Abstract
Deletions involving the long arm of chromosome 11 (11q) have been recently found as recurrent chromosome aberrations in mantle cell lymphoma (MCL). In the current study, the incidence and molecular extent of 11q deletions were analyzed in a series of 81 MCL by fluorescence in situ hybridization with probes from a contiguous set of yeast artificial chromosomes (YACs). Loss of chromosome 11 material was observed in 37 of 81 cases (46%). The minimally deleted segment comprised YAC 801e11 containing the ATM gene. To further narrow the minimal region of loss, P1-derived artificial chromosomes mapping to the critical region were isolated and used as probes in cases without aberrations detectable with YACs. This allowed the identification of an ATM deletion that was beyond the resolution of YAC probes. The identification of a minimally deleted segment affecting ATM suggests a pathogenic role of ATMas a tumor suppressor gene in MCL.
MANTLE CELL LYMPHOMA (MCL) is a specific subtype of non-Hodgkin’s lymphoma (NHL) characterized by distinct clinical, morphological, and genetic features.1 The cytogenetic hallmark of MCL is the translocation t(11;14)(q13;q32).2,3 This aberration leads to a rearrangement of the BCL1 (11q13) and IgH (14q32) loci resulting in the deregulation of the cyclin D1 gene (CCND1).4-6 The overexpression of CCND1 at the mRNA and protein levels is highly characteristic of MCL.6-8 Because the cyclin D1 protein plays a crucial role in the regulation of G1-S phase transition of the cell cycle, theCCND1 deregulation by the t(11;14)(q13;q32) is considered significant for MCL pathogenesis. However, transgenic mice overexpressing cyclin D1 do not spontaneously develop lymphoma, and other oncogenic factors such as c-MYC are necessary for tumor formation.9 10 Therefore, additional genetic events appear to be required for the malignant transformation of MCL cells.
By chromosome banding analyses, aberrations in addition to the t(11;14)(q13;q32) have been found in MCL.11,12 Deletions involving bands 11q22-q23 have been recently observed as recurrent aberrations in MCL13 (and Bentz et al, submitted), and are also frequent in other B-cell lymphoproliferative disorders.14 The current study was aimed at the identification of the minimally deleted segment in 11q likely harboring a tumor suppressor gene potentially involved in the pathogenesis of MCL.
MATERIALS AND METHODS
Tumor samples were prepared from 81 patients with MCL and molecular cytogenetic analysis was performed as previously described.14 The specimens of the 37 MCL with 11q deletions were derived from lymph node (n = 20), peripheral blood (n = 13), spleen (n = 1), tonsil (n = 1), stomach (n = 1), or conjunctiva (n = 1). Diagnoses were based on morphologic and immunophenotypic analyses.1 Among the 37 MCL with 11q deletions, the t(11;14)(q13;q32) corresponding to a CCND1/IgH rearrangement was present in all 35 cases tested by our interphase fluorescence in situ hybridization (FISH) assay.15 Molecular cytogenetic analysis was performed by dual-color FISH with a physically mapped probe set of yeast artificial chromosome (YAC) clones (obtained from the CEPH library, Généthon, Fondation Jean Dausset, Paris, France) spanning bands 11q14 to 11q24 as previously described (for YAC numbers, STS markers, and genes contained, see Fig1).14,16 P1-derived artificial chromosome (PAC) probes specific for ATM (PAC ATM-1, LLNLP704G18220Q19; PAC ATM-2, LLNLP704O01298Q19) and for YAC 755b11 (PAC 755b11, LLNLP704H1725Q13) were identified from a human PAC library (RPCI segments 1, 3-5) obtained from the Resource Center/Reference Library of the German Human Genome Project (Berlin, Germany) by hybridization with ATM cDNA clone pCEV7-9 and Alu-polymerase chain reaction (PCR) products derived from the YAC, respectively. TheATM exon content of the PACs was determined by PCR analyses.17
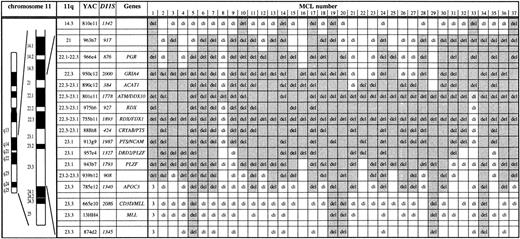
Mapping of deletions involving chromosome bands 11q14 to 11q24 in 37 MCL by FISH. Chromosome 11 ideogram, band designation, CEPH YAC address, DNA locus, and genes contained in the probe set are given. YACs 950c12 through 785e12 form a contig and therefore allowed a precise deletion mapping. del, deletion (1 FISH signal); di, disomy (2 FISH signals); 3, partial trisomy in MCL no. 1 (3 FISH signals). The extent of the deletion in each case is indicated by shading. The half shading of YAC 801e11 in MCL no. 33 indicates partial deletion of this probe. Three MCL (nos. 1, 3, 4) were included in a previous study.14 The minimal deletion region lost in all MCL analyzed is the centromeric part of YAC 801e11 to which the ATMgene maps. The centromeric border of the minimal deletion is defined by MCL nos. 8 and 21. The telomeric border is defined by MCL no. 33 exhibiting loss of PAC ATM-1 and -2 and retention of 2 signals for YAC 801e11.
Mapping of deletions involving chromosome bands 11q14 to 11q24 in 37 MCL by FISH. Chromosome 11 ideogram, band designation, CEPH YAC address, DNA locus, and genes contained in the probe set are given. YACs 950c12 through 785e12 form a contig and therefore allowed a precise deletion mapping. del, deletion (1 FISH signal); di, disomy (2 FISH signals); 3, partial trisomy in MCL no. 1 (3 FISH signals). The extent of the deletion in each case is indicated by shading. The half shading of YAC 801e11 in MCL no. 33 indicates partial deletion of this probe. Three MCL (nos. 1, 3, 4) were included in a previous study.14 The minimal deletion region lost in all MCL analyzed is the centromeric part of YAC 801e11 to which the ATMgene maps. The centromeric border of the minimal deletion is defined by MCL nos. 8 and 21. The telomeric border is defined by MCL no. 33 exhibiting loss of PAC ATM-1 and -2 and retention of 2 signals for YAC 801e11.
RESULTS AND DISCUSSION
Screening for 11q deletions was initially performed by interphase FISH with YAC 755b11 and YAC 801e11 mapping to previously identified deletion regions in lymphoid neoplasms.14,17,18 Deletions involving at least one of these probes were observed in 36 of the 81 MCL studied (46%). No biallelic deletion was found. The percentage of cells carrying a deletion ranged from 13% to 98.5% (median, 84%). The 46% incidence of 11q loss in MCL is remarkably higher than assumed from smaller series,13 and exceeds the frequency of 11q deletions in B-CLL.15 The high incidence of 11q22-q23 deletion, together with the frequent observation of 13q14 deletion in MCL and B-CLL,19 suggests common genetic mechanisms in the pathogenesis of the two diseases despite their different clinical behavior.
The extent of the 11q deletions was determined by FISH with a probe set consisting of a YAC contig spanning bands 11q14 to 11q24 (Fig 1). Among the 36 MCL with 11q deletions detected, a minimal deletion region affecting YAC 801e11 was established. The genomic segment corresponding to this YAC probe was lost in all MCL cases with 11q deletions studied and in one of the MCL was the sole fragment lost (no. 8 in Fig 1). YAC 801e11 is 1.2 Mb in size and contains the genomic region of theATM gene.
To further narrow the minimal deletion segment and to test whetherATM or an adjacent locus was affected by the 11q deletions in MCL, we isolated PACs containing ATM coding sequence (PAC ATM-1 and -2) and a PAC from the YAC 755b11 region (PAC 755b11). Exons 4 to 49 and exons 25 to 65 of the ATM gene were contained in PAC ATM-1 and -2, respectively, as detected by PCR analysis. Forty MCL cases showing no aberration on initial FISH screening with YACs 801e11 and 755b11 were subjected to analysis with these PAC probes. One MCL (no. 33, Fig 1) was found to carry a deletion of the genomic region corresponding to PAC ATM-1 and -2. Subsequent analysis with YAC probes showed that this MCL carried a deletion extending in centromeric direction (see Fig 1). Because PAC ATM-1 and -2 are located in the centromeric end of YAC 801e11, this further narrowed the minimal deletion region lost in all MCL with 11q deletions analyzed to the centromeric portion of YAC 801e11 where the ATM gene resides (Fig 1). Also affected by the minimal deletion is NPAT, which is located centromeric of ATM and shares the same promoter region. However, there are no data suggesting a tumor suppressor function for NPAT and it has not been linked to hematological neoplasms. Interestingly, our data are at variance to a concurrent report by Monni et al,20 who analyzed 20 MCL with 11q deletions by FISH. In this study, one MCL with an isolated deletion of the genomic region of YAC 755b11 was identified, which is located 2 to 3 Mb telomeric of ATM. In all other cases, larger deletions also affecting ATM were found.
Disruption of both ATM alleles in line with the two-hit model of tumor suppressor gene inactivation was previously demonstrated in T-PLL.17,18 In addition, recent studies showed loss ofATM protein expression and mutational disruption of ATMin B-cell chronic lymphocytic leukemia, suggesting a pathogenic role also in B-cell malignancies.21-24 The identification of an 11q22-q23 minimal deletion region in MCL tumor cells specifically affecting ATM points to a role ofATM as a tumor suppressor gene in MCL. This is supported by the fact that in a series of B-cell neoplasms recently analyzed there were two MCL cases showing ATM mutations.24 In one of these cases mutations were found in both ATM alleles in the absence of a genomic deletion.
Although the function of the ATM gene product is not fully established, studies of ataxia-telangiectasia cell lines andATM-deficient mice have shown that ATM is a key regulator in response to DNA strand breaks induced by mutagenic agents or physiological processes such as VDJ recombination.25 In this context, it is interesting to note that the hallmark of MCL is the rearrangement of the IgH locus with the proto-oncogeneCCND1 as a result of illegitimate VDJ recombination. Based on the current results, this aberrant VDJ recombination in MCL and the subsequent acquisition of additional genetic changes leading to complex karyotypes could result from a faulty surveillance of the genomic integrity by loss of the ATM gene product.
ACKNOWLEDGMENT
The excellent technical assistance of Kathrin Wildenberger and Traudel Weilguni is gratefully acknowledged. Yosef Shiloh generously provided the pCEV7-9 ATM cDNA clone. The PACs were isolated from libraries created in the laboratory of P.J. de Jong at Roswell Park Cancer Institute (Buffalo, NY) and generously provided by the Resource Center/Primary Database of the German Human Genome Project (Berlin, Germany).
Supported by grants from the Deutsche Krebshilfe (10-1289-St 1), the Wilhelm Sander-Stiftung (97.003.1), and the Tumorzentrum Heidelberg/Mannheim (I/I.1).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hartmut Döhner, MD, Department of Internal Medicine III, University of Ulm, Robert Koch Str 8, 89081 Ulm, Germany.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal