Abstract
Anaplastic large cell lymphoma (ALCL) is associated with the t(2;5)(p23;q35), which generates the NPM-ALK fusion gene encoding an 80-kD protein. Several studies have suggested that genes other than NPM may be fused to theALK gene. Here we have identified TRK-fused gene (TFG) as a new ALK partner in 2 ALCL, 1 of which exhibited a t(2;3)(p23;q21). In these cases, TFG was involved in 2 different fusion genes, TFG-ALKS andTFG-ALKL, coding respectively 85-kD and 97-kD chimeric proteins. The ALK breakpoint in these translocations was the same as in the classic t(2;5) translocation. These 2 proteins were both active in an in vitro tyrosine kinase assay showing that the new cloned cDNA sequences are translated into chimeric proteins with functional activity. These findings indicate thatTFG can provide an alternative to NPM as a fusion partner responsible for activation of the ALK and the pathogenesis of ALCL.
ANAPLASTIC LARGE cell lymphoma (ALCL) is associated with the (2;5)(p23;q35) translocation,1,2which fuses the nucleophosmin (NPM ) gene on chromosome 5q35 to the anaplastic lymphoma kinase (ALK ) gene on chromosome 2p23.3 This rearrangement leads to the production of a novel 80-kilodalton (kD) fusion protein with transforming properties in which the N-terminal region of nucleophosmin is linked to the C-terminal region of ALK containing the tyrosine kinase catalytic domain.3-6 Similarly to other genes responsible for activating receptor tyrosine kinase oncogenes, NPM is constitutively expressed in all tissues, and activates the catalytic domain of the ALK segment through dimerization.5,7,8 The function of normal NPM is to shuttle ribonucleoproteins from the cytoplasm to the nucleus,7,8 and heterodimerization between normal and chimeric NPM probably explains the immunohistochemical detection of ALK in the nucleus as well as in the cytoplasm of tumor cells harboring the t(2;5)(p23;q35). However, this nuclear localization of the protein is not required for malignant transformation.5 6
Several cytogenetic and molecular studies have now suggested that translocations other than the classic t(2;5)(p23;q35) may also activate the ALK gene and participate in the pathogenesis of ALCL.9-17 Interestingly, ALK protein expression in ALCL negative for NPM-ALK translocation is restricted to the cytoplasm of the tumor cells.10,12,13,18 Recently, biochemical study of ALK protein in NPM-ALK–negative ALK-positive ALCL has detected novel ALK proteins with kinase activity and different molecular weights ranging from 85 to 113 kD.19 These observations indicate that genes other than NPM may also deregulate ALK and participate in the pathogenesis of ALCL, and recently, Lamant et al20 have found nonmuscular tropomyosin (TPM3) gene as the partner fused with ALK gene in a t(1;2)(q25;p23) translocation present in a case of ALCL.
In this study, we have identified the TFG (TRK-fused gene) gene as a new fusion partner for ALK in 2 cases of ALCL. In these cases, 2 structurally different translocations created TFG-ALK fusion genes encoding chimeric proteins of different molecular weight (85 kD and 97 kD) that showed tyrosine kinase activity in vitro.
MATERIALS AND METHODS
Materials.
The material used for the initial cloning experiments was a diagnostic lymph node biopsy from a 19-year-old man (case #789), which showed the typical features of “common”-type ALCL. However, reverse transcription-polymerase chain reaction (RT-PCR) analysis provided no evidence for the NPM-ALK fusion gene, and ALK protein was expressed with a cytoplasmic restricted pattern.
After identification of the TFG-ALK chimeric gene (see below), 6 additional cases of ALCL that lacked NPM-ALK andTPM3-ALK were examined by RT-PCR. One of these later cases (case #862) exhibited the t(2;3)(p23;q21) described previously.17 Expression of normal TFG transcripts was also examined by RT-PCR (see below) in 3 non-neoplastic lymphoid tissues, 14 non-Hodgkin’s lymphomas (NHLs), and 11 malignant hematopoietic cancer cell lines (8402, CEM, U937, HL-60, Karpas 299, MOLT-4, K562, Raji, Ramos, Namalwa, and HSB2). The SU-DHL-1 and Karpas 299 cell lines (both of which carry the (2;5) translocation) were used as positive controls.
Immunohistochemistry and antibodies.
Immunohistological staining was performed using a panel of monoclonal antibodies for B- and T-cell antigens, together with antibodies to CD30 (Dako-BerH2), and EMA (Dako-EMA/E29). Antibodies against ALK protein (ALK1 and ALKc) and the N-terminal region of nucleophosmin (NA24) used in the immunohistochemical and Western blot analysis were produced in the authors’ laboratories.10,13 21
Western blotting and in vitro kinase assay.
Cryostat sections (6 μm) were cut from frozen samples of normal tonsil and the biopsy specimens from cases no. 789 and 862. Cytocentrifuge preparations were made of the cultured SU-DHL-1 cell line. Western blotting using the monoclonal anti-ALK (ALKc) or anti-NPM (NA24) and an in vitro kinase assay were performed as previously described.19
RNA extraction and 5′ RACE reaction.
Total RNA was isolated from a frozen sample of the diagnostic lymph node using guanidine isothiocyanate extraction and cesium chloride gradient centrifugation. cDNA was obtained from 1 μg of total RNA using 2.5 pmol/L of GSP1 ALK-specific primer (5′-ACCCCAATGCAGCGAACAA-3′) and Super Script II Reverse Transcriptase (Life Technologies Inc, Paisley, UK). The RACE technique was used to obtain the 5′ sequence fused with the ALK gene following the manufacturer’s recommendations. The PCR primers used for 5′ RACE reaction were AAP primer (included in the kit) and GSP2 ALKprimer (5′-CTGGTGGTTGAATTTGCTGAT-3′) for the first round and AUAP primer (included in the kit) together with GSP3 ALK primer (5′-CTTGGGTCGTTGGGCATTC-3′) for the second nested round of PCR amplification. The specificity of the PCR fragments obtained were confirmed by hybridization with the fluorescein-labeled internal oligonucleotide ALK 3 (5′-GTCGAGGTGCGGAGCTTGCTCAGC-3′).
DNA sequencing.
The PCR products were purified from the gel and sequenced in ABI PRISM automated sequencer (Applied Biosystems, Foster City, CA). Both DNA strands were completely sequenced using different upstream and downstream primers by primer walking.
RT-PCR expression analysis of NPM-ALK, TPM3-ALK, TFG andTFG-ALK genes.
cDNA from all the analyzed samples was obtained from 1 μg of total RNA, random hexamer priming and SuperScriptTM II Reverse Transcriptase (Life Technologies Inc) following the manufacturer’s recommendations. Amplification of the RPS14 ribosomal mRNA was performed as control.NPM-ALK and TPM3-ALK expression was examined using previously described methods.3 19 Expression analysis of wild-type TFG gene was performed using TFG2 (5′-AACATCCTGGAGTCCACCATG-3′) and TFG4D (5′-GCCCTGAAACCTGATCATCTG-3′) primers to amplify a 601 bp fragment from 2 μL of cDNA. The PCR conditions were 35 cycles consisting of 45 seconds at 94°C, 45 seconds at 65°C and 45 seconds at 72°C, followed by a final extension of 20 minutes at 72°C. The PCR mixture contained 1 U of Taq (Boehringer Mannheim, Mannheim, Germany), 0.8 mmol/L each primer, 100 mmol/L dNTPs, and PCR buffer in a final volume of 25 μL.TFG-ALK expression analysis was assessed using a set of primers containing the translocation breakpoint described in this study. The primers used were TFG1 (5′-AGCTTGGAACCACCTGGAGAACC-3′)/ALK3.
RESULTS AND DISCUSSION
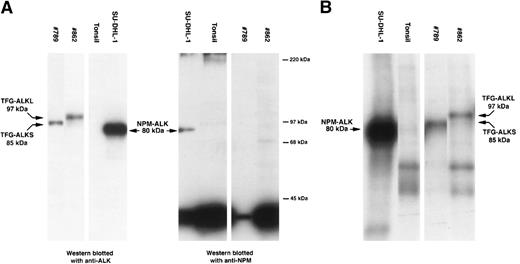
The lymph node biopsy specimen of case no. 789 showed a typical ALCL of “common” morphology with a T-cell phenotype (CD3-positive, CD5-positive) coexpressing CD30 and EMA. ALK protein was detected in all tumor cells but was restricted to the cytoplasm. RT-PCR studies forNPM-ALK and TPM-ALK chimeric products were negative. Western blot analysis (using ALKc antibody) detected an ALK protein with an apparent molecular weight of 85 kD, higher than the 80 kD of the classic NPM-ALK fusion protein.19 Immunohistochemical studies using a monoclonal antibody against the N-terminal portion of NPM only detected nuclear NPM. This was in contrast to the cytoplasm and nuclear distribution of NPM observed in cases of t(2;5)-positive NPM-ALK–positive ALCL.6 10 Furthermore, Western blotting studies only detected the presence of wild-type 38-kD NPM (Fig1A). These observations confirmed at the protein level that NPM was not fused to ALK in this tumor. However, an in vitro kinase assay showed that the ALK portion of the novel protein present in this tumor possessed tyrosine kinase activity (Fig 1B).
Biochemical assays of proteins extracted from cytocentrifuge preparations and cryostat tissue sections. (A) Western blotting. Anti-ALK (ALKc) identifies the 80-kD NPM-ALK protein present in lysates of SU-DHL-1 cells. This protein is also detected by anti-NPM (NA24). In contrast, anti-ALK detects a protein of 85 kD in lysates from case #789 and a protein of 97 kD in lysates from case #862 but only wild-type 38-kD NPM are present in these cases. Normal tonsil contains no ALK protein and only wild-type NPM. (B) In vitro kinase assay. Kinase activity is present in the 80-kD NPM-ALK protein present in the SU-DHL-1 cell line, in an 85-kD ALK protein present in case #789, and in a 97-kD ALK protein present in case #862. No corresponding band is detected in the tonsil used as negative control.
Biochemical assays of proteins extracted from cytocentrifuge preparations and cryostat tissue sections. (A) Western blotting. Anti-ALK (ALKc) identifies the 80-kD NPM-ALK protein present in lysates of SU-DHL-1 cells. This protein is also detected by anti-NPM (NA24). In contrast, anti-ALK detects a protein of 85 kD in lysates from case #789 and a protein of 97 kD in lysates from case #862 but only wild-type 38-kD NPM are present in these cases. Normal tonsil contains no ALK protein and only wild-type NPM. (B) In vitro kinase assay. Kinase activity is present in the 80-kD NPM-ALK protein present in the SU-DHL-1 cell line, in an 85-kD ALK protein present in case #789, and in a 97-kD ALK protein present in case #862. No corresponding band is detected in the tonsil used as negative control.
To identify the gene involved in this new ALK translocation, we used a 5′RACE strategy. The primers for this technique were designed within the known 3′ catalytic domain of the ALK. The result of the 5′ amplification yielded a major product of approximately 800 bp. Hybridization with an internal ALK primer confirmed the specificity of this product, which was subsequently purified, cloned, and sequenced.
The sequence of this fragment confirmed that a gene other thanNPM was fused to the 5′ region of ALK. Comparison of this sequence with the Genbank database showed 99.7% homology with the 5′ region of the TFG gene (TRK-fused gene), which has been mapped to chromosome 3q11-12.22 This gene was initially identified as the rearranged partner of the NTRK1gene in a thyroid papillary carcinoma generating the TRK-T3fusion gene that has transforming activity in different models.23 Interestingly, the TFG gene, in common with other genes which can cause oncogenic activation of receptor tyrosine kinase genes, is constitutively expressed in many different tissues.23 To determine whether the TFG gene is also expressed in lymphoid cells, we analyzed different lymphoid samples by RT-PCR and could show that wild-type TFG was constitutively expressed in all non-neoplastic lymphoid tissues, in 14 NHLs, and in 11 neoplastic hematopoietic cell lines.
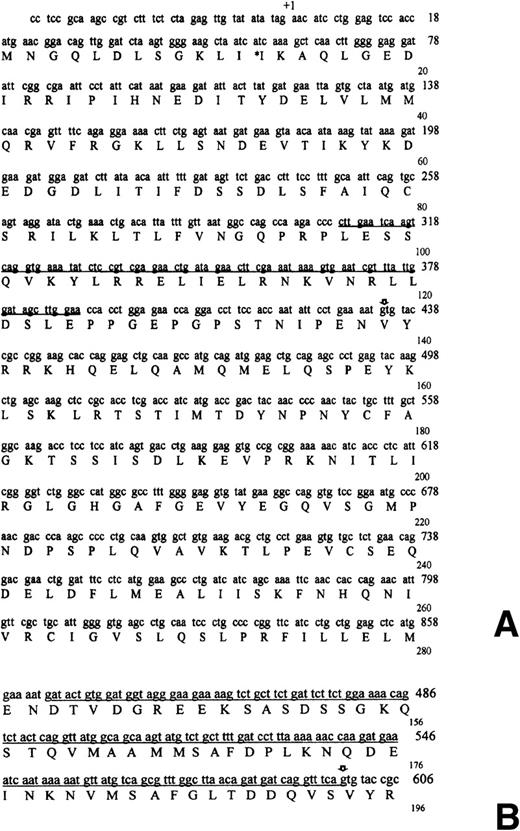
Using primers from the 5′ TFG and 3′ ALK distal regions we could amplify by RT-PCR a 2.5-Kb fragment containing the full-lengthTFG-ALK coding sequence. The 5′ TFG segment was composed of 459 bp containing 44 bp of the 5′ untranslated region and 415 bp of the coding sequence. This region included the predicted coiled-coil oligomerization domain of the protein (Fig2A). The breakpoint was located at 415 bp from the initial methionine. The only difference between theTFG coding sequence obtained in our case and that previously described (GenBank accession number Y07968) was a single nucleotide change in codon 13 (GTC → ATC; Val → Ile). This change seems to be a polymorphic variant because it was also present in genomic DNA obtained from normal epithelial cells of the patient.
Nucleotide and deduced amino acid sequences ofTFG-ALK cDNA in short TFG-ALKS (A) and long TFG-ALKL (B) forms. (A) Nucleotide sequence was numbered according to the previously described sequences. The putative coiled-coil domain of the TFG sequence is underlined. The asterisk marks a previously undescribed polymorphism (gtc → atc; Val → Ile). The translocation breakpoint in the cDNA is marked with an arrow. (B) The additional 165 bp TFG sequence is underlined. The translocation breakpoint in the cDNA is marked with an arrow.
Nucleotide and deduced amino acid sequences ofTFG-ALK cDNA in short TFG-ALKS (A) and long TFG-ALKL (B) forms. (A) Nucleotide sequence was numbered according to the previously described sequences. The putative coiled-coil domain of the TFG sequence is underlined. The asterisk marks a previously undescribed polymorphism (gtc → atc; Val → Ile). The translocation breakpoint in the cDNA is marked with an arrow. (B) The additional 165 bp TFG sequence is underlined. The translocation breakpoint in the cDNA is marked with an arrow.
The ALK sequence in our case was identical to that previously described,3 and it was in frame with the upstreamTFG open reading frame. Noticeably, the ALK breakpoint in this TFG-ALK cDNA was the same as that found in theNPM-ALK fusion gene. The predicted chimeric TFG-ALK protein was composed of 701 aminoacids, 138 of which were encoded by theTFG gene and 562 by the ALK gene. The valine in position 139 is encoded by a new codon created by the rearrangement. The predicted molecular weight of the new TFG-ALK fused protein is 83 kD, which is very similar to the value obtained by Western blot analysis (85 kD).19
To determine if this new TFG-ALK translocation was expressed in other ALCLs, we analyzed 6 additional tumors with ALK protein expression restricted to the cytoplasm and which were negative forNPM-ALK and TPM3-ALK translocations. In addition, 3NPM-ALK-positive and 1 TPM3-ALK–positive ALCL, and 5 thyroid carcinomas were also analyzed. Expression of theTFG-ALK chimeric product was examined with a set of primers from TFG and ALK genes spanning 141 bp of the breakpoint region. The TFG-ALK chimeric transcript could be amplified only in our control tumor and also in 1 ALCL (case #862) that was negative for NPM and TPM3-ALK translocations. The cytogenetic analysis of this tumor had shown the presence of a (2;3)(p23;q21) translocation.17 Interestingly, however, the amplified product in this case showed an unexpected larger size (306 bp) than that obtained (141 bp) in our control case. Sequencing of this product confirmed a TFG-ALK rearrangement in which theALK breakpoint was the same as in the other ALKtranslocations. However, the TFG fragment contained an additional 165 bp sequence from the TFG gene not present in the hybrid gene in our previous case (Fig 2B). However, the TFGbreakpoint region in this second case was the same as in theTFG-NTRK1 translocation in thyroid carcinomas.23 In this new rearrangement creating a larger TFG-ALK(TFG-ALKL) gene, the additional 165 bpTFG sequence was also in frame with the 5′ TFG fragment of our previous shorter TFG-ALK(TFG-ALKS) translocation, and with the distal 3′ ALK sequence. The predicted molecular weight of this new chimeric TFG-ALKL protein was 96 kD, in keeping with the value of 97 kD determined by Western blot and in vitro kinase analysis of this case (Fig 1).
These findings indicate that TFG is an alternative toNPM as a partner for ALK in ALCL, and that it can be involved in 2 structurally different translocations,TFG-ALKL, encoding 85-kD and 97-kD chimeric proteins, respectively. An in vitro kinase assay showed that these proteins also had tyrosine kinase activity (Fig 1) indicating that the new cloned cDNA sequences are translated into chimeric proteins with functional activity. The 5′ TFG segment included in this translocation contains the full predicted coiled-coil oligomerization domain of the gene. Similarly to the 5′ region of NPM in theNPM-ALK translocation, this TFG domain is absolutely required for oligomerization and transforming activity ofTFG-NTRK (TRK-T3) oncogene.24 Therefore, it is also possible that TFG may play a similar role in the activation of ALK in ALCL. However, in distinction toNPM, the TFG gene lacks nuclear-signaling domains, which is in keeping with the restricted cytoplasmic pattern ofALK expression observed in our tumors. These findings support previous in vitro experiments, indicating that the oncogenic activity of rearranged ALK is independent of the nuclear localization.5 6
L.H. and M.P. contributed equally to this study.
Supported by The Leukemia Research Found (Grant No. #94/46); the Comision Interministerial de Ciencia y Tecnologia (SAF 99/20), Asociacion Española Contra el Cancer, CIRIT, Generalitat de Catulunya (98SGR21); and the AIRP (Associasione Italiana per la Ricerca sul Cancro). Genbank accession numbers for the TFG-ALKS and TFG-ALKL sequences are AF125093 and AF143407, respectively.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Elias Campo, MD, Laboratory of Pathology, Hospital Clinic, Villarroel 170, 08036-Barcelona, Spain; e-mail: campo@medicina.ub.es.