Abstract
In acute myeloid leukemia (AML), granulocyte colony-stimulating factor receptor (G-CSFR) proliferative and maturational signaling pathways are uncoupled. Seven human G-CSFR mRNA isoforms exist, named class I through class VII. The 183-amino acid cytosolic domain of the class I isoform provides all signaling activities. The class IV isoform is “differentiation defective” because the carboxy-terminal 87 amino acids are replaced with 34 amino acids of novel sequence. In more than 50% of AML samples, the class IV/class I G-CSFR mRNA ratio is aberrantly elevated compared to normal CD34+ bone marrow cells. We hypothesized that the increased relative expression of class IV G-CSFR in AML uncouples proliferative and maturational G-CSFR signaling pathways. To test this, we transfected the G-CSF–responsive murine cell line 32Dcl3 with class IV G-CSFR cDNA. After 10 days of G-CSF stimulation, clones expressing class IV G-CSFR had greater percentages of myeloblasts and promyelocytes than controls (53% ± 13% versus 3% ± 2%). Differential counts over time demonstrated delayed G-CSF–driven maturation in 5 class IV-expressing clones, with 2 clones demonstrating a subpopulation that completely failed to differentiate. Heterologous class IV expression did not affect G-CSF–dependent proliferation. Class IV/murine G-CSFR mRNA ratios after 24 hours of G-CSF stimulation for 3 of the 5 clones (range, 0.090 to 0.245; mean, 0.152 ± 0.055) are within the range of class IV/class I mRNA ratios seen in patients with AML. This indicates that aberrantly increased relative class IV G-CSFR expression seen in AML can uncouple G-CSFR proliferative and maturational signaling pathways.
Granulocyte colony-stimulating factor (G-CSF), the critical growth factor for the normal production of mature circulating neutrophils, acts on myeloid cells during all stages of development.1-6 Because of the ability of G-CSF to promote the maturation of myeloid cells, it was tested as a potential means of differentiation therapy for acute myeloid leukemia (AML).7-10 However, the typical effect was to stimulate proliferation in blasts of patients with AML, with maturation either aberrant or absent.11-13 This indicated that the proliferative and maturational responses to G-CSF are somehow uncoupled in AML. Mutation of the G-CSF receptor (G-CSFR) or alterations in the levels or functions of the downstream signaling molecules have been proposed as hypotheses to explain this aberrant response in AML.14-19
The human G-CSFR is known to occur in 7 isoforms (class I through class VII), all produced by alternative splicing of the single gene transcript.20-24 Only the class I and class IV G-CSFR mRNA isoforms are detected at significant levels in myeloid cells.25 Functional mapping studies have shown that the carboxy-terminal 87 amino acids of the class I G-CSFR are crucial to the maturational signaling function of the G-CSFR, whereas the membrane proximal 96 amino acids are fully competent for driving proliferation.14,22,26-31 The class IV G-CSFR isoform replaces the carboxy-terminal 87-amino acid maturation domain of the full-length class I G-CSFR with 34 amino acids of novel peptide sequence, thus leaving the proliferative capacity of this isoform intact. However, when class IV G-CSFR is expressed in isolation, it fails to mediate G-CSF–driven myeloid differentiation.21,22,28 We have previously demonstrated that in blasts of patients with AML, the class IV G-CSFR isoform is coexpressed at relatively increased levels compared with normal immature myeloid cells.25 Given the “differentiation-defective” nature of the class IV G-CSFR isoform, we have examined whether this alteration in the relative levels of G-CSFR isoform coexpression is sufficient to uncouple the proliferative and maturational responses to G-CSF that typify AML.
Materials and methods
Cells
The murine myeloblastic cell line 32Dcl3 expresses endogenous murine G-CSFR and differentiates into phenotypic neutrophils in response to G-CSF stimulation.32 The parental cell line is maintained in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin G, 100 μg/mL streptomycin, 1 mmol/LL-glutamine, and 10% WEHI 3B D+cell-conditioned medium as a source of murine interleukin-3 (mIL-3).
Expression vector constructs and transfection
The cDNA clone D-7 (generous gift from S. Zeigler) in pBluescript (Stratagene, La Jolla, CA) was isolated after digestion withEcoRI and NotI. This fragment was then cloned into theEcoRI/NotI digested expression vector pcDNA3.1(−)/NEO (Clontech, Palo Alto, CA). 32Dcl3 cells were then electroporated as described elsewhere33 with either the pcDNA3.1(−)/NEO/D-7 (D-7) or the pcDNA3.1(−)/NEO (NEO) construct, and stable transfectants were selected with 600 μg/mL Geneticin (Life Technologies, Grand Island, NY). Limiting dilution was used to isolate individual clones from each bulk population, and their expression of murine G-CSFR and D-7 cDNA was determined using the polymerase chain reaction (PCR)-based method described below.
Reverse transcription–polymerase chain reaction and quantitative polymerase chain reaction
Total RNA was isolated from each clone using the TRIzol reagent (Life Technologies, Grand Island, NY). Two micrograms RNA was then used in a reverse transcription–polymerase chain reaction (RT-PCR) described elsewhere.25 The PCR primers used for amplification of D-7 cDNA sequence (HGR5, HGR3) have been published.25 The PCR primers used for amplification of the murine G-CSFR were MGR5 5′-CCACTACACCATCTTCTG-3′ and MGR3 5′-CCAAGAGGGGCTGAGTGG-3′. The primer combination for quantitative (Q)-PCR measurement of the D-7/murine G-CSFR mRNA ratio is MGR5 (200 nmol) plus HGR3 (100 nmol) and MGR3 (100 nmol). Data were discarded unless equivalent PCR amplification efficiency was confirmed for the murine G-CSFR and D-7 PCR fragments over a minimum of 3 cycles during the linear phase of amplification.
Maturation assay
32Dcl3 clones were stimulated with rhG-CSF (G-CSF; Amgen, Thousand Oaks, CA) 100 ng/mL, and differential counts were made from preparations of Wright–Giemsa-stained cells at days 5, 7, and 10 of culture. To delay apoptosis and allow up-regulation of endogenous G-CSFR expression, recombinant murine IL-3 (Peprotech, Rocky Hill, NJ) was added at a concentration of 0.1 ng/mL during the initial 72 hours of G-CSF stimulation. This dose of IL-3 was determined to be adequate to suppress apoptosis but not to induce more than 1 cycle of cell division over 72 hours, as measured by the proliferation assay discussed below (data not shown).
Proliferation assay
The proliferative response of D-7 and NEO clones was measured using the Cell Census System (Sigma, St Louis, MO). One million cells were labeled with PHK26 and divided into 3 equal aliquots. Immediately, 1 aliquot was fixed in 2% paraformaldehyde (Sigma) in phosphate-buffered saline (PBS). The remaining 2 aliquots were stimulated with either 100 ng/mL G-CSF or 30 ng/mL recombinant mIL-3. After 72 hours of cytokine stimulation, the cells were fixed in 2% paraformaldehyde in PBS for subsequent flow cytometric evaluation according to the manufacturer's protocol in comparison to unstimulated cells fixed immediately after staining. Flow cytometric data were then analyzed using Cell Census System (Sigma) software, and a “proliferation index” was computed that represented the average number of generations the initial population of cells produced, with the initial population having a proliferation index of 1.00. The proliferation index after G-CSF stimulation for an individual clone was divided by the proliferation index after IL-3 stimulation for that clone as a means of normalizing for the clone's baseline proliferative capacity.
Statistics
The Student t test was used to analyze results from the maturation and proliferation studies (P values). The Pearson product moment correlation coefficient was used to test the correlation between the maturation data and class IV/murine G-CSFR mRNA ratios (r value).
Results
Differentiation-defective G-CSFR expression in 32Dcl3 cells at levels observed in AML cells blocks G-CSF–mediated maturation
To assess the ability of the class IV G-CSFR isoform to uncouple the proliferative and maturational signaling functions of the full-length G-CSFR, we expressed the class IV G-CSFR cDNA (D-7) in the IL-3–dependent murine myeloblastic cell line 32Dcl3. This cell line expresses the murine G-CSFR, a structural and functional homologue of the human class I G-CSFR, and differentiates into phenotypic neutrophils with G-CSF stimulation. Individual clones expressing either the class IV G-CSFR (D-7 number) or empty expression vector (NEO number) were stimulated with G-CSF 100 ng/mL, and their phenotypic maturation was assessed over 10 days. Initial screening demonstrated that all the D-7 clones examined demonstrated a significant subpopulation that failed to enter the postmitotic phase of maturation (46.9% ± 13.4%) in comparison to the NEO clones (97.0% ± 2.2%; P ≤ .001) (Figure1 and Table1). All clones were cytokine dependent based on the observation that more than 99% of cells for each clone were nonviable by trypan blue staining after 72 hours of withdrawing cytokine (data not shown).
Photomicrographs of 32Dcl3 cell clones following 10 days of G-CSF stimulation (100 ng/mL).
(A) Clone NEO.1, (B) Clone D-7.1, (C) Clone D-7.3, and (D) Clone D-7.4. (Wright-Giemsa stain, original magnification × 1000 [with oil immersion lens].)
Photomicrographs of 32Dcl3 cell clones following 10 days of G-CSF stimulation (100 ng/mL).
(A) Clone NEO.1, (B) Clone D-7.1, (C) Clone D-7.3, and (D) Clone D-7.4. (Wright-Giemsa stain, original magnification × 1000 [with oil immersion lens].)
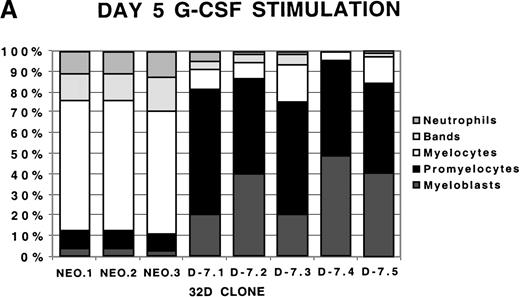
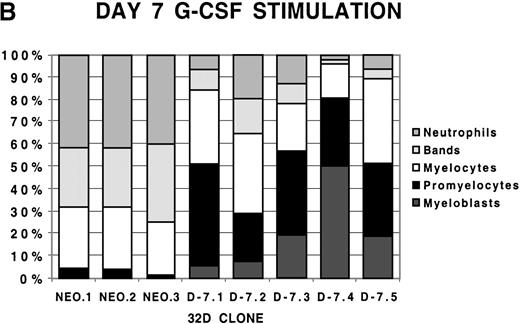
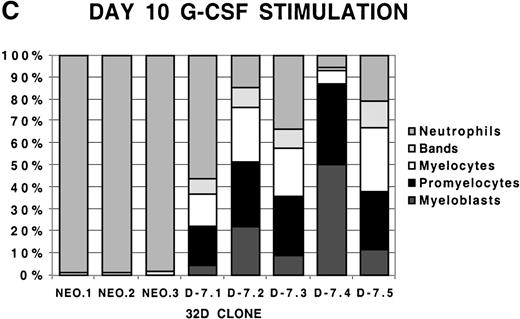
To examine more closely the dynamics of the effect of class IV G-CSFR coexpression on G-CSF–driven maturation, differential counts were performed at days 5, 7, and 10 of G-CSF stimulation on select D-7 and NEO clones; results are shown in Figure 2. When all clones were compared at day 5 of G-CSF stimulation, it was clear that in the NEO clones, 87.3% ± 0.85% of cells were in the postmitotic phase of differentiation, whereas in the D-7 clones, only 15.8% ± 6.5% (P ≤ .001) of cells were postmitotic. By day 7, this increased to between 94.0% ± 4.4% for the NEO clones but only to 45.0% ± 16.7% (P = .001) for D-7 clones. By 10 days of G-CSF stimulation, 100% ± 0% of the cells in each of the NEO clones were postmitotic. However, only 54.0% ± 21.9% of cells in the D-7 clones were in the postmitotic phase of maturation. When individual clones were examined over time, it was observed that for NEO clones there was a consistent progression of all cells forward along the myelopoietic pathway. In the D-7 clones, this progression was dramatically delayed. Even more intriguing was that in 2 of the 5 D-7 clones studied (clones D-7.2 and D-7.4), a sizable myeloblastic subpopulation expanded between days 7 and 10 of G-CSF stimulation. This demonstrated that coexpression of the class IV G-CSFR isoform delays, and in some circumstances blocks, G-CSF–driven differentiation.
Differential counts of 32Dcl3 cell clones over time during G-CSF stimulation.
Clones NEO.1-NEO.3 and D-7.1-D-7.5 were stimulated with G-CSF 100 ng/mL and differential counts performed on Wright-Geimsa stained samples at day 5 (A), day 7 (B), and day 10 (C) of culture.
Differential counts of 32Dcl3 cell clones over time during G-CSF stimulation.
Clones NEO.1-NEO.3 and D-7.1-D-7.5 were stimulated with G-CSF 100 ng/mL and differential counts performed on Wright-Geimsa stained samples at day 5 (A), day 7 (B), and day 10 (C) of culture.
We have previously observed ratios of class IV/class I G-CSFR mRNA in patients with AML, ranging from 0.057 to 0.158 (mean, 0.106 ± 0.035)25 (and unpublished data). These values are significantly greater than the ratios observed in the CD34+ fraction of normal bone marrow cells (range, 0.050 to 0.080; mean, 0.065 ± 0.009).25 Coexpression of the differentiation-defective class IV G-CSFR isoform in 32Dcl3, which expressed a full-length G-CSFR, acted to disrupt the G-CSFR maturational signaling activities. To determine whether relative overexpression of the class IV G-CSFR isoform could have been responsible for the disruption of G-CSFR maturational signaling that occurred in patients with AML, we proceeded to measure the ratio of class IV/murine G-CSFR mRNA to compare to the class IV/class I mRNA ratios previously observed in patients with AML. This ratio, determined using Q-PCR, was measured on mRNA harvested from 2 different populations—cells maintained in IL-3 and cells stimulated with G-CSF 100 ng/mL for 24 hours. The latter group was included to account for the G-CSF–induced up-regulation of murine G-CSFR mRNA. The results of this analysis are shown in Table 2. At baseline, the class IV/murine G-CSFR mRNA ratios ranged from 0.225 to 0.560 (mean, 0.377 ± 0.123). After 24 hours of G-CSF stimulation and the expected up-regulation of the murine G-CSFR mRNA, these ratios changed to 0.090 to 0.245 (mean, 0.152 ± 0.055). The ratios observed here overlapped significantly with those previously observed in patients with AML (P = .14) and not with those observed for normal immature myeloid cells (P = .03).
The degree of maturation delay or block varied among the D-7 clones examined, as did the class IV/murine G-CSFR mRNA ratio. It was considered that an increasing relative level of class IV G-CSFR mRNA might result in an increase in the degree of maturation delay. Statistical analysis looking for such a correlation between the class IV/murine G-CSFR mRNA ratio and the percentage of cells failing to differentiate terminally after 10 days of G-CSF stimulation found no correlation between these values (r = −0.329) (Figure3).
Plot of percent mitotic cells versus human class IV/murine G-CSFR mRNA ratio.
A plot of percent mitotic cells (myeloblasts + promyelocytes) versus the class IV/murine G-CSFR mRNA ratio is shown. The Pearson product moment correlation coefficient [r] for this plot is -0.329 indicating a poor correlation between the class IV/murine G-CSFR mRNA ratio and the fraction of cells that remain in the mitotic phase of myelopoiesis for each clone.
Plot of percent mitotic cells versus human class IV/murine G-CSFR mRNA ratio.
A plot of percent mitotic cells (myeloblasts + promyelocytes) versus the class IV/murine G-CSFR mRNA ratio is shown. The Pearson product moment correlation coefficient [r] for this plot is -0.329 indicating a poor correlation between the class IV/murine G-CSFR mRNA ratio and the fraction of cells that remain in the mitotic phase of myelopoiesis for each clone.
Differentiation-defective G-CSFR expression in 32Dcl3 cells does not affect G-CSF–mediated proliferation
Coexpression of the class IV G-CSFR with a full-length G-CSFR at levels seen in patients with AML disrupted maturational signaling in a manner similar to that seen in AML. If our model system correctly represented the physiology of class IV G-CSFR activity in AML, then our D-7 clones should have demonstrated a proliferative response to G-CSF that was at least as vigorous as the NEO control clones.
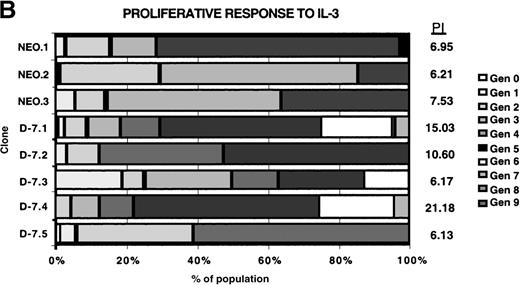
Proliferation was measured using the Cell Census System (Sigma) to generate a proliferation index (PI) as outlined in “Materials and methods,” with the results shown in Figure4. Each clone was stimulated with 30 ng/mL rmIL-3 to determine the baseline proliferative capacity (PI [IL-3]), and then the response to 100 ng/mL G-CSF stimulation for each clone was measured (PI [G-CSF]). For comparison between clones, the PI [G-CSF] was divided by the PI [IL-3] for each clone to normalize for the baseline proliferative capacity of that clone, with the result expressed as the percentage of IL-3 response. The normalized proliferative response to G-CSF was unchanged in the D-7 clones (16.5% to 22.5%; mean, 19.1% ± 0.02%) compared with the NEO clones (15.1% to 3.9%; mean, 18.63% ± 0.04%; P = .88). This clearly demonstrated that coexpression of the class IV G-CSFR isoform with the full-length G-CSFR selectively disrupted the maturational signaling pathway; a pattern identical to that seen in most patients with AML.
Proliferative response of 32Dcl3 cell clones to G-CSF or IL-3.
Using the Cell Census Plus System (Sigma) the proliferative response of clones NEO.1-NEO.3 and D-7.1-D-7.5 was measured following stimulation with either G-CSF (100 ng/mL) or IL-3 (30 ng/mL) for 72 hours. The raw data indicating the percent of cells in each generation (Gen 0-Gen 9) and the “proliferation index” (see “Materials and methods”) is shown at the right for clones stimulated with G-CSF (A) or IL-3 (B). The proliferation index for G-CSF stimulation was normalized to the proliferation index for IL-3 stimulation for each clone and this value (% IL-3 response) is presented in C.
Proliferative response of 32Dcl3 cell clones to G-CSF or IL-3.
Using the Cell Census Plus System (Sigma) the proliferative response of clones NEO.1-NEO.3 and D-7.1-D-7.5 was measured following stimulation with either G-CSF (100 ng/mL) or IL-3 (30 ng/mL) for 72 hours. The raw data indicating the percent of cells in each generation (Gen 0-Gen 9) and the “proliferation index” (see “Materials and methods”) is shown at the right for clones stimulated with G-CSF (A) or IL-3 (B). The proliferation index for G-CSF stimulation was normalized to the proliferation index for IL-3 stimulation for each clone and this value (% IL-3 response) is presented in C.
Discussion
In this study, we have demonstrated that coexpression of the class IV G-CSFR isoform with the full-length murine G-CSFR is able to block completely G-CSF–driven myeloid differentiation in a cell line capable of terminal phenotypic myeloid maturation. However, this coexpression leaves G-CSF–driven proliferation unaffected. The maturation block is observed at ratios of class IV/murine G-CSFR mRNA in the range of 0.090 to 0.245, values similar to the class IV/class I G-CSFR mRNA ratios we previously demonstrated in patients with AML.25 This implicates overexpression of the class IV G-CSFR isoform in the uncoupling of the proliferative and maturational responses to G-CSF typical of patients with AML.
How coexpression of the class IV G-CSFR uncouples the proliferative and maturational signaling pathways of the full-length G-CSFR is unclear. It is presumed that the class I and class IV G-CSFR isoforms can freely lead to homodimerization and heterodimerization after ligand binding. Based on this assumption, 2 general mechanisms can be hypothesized. One would be that the class IV G-CSFR acts as a dominant-negative to the class I G-CSFR and would then interfere with the activation of all the necessary signaling molecules required to halt cell cycling, initiate granule formation, and promote nuclear segmentation. The other mechanism would involve the activation of novel signaling molecules by the class IV G-CSFR. Both mechanisms would depend on the total number of class IV receptors at the cell surface; however, the dominant-negative mechanism would be more sensitive to the ratio of receptor isoforms coexpressed. We found no correlation between the G-CSFR isoform ratio and the degree of maturation block in our study, but the ratio varied over a narrow range for the isolated clones.
Studies of the comparatively rare severe congenital neutropenia (SCN)-related G-CSFR mutations provide insight into how aberrant relative class IV G-CSFR expression could uncouple the proliferative and maturational G-CSFR signaling pathways in most patients with AML by a dominant-negative mechanism. The mutant G-CSFR isoforms found in a subset of patients with SCN are the result of nonsense mutations that truncate the C-terminal 83 to 98 amino acids of the receptor.14,19,34-37 This results in a receptor structure similar to that of class IV G-CSFR in that they all lack the C-terminal amino acid sequence critical to driving myeloid maturation. However, the SCN mutants cannot activate additional signaling molecules, whereas the class IV G-CSFR isoform, with its novel carboxy-terminal peptide sequence, may indeed activate yet unidentified signaling molecules. Functionally, the SCN-related G-CSFR mutants, when expressed alone or in combination with class I G-CSFR, act to delay markedly the ligand-induced G-CSFR internalization, caused in part by the absence of a di-leucine motif, and they produce a hyperproliferative response to G-CSF stimulation.15,18,19 38 The class IV G-CSFR also lacks the di-leucine internalization motif and could be expected to behave in a manner similar to the SCN-related G-CSFR mutant isoforms. However, in our clones, coexpression of the class IV isoform did not impart a hyperproliferative response to G-CSF stimulation. This might have been caused by a difference in the receptor isoform ratio in our clones versus those in the SCN-related G-CSFR mutant studies, in which relative mRNA levels were not determined. It also might have been caused by signaling activity derived from the unique carboxy-terminus of the class IV G-CSFR. In either setting, it is still possible that the same mechanisms that supported the hyperproliferative response to G-CSF in the studies of SCN-related G-CSFR mutants also participated in the maturation block observed in our class IV G-CSFR–expressing clones.
The delay in G-CSFR internalization seen with the SCN-related G-CSFR mutants is associated with prolonged-activation Stat5 and, to a lesser extent, Stat3.15,16 The relevance of this observation has been discussed by other authors.15 16 Preliminary data from our laboratory (not shown), however, suggest that prolonged G-CSF–mediated Stat5 or Stat3 activation does not occur in the setting of class IV G-CSFR/murine G-CSFR coexpression. If this observation holds true, it may be that it is not the duration of activation but rather the balance between the activation of Stat5 and Stat3 that is most critical to disrupting the myeloid maturation process. Therefore, the prolonged activation of Stat5 may be more relevant to the hyperproliferative response to G-CSF imparted by the SCN-related mutant G-CSFR isoforms. This would also explain why we observed a maturation block without a hyperproliferative response to G-CSF stimulation in our class IV/murine G-CSFR coexpression model system.
Delayed mutant G-CSFR internalization may also contribute to the prolonged activity of the mitogenic inositol triphosphate products of phosphatidyl inositol 3′-kinase (PI-3 kinase) caused by the failure to activate SH-2–containing inositol phosphatase (SHIP).39 Inositol triphosphates are known to support cell survival, prevent apoptosis, and augment G-CSF–mediated proliferation.39 This is another possible mechanism that allows the SCN-related mutant G-CSFR isoforms, and potentially the class IV G-CSFR, to override the maturational signals emanating from the full-length G-CSFR.
An alternative to the dominant-negative mechanism of the SCN-related G-CSFR mutants has the novel carboxy-terminal 38 amino acid sequence of the class IV G-CSFR recruiting a distinct signaling molecule (or molecules) that act to suppress maturation. This mechanism would be independent of the duration of surface expression and potentially would be less sensitive to the relative levels of full-length G-CSFR coexpressed as long as sufficient numbers of class IV G-CSFR molecules were present to signal. Such a mechanism would explain the lack of correlation between the ratio of class IV/murine G-CSFR and the degree of maturation delay or block observed in the D-7 clones studied here. We are currently investigating this possibility.
Supported by National Institutes of Health/National Cancer Institute grants K11 CA68480 (S.M.W.) and R01 CA72261 (D.J.T.).
Reprints:Scott M. White, Division of Infectious Diseases, Department of Medicine, University of Pittsburgh School of Medicine, Suite 501 Lillian S. Kaufmann Bldg, 3471 Fifth Ave, Pittsburgh, PA 15213.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 1. Photomicrographs of 32Dcl3 cell clones following 10 days of G-CSF stimulation (100 ng/mL). / (A) Clone NEO.1, (B) Clone D-7.1, (C) Clone D-7.3, and (D) Clone D-7.4. (Wright-Giemsa stain, original magnification × 1000 [with oil immersion lens].)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/11/10.1182_blood.v95.11.3335/6/m_bloo01123001y.jpeg?Expires=1766353499&Signature=YaSdSlhAyxI5izyX4KIrs5rl~ZXEz9pu45LAgW6-Rc7DfGHbvEFPC2CgmmzyIUMBL4Gx353vv9oqnyZ01Sx7EG5AtjM3kYLB~05m2WmouKJzT1GgI0dWhFtI6iG~nu4q8Hu9x8FPyra9XD1KWuGKLTd3VhHsVSob2a6CT4tLHRc7PPLcoG5m7t2Bg~gtPuXoOiIGuDUBjyBxg3H5lSS5aoGJhihA0lwSVjaZ5t-n-EHZvmqfeDmajrppuqpiplSEqdEB8-l2Ibafjxa6aNVhfDiJ~H~H8Evov5L6pQnWIRpy4tUkmupk9me3XQ~rQBYDK32bTCb5r4wp85boS2sv7g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Plot of percent mitotic cells versus human class IV/murine G-CSFR mRNA ratio. / A plot of percent mitotic cells (myeloblasts + promyelocytes) versus the class IV/murine G-CSFR mRNA ratio is shown. The Pearson product moment correlation coefficient [r] for this plot is -0.329 indicating a poor correlation between the class IV/murine G-CSFR mRNA ratio and the fraction of cells that remain in the mitotic phase of myelopoiesis for each clone.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/11/10.1182_blood.v95.11.3335/6/m_bloo01123003x.jpeg?Expires=1766353499&Signature=yUs-b-ioi8rzfcsaKS-Ao0gSm0~T~f9y1vBJeIgTCyHUdzekbh3tqNM~MMqXb33Hp3PBSi9jGW0sFjUlv9Enj8PEIcLZUQqYIaEoJQZQM5wb90uUPXMdR72xzGWSv44GzmFFhizejSZCDMBMt35xw2oM3c1nGf0uMI~Ap8zcUoCNhK3qzN7A21f2WUY6xGzqga2-5-Lmx1cldiY2-SLr6SuOZJhIh6T6COsc~5i6N8uBylzoOfxCTlXXZTYzP4X6tEQAMO-Ihc4-N~oOL4c7aY6848kwYzxp76o4uOHTYCGpvppwNj8OfeFwW3gaQnowZI9nLB444LGmiOxkzVmO5g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)