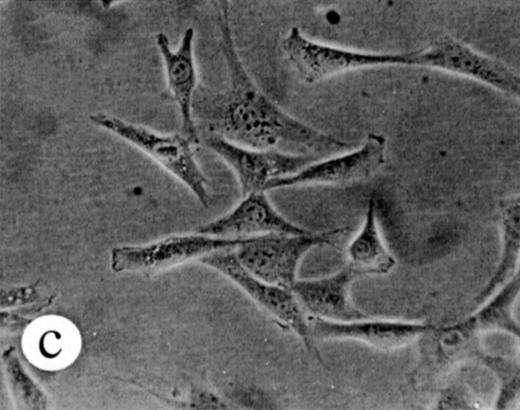
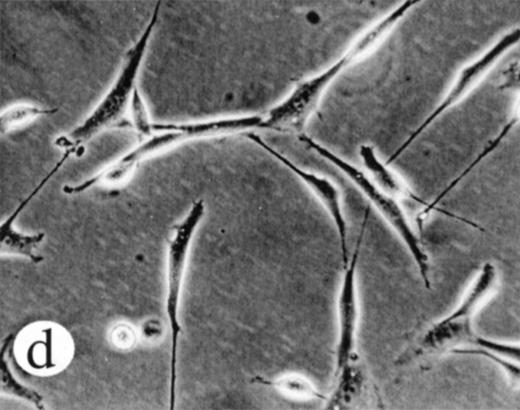
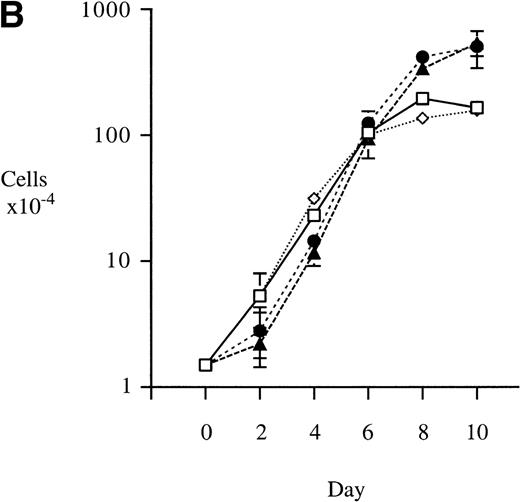
We used retroviral vectors to express wild-type M-Ras, M-Ras G22V, or N-Ras Q61K in NIH-3T3 fibroblasts. As shown in Fig 7A, NIH 3T3 cells expressing M-Ras G22V, but not those expressing wild-type M-Ras, exhibited a refractile, transformed morphology, strongly resembling that of the cells expressing N-Ras Q61K. Cells expressing M-Ras G22V, but not those expressing wild-type M-Ras, also resembled cells expressing N-Ras Q61K in gaining the ability to grow to higher saturation densities (Fig 7B).
Transformation of NIH 3T3 fibroblasts by constitutively active M-Ras G22V. (A) NIH 3T3 fibroblasts were retrovirally transduced with (a) empty vector, (b) N-Ras Q61K, (c) M-Ras wt, or (d) M-Ras G22V. Shown are phase-contrast photomicrographs of cells grown in medium containing 10% calf serum. (B) Cell densities reached by polyclonal populations of NIH3T3 cells expressing vector alone (◊), M-Ras wt (□), M-Ras G22V (•), and N-Ras Q61K (▴). Cells were grown in 10% calf serum and figures represent data from 2 independent experiments, each performed in triplicate, and are shown as means ± SD.
Transformation of NIH 3T3 fibroblasts by constitutively active M-Ras G22V. (A) NIH 3T3 fibroblasts were retrovirally transduced with (a) empty vector, (b) N-Ras Q61K, (c) M-Ras wt, or (d) M-Ras G22V. Shown are phase-contrast photomicrographs of cells grown in medium containing 10% calf serum. (B) Cell densities reached by polyclonal populations of NIH3T3 cells expressing vector alone (◊), M-Ras wt (□), M-Ras G22V (•), and N-Ras Q61K (▴). Cells were grown in 10% calf serum and figures represent data from 2 independent experiments, each performed in triplicate, and are shown as means ± SD.
Expression of activated M-Ras activated the c-fospromoter.
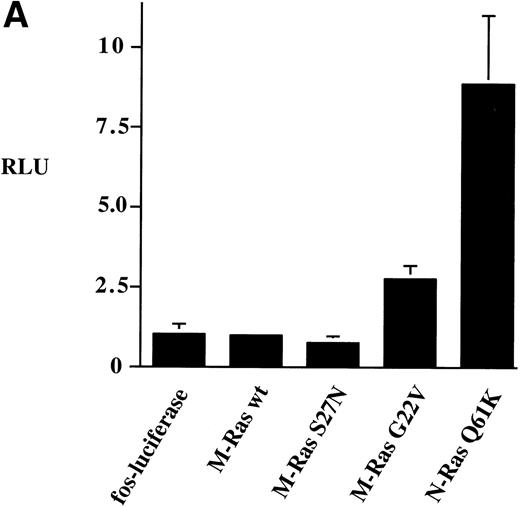
We tested the ability of M-Ras and mutants of M-Ras to activate thec-fos promoter using the luciferase reporter gene assay. Expression of wild-type M-Ras did not activate the c-fospromoter (Fig 8A). However, expression of the G22V mutant of M-Ras resulted in 3-fold activation of thec-fos promoter. This was reproducibly less than the 9-fold activation of the c-fos promoter induced by N-Ras Q61K (Fig8A).
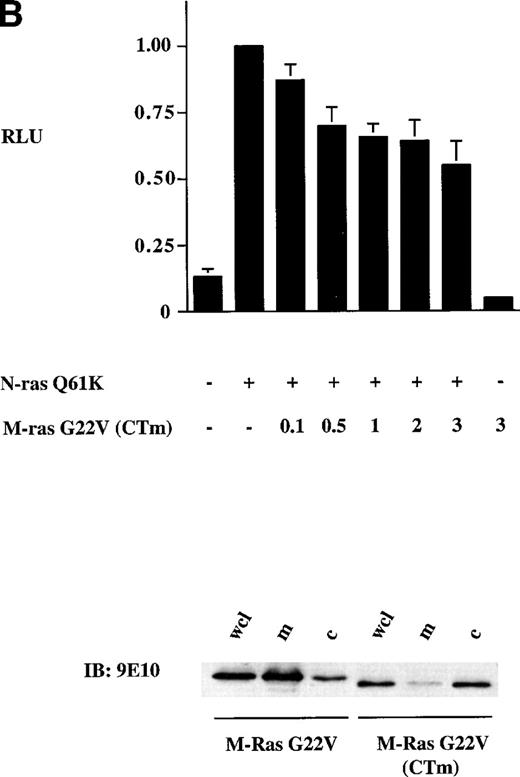
Activation of the c-fos promoter by M-Ras; evidence for sharing of effectors with p21 Ras. Shown are relative luciferase units derived from assays of lysates of 293-cells transiently transfected with 0.25 μg of the c-fos luciferase reporter plasmid and: (A) 3 μg of the indicated plasmids; (B) 0.25 μg of a N-Ras Q16K plasmid and the indicated amounts of M-Ras G22V plasmids. Results represent data from 6 (A) or 3 (B) independent experiments and are shown as mean ± SD. All transient transfections contained equal amounts of total plasmid DNA, normalized with empty vector containing the same promoter. The lower panel of (B) shows results of a cell-fractionation experiment. Lysates of N-terminally tagged M-Ras G22V or C-terminally tagged M-Ras G22V (CTm) were subjected to ultra-centrifugation. Samples of whole-cell lysate (wcl), the membrane-enriched fraction (m) and the cytosolic fraction (c) were separated on SDS-PAGE and immunoblotted with anti–myc-epitope antibody 9E10.
Activation of the c-fos promoter by M-Ras; evidence for sharing of effectors with p21 Ras. Shown are relative luciferase units derived from assays of lysates of 293-cells transiently transfected with 0.25 μg of the c-fos luciferase reporter plasmid and: (A) 3 μg of the indicated plasmids; (B) 0.25 μg of a N-Ras Q16K plasmid and the indicated amounts of M-Ras G22V plasmids. Results represent data from 6 (A) or 3 (B) independent experiments and are shown as mean ± SD. All transient transfections contained equal amounts of total plasmid DNA, normalized with empty vector containing the same promoter. The lower panel of (B) shows results of a cell-fractionation experiment. Lysates of N-terminally tagged M-Ras G22V or C-terminally tagged M-Ras G22V (CTm) were subjected to ultra-centrifugation. Samples of whole-cell lysate (wcl), the membrane-enriched fraction (m) and the cytosolic fraction (c) were separated on SDS-PAGE and immunoblotted with anti–myc-epitope antibody 9E10.
To explore whether M-Ras also shared effector molecules with members of the p21 Ras family, we expressed an activated M-Ras G22V that was modified at the C-terminus so that it was no longer localized at the plasma membrane and remained in the cytoplasm (Fig 8B, bottom panel). Expression of this cytoplasmic form of M-Ras G22V (CTm) failed to activate the c-fos reporter gene construct (Fig 8B, top panel). However, expression of M-Ras G22V (CTm) inhibited activation of thec-fos promoter induced by coexpressed N-Ras Q61K. This inhibition was dose-dependent but partial, reaching a maximum of about 40% when the ratio of plasmid DNA encoding M-Ras G22V (CTm) to that encoding N-Ras Q61K reached 2:1 (Fig 8B, top panel). This inhibitory effect required that M-Ras was in the active, GTP-bound conformation, as expression of carboxy-terminally modified M-Ras S27N failed to inhibit activation of the c-fos promoter by N-Ras Q61K (data not shown).
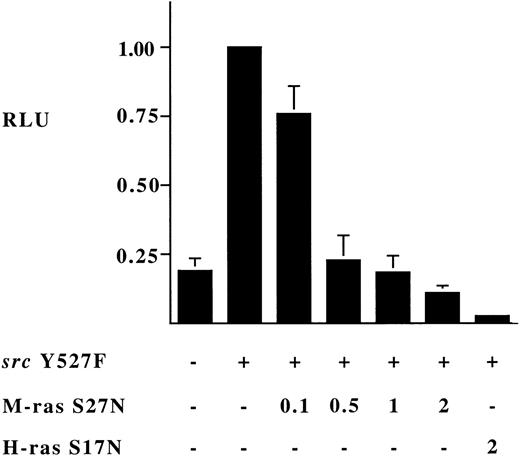
Expression of dominant negative M-Ras S27N completely inhibits activation of the c-fos promoter by activated Src.
To investigate the possibility that M-Ras shared exchange factors with p21 Ras proteins, we generated an S27N mutant of M-Ras, analogous to the S17N dominant inhibitory mutants of p21 Ras. Expression of M-Ras S27N itself failed to activate transcription of the c-fospromoter (Fig 8A). However, when coexpressed it completely suppressed activation of the c-fos promoter induced by expression of constitutively active Src Y527F (Fig 9). Thus, M-Ras S27N acted, like H-Ras S17N, as a dominant inhibitor of the Src pathway to the c-fos promoter (Fig 9).
Dominant inhibitory effect of expression of M-Ras S27N on activation of c-fos promoter by activated Src Y527F. 293-cells were transfected as described in the legend to Fig 8 with 0.25 μg of the c-fos luciferase reporter plasmid and 0.25 μg of a Src Y527F plasmid and the indicated amount in micrograms of the indicated plasmids. Shown are relative luciferase units expressed as the mean ± SD of 3 independent experiments.
Dominant inhibitory effect of expression of M-Ras S27N on activation of c-fos promoter by activated Src Y527F. 293-cells were transfected as described in the legend to Fig 8 with 0.25 μg of the c-fos luciferase reporter plasmid and 0.25 μg of a Src Y527F plasmid and the indicated amount in micrograms of the indicated plasmids. Shown are relative luciferase units expressed as the mean ± SD of 3 independent experiments.
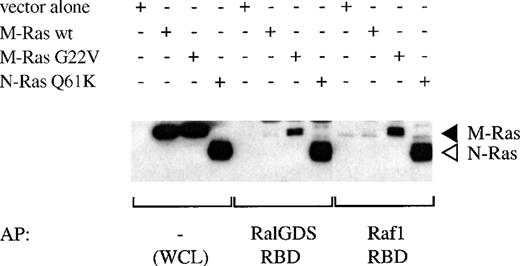
Interaction of activated M-Ras with Raf-1 and Ral-GDS.
We investigated whether constitutively active M-Ras interacted directly with two known effectors of p21 Ras, Raf-1 and Ral-GDS. The binding of M-Ras G22V to the Ras-binding domain (RBD) of Raf-1 was readily detectable, although the amounts of M-Ras G22V present in eluates were significantly less than those of Q61K N-Ras observed in parallel experiments (Fig 10). This indicated that M-Ras G22V interacted with the Raf-1-RBD with lower affinity than did N-Ras Q61K. Likewise, M-Ras G22V bound detectably to RalGDS-RBD. Again, comparison of the amount of M-Ras G22V bound with that of N-Ras Q61K indicated that M-Ras G22V had a lower affinity for Ral-GDS. Nevertheless, both the binding of M-Ras G22V to the Raf-1-RBD and to the RalGDS-RBD were inhibited by Y13-259, showing that in both cases binding was specific (data not shown). As expected, neither the RBD of Raf-1, nor that of RalGDS, bound to wild-type M-Ras.
Binding of M-Ras to the Ras binding domains (RBD) of Raf-1 and Ral-GDS. Lysates of 293-cells transiently transfected with HA-tagged M-Ras wt, M-Ras G22V, N-Ras Q61K, or vector alone were mixed as indicated with GST-fusion proteins of the RBD of Raf-1 or Ral-GDS precoupled to glutathione beads. Proteins were eluted from the beads by boiling and, in parallel with samples of the original lysates (WCL), were subjected to SDS-PAGE and immunoblotting with anti-HA antibody 12CA5. (◂), HA-M-Ras; (◃), HA-N-Ras.
Binding of M-Ras to the Ras binding domains (RBD) of Raf-1 and Ral-GDS. Lysates of 293-cells transiently transfected with HA-tagged M-Ras wt, M-Ras G22V, N-Ras Q61K, or vector alone were mixed as indicated with GST-fusion proteins of the RBD of Raf-1 or Ral-GDS precoupled to glutathione beads. Proteins were eluted from the beads by boiling and, in parallel with samples of the original lysates (WCL), were subjected to SDS-PAGE and immunoblotting with anti-HA antibody 12CA5. (◂), HA-M-Ras; (◃), HA-N-Ras.
DISCUSSION
Our data show that M-Ras is a 29-kD protein that is present in lymphohematopoietic cells and in many other tissues and cells, including breast epithelial lines and fibroblasts. In the spleen (and in 3T3 fibroblasts), the levels of expression of M-Ras protein exceeded those of members of the p21 Ras family. In the thymus, this ratio was reversed, but the levels of M-Ras protein were comparable with those in spleen, brain, and heart. Importantly, M-Ras could not be distinguished from p21 Ras in assays based on the use of the anti-Ras monoclonal antibody Y13-259 and dominant negative Ras mutants. Our observations that expression of a constitutively active mutant of M-Ras in an IL-3–dependent mast cell/megakaryocyte cell line increased survival and resulted in factor-independent growth and in 3T3 fibroblasts in transformation indicate that mutants of M-Ras have a role in oncogenesis. Collectively, these data suggest that M-Ras and p21 Ras are activated by common exchange factors and can regulate growth via both shared and distinctive effectors.
Comparison of the C-termini of M-Ras and those of other members of the Ras family show that, although sharing the terminal CAAX-motif, M-Ras lacks the cysteine residues characteristic of p21 Ras, R-Ras, and R-Ras2, which are involved in palmitoylation. M-Ras also lacks the proline-rich region characteristic of R-Ras and Tc21, which we term the R-Ras box (Fig 1), or a motif found in the C-termini of the classical p21 Ras proteins, H-, N-, and K-Ras (shaded light gray in Fig 1). Instead, M-Ras, in both mammals and C elegans, exhibits a conserved carboxy terminal motif (Fig 1) that is rich in basic residues and has a conserved threonine. These features
may govern the interaction of M-Ras with other molecules or with other stuctures, such as subregions of the membrane.
The best-characterized signaling pathway downstream of p21 Ras involves activation of a cascade of ser/thr-kinases, Raf-1, MEK 1/2 and Erk 1/2, which ultimately leads to transcriptional activation of genes such asc-fos.39 We observed that constitutively active M-Ras was able to activate transcription of a reporter gene under the control of the c-fos promoter (Fig 8A). Membrane localization was crucial because a constitutively active mutant that was modified at the carboxy-terminus so that it remained in the cytoplasm (Fig 8B, bottom panel) was no longer able to activate the c-fos promoter (Fig 8B). The moderate degree of activation of the c-fospromoter induced by coexpression of M-Ras G22V relative to that of N-Ras Q61K correlated with their relative affinities for the Ras-binding domain of Raf-1 (Fig 10) and their relative abilities to induce activation of Erk2.32 It remains to be established whether M-Ras activates the Raf-MAPK pathway under physiological conditions. Our observation that coexpression of a cytoplasmically localized, constitutively active mutant of M-Ras partially inhibited N-Ras–mediated activation of the c-fospromoter (Fig 8B) suggests that M-Ras shares some, but not all, of the pathways downstream of p21 Ras. The presence of orthologues of both p21 Ras and M-Ras in C elegans suggests that these molecules are likely to have distinctive functions and use distinctive effectors. The assignment of functions to M-Ras, however, is complicated by the fact that characterization of the function of p21 Ras proteins in their various physiological and pathological processes has relied heavily on the monoclonal antibody Y13-259 and dominant inhibitory mutants such as H-Ras S17N. As discussed below, these two experimental tools fail to discriminate between M-Ras and p21 Ras.
Y13-259 binds to a highly conserved epitope on the switch II region of p21 Ras23 and blocks interaction of Raf-1 both with p21 Ras and with M-Ras (data not shown). The demonstration that Y13-259 recognizes M-Ras and N-Ras with equivalent efficiency (Figs 2C and 3) is consistent with the overall similarities in the sequences of M-Ras and p21 Ras in the switch II region, and the fact that those residues known to comprise the epitope are identical. Given our evidence that M-Ras is expressed ubiquitously, and is more abundant than p21 Ras intissues like spleen and in cells like NIH 3T3 fibroblasts (which have been used widely to study p21 Ras), it will be necessary to reevaluate many published experiments to determine whether the experimental observations reflected the function of p21 Ras, M-Ras, or both. For example, the biological effects of micro-injection of Y13-259 into 3T3 cells expressing oncogenes,25 which have been interpreted as reflecting interference with the function of p21 Ras, could equally reflect inhibition of M-Ras function. Moreover, experiments designed to assess the activation of p21 Ras by measuring ratios of GTP:GDP in Y13-259 immunoprecipitates in fibroblasts40 and in hematopoietic cells stimulated by hematopoietic growth factors16,17 or antigens18 19 also may have been monitoring the activation of M-Ras.
Dominant-negative p21 Ras mutants such as H-Ras S17N26 act by sequestrating GEFs involved in the activation of p21 Ras proteins41,42 and have been widely used to implicate p21 Ras in paths downstream of cell-surface receptors and oncogenes20,26-28,40 and in the development of cells of hematopoietic origin.21,22 The demonstration that M-Ras S27N also acted as a dominant inhibitor of activation of thec-fos promoter by an activated Src Y527F (Fig 9) suggests that M-Ras shares GEFs with p21 Ras. One is likely to be mSos-1, which is known to be translocated to the membrane in cells expressing v-src.43 It is likely that M-Ras not only binds GEFs like mSos-1, but is also activated by them. Thus, M-Ras is likely to be activated by most stimuli that bring GEFs to the membrane and have been thought to activate p21 Ras. This failure of dominant negative mutants to discriminate between p21 Ras and M-Ras and the high levels of expression of M-Ras in tissues such as the spleen means that the conclusions of experiments based on their use—for example, concerning the role of p21 Ras in the action of hematopoietic growth factors20 or in the development of the thymus21 or B lymphocytes22—will require reevaluation.
The demonstration that expression of a constitutively active mutant of M-Ras in an IL-3–dependent mast cell/megakaryocyte line resulted in enhanced survival and factor-independent growth (Figs 4 through 6) point to the potential significance of M-Ras in leukemogenesis. The strong correlation between the ability of cells to grow in the absence of IL-3 and levels of expression of M-Ras G22V (Fig 5) indicates that when expressed at high levels, active M-Ras was able to activate to some degree all those signal transduction paths required for growth. We looked carefully for signs of the operation of an autocrine mechanism, because this would provide the simplest explanation of how expression of M-Ras G22V could trigger multiple intracellular signal tranduction paths. However, growth and survival of R6-X cells expressing M-Ras G22V in the absence of IL-3 was not density-dependent (Fig 5 and data not shown). When expressed at lower levels, expression of M-Ras G22V resulted in prolonged survival but did not result in continuous, factor-independent growth. These results are qualitatively similar to those seen when activated p21 Ras is expressed in factor-dependent hematopoietic cells.13 The synergy observed between active M-Ras and IL-4 (Fig 4) was also reminiscent of that seen between activated p21 Ras and IL-4.38 We have shown that this role of active p21 Ras in synergizing with IL-4 to promote the growth of IL-3–dependent Baf/3 cells can be replaced by activated Raf-1.38 It is thus conceivable that the observed synergy between M-Ras G22V and IL-4 (Fig 5) reflects activation of Raf-1 by M-Ras G22V through the weak interaction we showed (Fig10). The enhancing effect of IL-4 is likely to reflect its ability to upregulate levels of c-myc mRNA (J.S.W., J.W.S., submitted for publication) because IL-4 cannot stimulate activation of p21 Ras16,17 or the Erk,44 JNK,45 or p38 MAP kinases.46
Our results clearly show that active M-Ras also could, in some circumstances, negatively influence the growth of hematopoietic cells. Thus, although cells expressing high levels of M-Ras G22V were capable of factor-independent growth, they also exhibited slower growth and reduced cloning efficiency in the presence of IL-3 (Fig 6). We have seen qualitatively similar negative effects on growth when activated p21 Ras is expressed in hematopoietic cells (unpublished observations, May 1999). The mechanisms may be similar to those reported in fibroblasts which involve overactivation of Raf-147 and consequent upregulation of p21WAFand cell-cycle arrest.48
It will be important to determine the frequency of activating mutations of M-Ras in leukemia and other neoplasias of lympho-hematopoietic origin. Likewise, given evidence (Fig 7A and B, ref 32) that activated mutants of M-Ras can transform 3T3 fibroblasts, it will be important to determine whether mutations of M-Ras occur in solid tumors, particularly those involving tissues such as breast or prostate, where mutations in p21 Ras occur at a relatively low frequencies.14
In summary, we have presented evidence that M-Ras is widely expressed, with the spleen and thymus being major sites of expression, interacts with at least some of the GEFs acting on p21 Ras, and when constitutively active can result in enhanced survival and factor-independent growth of a hematopoietic cell line and transformation of 3T3 fibroblasts. Collectively, these observations suggest that in many cells, including those of lympho-hematopoietic origin, M-Ras and p21 Ras will be activated coordinately and will activate parallel, distinctive signaling pathways. Future work will determine the molecular nature and degree of overlap of these paths and their role in the growth and differentiation of the lympho-hematopoietic system. Finally, some of the functions of the M-Ras pathway may have been misattributed to the classical p21 Ras pathway on the basis of the use of Y13-259 and S17N mutants of p21 Ras, and it will be important to develop more selective reagents.
ACKNOWLEDGMENT
We thank Wendy E. Lamson for excellent technical assistance, Annette B.I. Schallhorn for RNA-preparations, and Ruth A. Salmon and Ian N. Foltz for helpful discussions throughout the project and critical reading of the manuscript.
Supported by a grant from the Medical Research Council, Canada.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to John W. Schrader, MB, PhD, The Biomedical Research Centre, University of British Columbia, 2222 Health Sciences Mall, Vancouver, BC V6T 1Z3, Canada; e-mail: john@brc.ubc.ca.