Abstract
The tyrosine kinase activity of the Bcr/Abl oncogene is required for transformation of hematopoietic cells. The tyrosine kinase inhibitor STI571 (formerly called CGP57148B, Novartis Pharmaceuticals) inhibits BCR/ABL, TEL/ABL, and v-ABL kinase activity and inhibits growth and viability of cells transformed by any of these ABL oncogenes. Here we report the generation of 2 BCR/ABL–positive cell lines that have developed partial resistance to STI571. BCR/ABL–transformed Ba/F3 hematopoietic cells and Philadelphia-positive human K562 cells were cultured in gradually increasing concentrations of STI571 over a period of several months to generate resistant lines. Resistant Ba/F3.p210 cells were found to have an increase in Bcr/Abl messenger RNA, amplification of the Bcr/Abl transgene, and a greater than tenfold increase in the level of BCR/ABL protein. In contrast to Ba/F3.p210 cells, drug-resistant K562 cells did not undergo detectable amplification of the BCR/ABL gene, although they displayed a 2-fold to 3-fold increase in p210BCR/ABL protein. The addition of STI571 to both resistant Ba/F3.p210 and K562 cells resulted in a rapid reduction of tyrosine phosphorylation of cellular proteins, similar to that observed for nonresistant cells. However, the inhibition of kinase activity was transient and partial and was not accompanied by apoptosis. The results suggest that resistance to STI571 may be multifactorial. Increased expression of the target protein BCR/ABL was observed in both lines, and resulted from oncogene amplification in one line. However, altered drug metabolism, transport, or other related mechanisms may also contribute to drug resistance.
Chronic myelogenous leukemia (CML) is a clonal myeloproliferative disorder that is characterized by excessive production, and premature release, of both mature and immature myeloid cells. CML is caused by the 210-kd BCR/ABL oncoprotein, a chimeric fusion of either exon 2 or exon 3 of c-Bcr and exon 2 of c-Abl, which results from the Philadelphia (Ph) chromosome translocation t (9,22).1-3 One of the hallmarks of BCR/ABL transformation is the greatly elevated ABL tyrosine kinase activity, which is possibly related to oligomerization of BCR/ABL owing to the presence of a coiled-coil motif in BCR.4-7
Since transformation by BCR/ABL is absolutely dependent on tyrosine kinase activity,6 it has been evident for some time that a drug that inhibited ABL tyrosine kinase activity could be of therapeutic value. Synthesis of a class of tyrosine kinase–specific inhibitors called tyrphostins8 led to the discovery of an inhibitor of ABL tyrosine kinase toxic to the human CML cell line K562.9,10 More recently, STI571, a more potent and highly selective ABL tyrosine kinase inhibitor of the 2-phenyl-amino pyrimidine class, was developed.11,12 This compound, synthesized using the structure of the ATP binding site of protein kinases, selectively inhibits the anchorage-independent growth and tumorigenicity of v-Abl–transformed cells,11 inhibits the ability of BCR/ABL–positive cells to proliferate and develop tumors, and suppresses the formation of BCR/ABL colony formation in cells derived from peripheral blood or bone marrow from CML patients.12,13 STI571 induces apoptosis in a variety of BCR/ABL–positive cell lines and Ph+ cells.14-16 In addition to the p210 form of BCR/ABL (B2A2), STI571 also inhibits the p190 form of BCR/ABL (B1A2).16,17 STI571 is also effective in inhibiting the kinase activity of platelet-derived growth factor receptor (PDGFR)11 and TEL-PDGFR, an activated PDGFR kinase.17 Preliminary results from a phase I dose-escalating clinical trial suggest that STI571 has significant activity in stable and advanced phases of CML.18
A recent study demonstrated the need for a continuous block of BCR/ABL tyrosine kinase activity by STI571 to effectively cure BCR/ABL–positive tumor-bearing nude mice treated with the inhibitor.19 Since chronic or repeated administration of STI571 to CML patients may be necessary for successful treatment, we were interested in identifying potential mechanisms of resistance to this compound. Two cell lines were selected for study. K562 is a human Ph+ cell line derived from a patient with CML. Ba/F3.p210 was derived in our laboratory by transfecting a BCR/ABL complementary DNA (cDNA) into the nontransformed murine hematopoietic cell line Ba/F3. In this study, we have compared the mechanisms of resistance to STI571 in these 2 cell lines and find both similarities and distinct differences.
Materials and methods
Cells and cell culture
Ba/F3.p210 cells were generated by transfection of the interleukin (IL)–3–dependent murine hematopoietic cell line, Ba/F3, with a pGD vector containing p210Bcr/Abl (B2A2) cDNA,5 as previously described.20The human chronic myelogenous leukemia cell line, K562, was purchased from ATCC (Rockville, MD). Cells were cultured in RPMI 1640 medium (Mediatech, Herndon, VA) with 10% fetal calf serum and supplemented with 1% glutamine. Cells were maintained at a concentration of 2 to 5 × 105 cells/mL at 37°C with 5% CO2.
Antibodies
Anti-pTyr, monoclonal antibody (mAb) #4G10, was a gift from Dr Brian Druker, University of Oregon Health Sciences Center (Portland, OR) and was used at 1:2500 for immunoblotting. Anti-SHP2, mAb #79 from Transduction Laboratories (Lexington, KY), was used at 1:2000 for immunoblot. Anti-ABL (clone 3F12), which is directed against the SH2 domain of c-Abl, was developed by Dr Ravi Salgia and Dr James Griffin, Dana Farber Cancer Institute (Boston, MA) and was used at 1:500 for immunoblot. Anti-BCR, pAb N-20, directed against the amino terminus of human c-Bcr, was purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA), and was used at 1:200 for immunoblot. Anti-BCLXL, mAb #4 from Transduction Laboratories, was used at 1:2000 for immunoblot. Anti–Mdr-1, pAb H-241, directed against the carboxy terminus of human Mdr-1, was purchased from Santa Cruz Biotechnology, Inc, and used for fluorescence-activated cell sorter (FACS) analysis. Anti-MRP1, pAb N-19, directed against the amino terminus of human MRP-1, was purchased from Santa Cruz Biotechnology, and used for FACS analysis.
Proteasome/protease inhibitors
Calpain Inhibitor I (MG101; ALLN; N-Ac-Leu-Leu-norleucinal) was purchased from Calbiochem (La Jolla, CA), resuspended as a 200-mmol/L stock in dimethyl sulfoxide (DMSO), and stored at −20°C. AEBSF, hydrochloric acid (4-[2-Aminoethyl] benzenesulfonylfluoride, HCl), was purchased from Calbiochem, resuspended as a 100-mmol/L stock in water, and stored at −20°C. Proteasome Inhibitor I (PSI; z-Ile-Glu [OtBu]-Ala-Leu-CHO) was purchased from Calbiochem, resuspended as a 50-mmol/L stock in DMSO, and stored at −20°C. Clasto-Lactacystin β-Lactone was purchased from Calbiochem, resuspended as a 10-mmol/L stock in DMSO, and stored at −20°C. MG132 (z-Leu-Leu-Leu-H [aldehyde]) was purchased from Peptide Institute (Osaka, Japan), resuspended as a 10-mmol/L stock in DMSO, and stored at −20°C.
Immunoblotting
Cells were lysed in lysis buffer containing 0.02 mol/L Tris, pH 8.0, 0.15 mol/L NaCl, 10% glycerol, 1% NP-40 (wt/vol), 0.1 mol/L NaF, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L sodium orthovanadate, 40 μg/mL leupeptin, and 20 μg/mL aprotinin. Cell lysates were incubated on ice for 25 minutes, with vortexing every 5 minutes, and then centrifuged at 12 000g for 15 minutes. Protein yields were determined in the supernatants by Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA), and values were normalized accordingly. Protein lysates were dissolved in Laemmli's sample buffer by boiling for 5 minutes. Whole cell lysates were resolved on a sodium dodecyl sulfate (SDS) 7.5% polyacrylamide gel. Protein was then electrophoretically transferred to a Protran nitrocellulose transfer and immobilization membrane (Schleicher and Schuell, Dassel, Germany). The membrane was blocked either for 1 hour at 25°C or overnight at 4°C with 5% nonfat dry milk in 1 × TBS (10 mmol/L Tris-HCl, pH 8.0, 150 mmol/L NaCl), and then probed for 2 hours at 25°C or overnight at 4°C with antibody in 1 × TBST buffer (10 mmol/L Tris-HCl, pH 8.0, 150 mmol/L NaCl, 0.05% Tween20). Following 3 washes with 1 × TBST, membranes were incubated for 1 hour at 25°C with antimouse immunoglobulin (horseradish peroxidase linked whole antibody from sheep) or antirabbit immunoglobulin (horseradish peroxidase linked whole antibody from donkey) (Amersham Life Science, Arlington Heights, IL). The membrane was washed 5 times for 5 minutes/wash in 1 × TBST buffer, and bound antibodies were detected with enhanced luminol and oxidizing reagent as specified by the manufacturer (NEN Life Science Products, Boston, MA). Filters were stripped with stripping buffer (2% SDS, 0.0625 mol/L Tris, pH 6.8, and 0.7% 2-mercaptoethanol) for 30 minutes at 50°C prior to probing with additional antibodies.
Northern blotting
Total RNAs were extracted from 50 to 100 × 106cells with the use of Trizol Reagent (Gibco BRL Life Technologies, Gaithersburg, MD), and then messenger RNA (mRNA) was extracted with the use of the Messagemaker Reagent Assembly Kit (Gibco BRL Life Technologies). Approximately 2 μg of mRNA per lane were electrophoresed overnight at 20 V on an ethidium-bromide–stained formaldehyde gel, denatured at room temperature for 30 minutes in 50 mmol/L NaOH, neutralized in 50 mmol/L Tris (pH 7.0) for 30 minutes, and transferred to a Genescreen Plus nylon membrane (NEN Life Science Products) overnight via capillary action. Messenger RNA was fixed to the membrane by UV crosslinking with a Stratalinker (Stratagene, La Jolla, CA), and prehybridized overnight in 50% deionized formamide, 4 × SSC, 0.02 mol/L Tris, 1 × Denhardt's Reagent (50 × Denhardt's Reagent contains 1% Ficoll, Type 400, 1% polyvinylpyrrolidone, and 1% bovine serum albumin [BSA], Fraction V), 0.5% SDS, and 10% dextran sulfate. 32P-labeled cDNA probe (1 to 2 × 106 cpm/mL), labeled by a Random Primed DNA Labeling Kit (Boehringer Mannheim GmbH, Germany) and purified by ProbeQuant G-50 Micro Columns (Amersham Pharmacia Biotech, Piscataway, NJ), was added to the prehybridization solution along with 10 μg/mL denatured salmon sperm DNA (Gibco BRL Life Technologies). Following overnight hybridization at 42°C, the membrane was washed for 20 minutes at 25°C with low-stringency wash buffer (2 × SSC, 0.1% SDS) and washed for 15 minutes at 25°C with high-stringency wash buffer (0.2 × SSC, 0.1% SDS). Filters were exposed to film for varying lengths of time.
Southern blotting
Genomic DNA (100 to 150 kilobases [kb]) was extracted from approximately 50 × 106 cells with the use of extraction buffer (10 mmol/L Tris-Cl, pH 8.0, 0.1 mol/L EDTA, pH 8.0, 20 μg/mL pancreatic RNAase, and 0.5% SDS) and treated with 100 μg/mL of proteinase K for 3 hours at 50°C. The solution was then extracted 3 times with phenol:chloroform:isoamyl alcohol (25:24:1), and genomic DNA was precipitated with 0.3 mol/L sodium acetate, pH 5.2, and 2 volumes of 100% ethanol. Pure genomic DNA was spooled out and washed twice with 70% ethanol. DNA was finally resuspended in 1 × TE buffer (10 mmol/L Tris-Cl, pH 8.0, 1 mmol/L EDTA, pH 8.0). DNA was digested with EcoRI, HindIII, StuI, and XhoI, and restriction fragments were separated by electrophoresis overnight at 20 V on a 0.8% ethidium bromide-stained agarose gel. Following electrophoresis, DNA was treated for 15 minutes at 25°C with 0.2 N HCl (for partial depurination), 30 minutes at 25°C with 0.4 N NaOH-0.6 mol/L NaCl (for hydrolysis of phosphodiester backbone), and 30 minutes at 25°C with 1.5 mol/L NaCl-0.5 mol/L Tris-Cl, pH 7.4 (for neutralization). Capillary transfer of DNA to a nylon filter, hybridization, and washing steps were carried out as described for Northern blotting.
Probes
Probes used in Northern and Southern analysis were generated by polymerase chain reaction (PCR) amplification of a BCR/ABL cDNA cloned in a pBluescript vector. PCR primers used for amplification of c-Bcr were 5′GGCGGCGCTCAGGTCCAACTTC3′ (upper primer) and 5′CCAGCTCCTCATCGGGCACCAG3′ (lower primer), which yield a product spanning the coding region of the c-Bcr gene from base pair (bp) 453 to bp 2332. PCR primers used for amplification of c-Abl were 5′TCGGAAGAGGGCAGGGGAGAAC3′ (upper primer) and 5′GCCGGACCACTGCCTGCTGACG3′ (lower primer), which yield a product spanning the C-terminus of the c-Abl gene coding region from bp 2289 to bp 3305. A human glyceraldehyde phosphodehydrogenase cDNA probe was generated by reverse transcriptase (RT)–PCR, subcloned into a TA vector (Invitrogen, Carlsbad, CA), and gel-purified. Primers used for amplification of a 332-bp portion of the 3′ end of the gene were 5′TTCAAGGGGTCTACATGGCAACTG3′ (upper primer) and 5′GGG- CATCCTGGGCTACACTG3′ (lower primer).
Quantitative PCR for Bcr/Abl
Total RNA was isolated from DMSO-treated and STI571-resistant Ba/F3.p210 cells with the use of Trizol Reagent (Gibco BRL Life Technologies).Reverse transcription of 1 μg of total RNA isolated from DMSO-treated and STI571-resistant Ba/F3.p210 cells (see discussion of Northern blotting under “Results”) was achieved by mixing the RNA with 2 μL of 10 × PCR buffer, 0.4 μL of deoxynucleotides (Perkin Elmer, Norwalk, CT), 0.65 μL of Random Hexamers (50 A260 Units) (Pharmacia, Piscataway, NJ), 0.25 μL RNAase Inhibitor (10 U/μL) (Gibco BRL Life Technologies), and 0.5 μL of Superscript II Reverse Transcriptase (200 U/μL) (Gibco BRL Life Technologies) in a 20-μL volume. This was followed by incubation of the mixture at 42°C for 60 minutes, and then for 5 minutes at 95°C.
Two microliters of product from the RT reaction were added to 48 μL of reaction mix (Taqman Core Reagent Kit, Perkin Elmer), consisting of 20.2 μL H2O, 0.3 μL Taq polymerase, 1 μL deoxyadenosine triphosphate, 1 μL deoxycytidine triphosphate, 1 μL deoxyguanosine triphosphate, 1 μL deoxyuridine triphosphate, 12 μL MgCl2, 1 μL B2A2 forward primer, 1 μL B2A2 reverse primer, 1 μL Bcr/Abl probe, 1 μL G3PDH forward primer, 1 μL G3PDH reverse primer, 1 μL G3PDH probe, 5 μL Taqman buffer, and 0.5 μL uracil N-glycosylase, and placed in a 96-well plate. The plate was transferred to an ABI 7700 thermocycler (Perkin Elmer), and samples were passed through denaturation cycles (50°C, 2 minutes; 95°C, 10 minutes). These were followed by 40 amplification cycles (95°C, 15 seconds; 60°C, 1 minute), during which fluorescent signals of Bcr/Abl and G3PDH probes were measured as described previously.21
Serial dilutions 106 to 10−1 of Bcr/Abl plasmid DNA were used as reference standards of fluorescent signals for copy number as described previously.21 Negative controls lacking reverse transcription product were used, as well as reverse-transcribed total RNA extracted from the IL-3–dependent, BCR/ABL–minus, murine hematopoietic cell line Ba/F3. G3PDH primers and probe placed in the same reaction mixture as Bcr/Abl primers and probe were used to assess RNA integrity.
G3PDH primers (forward primer: 5′GAAGGTGAAGGTCGGAGTC3′; reverse primer: 5′GAAGATGGTGATGGGATTC3′) and probe (5′CAA- GCTTCCCGTTCTCAGCC3′) were provided by the G3PDH Control Reagent Kit (Perkin Elmer). The Taqman G3PDH probe carried a 5′ JOE reporter label and a 3′ TAMRA quencher group. PCR primers for the amplification of B2A2 cDNA (forward primer: 5′GCATTCCGCTGACCATCAATA3′; reverse primer: 5′GGCTTCACTCAGACCCTGAGG3′) and the Bcr/Abl probe (5′AGCGGCCAGTAGCATCTGACTTTGAGC3′) were designed with the use of the Primer Express Software Program (Perkin Elmer). The Taqman Bcr/Abl probe, synthesized by PE-Applied Biosystems (Perkin Elmer), carried a 5′FAM reporter label and a 3′TAMRA quencher group.
FACS analysis
Approximately 1 × 105 cells were incubated with Mdr-1 antibody (diluted 1:100) for 1 hour at 4°C. Cell/antibody mixtures were diluted with 1% BSA in 1 × PBS (PBSA) and centrifuged for 10 minutes at 2000 rpm. Cell pellets were then resuspended in fluorescein isothiocyanate (FITC)–conjugated antirabbit immunoglobulin (Ig)–G (H+L) (0.5 μg/mL) (Zymed, San Francisco, CA) and incubated for 45 minutes at 4°C. Samples were diluted with PBSA and centrifuged for 10 minutes at 2000 rpm. Cell pellets were resuspended in 3% formaldehyde in PBSA and analyzed by FACS analysis. As controls, cells were incubated at 4°C in the absence of Mdr-1 and FITC-conjugated antirabbit IgG antibodies, and cells were incubated at 4°C in the presence of only FITC-conjugated antirabbit IgG antibody. FACS analysis was performed as described above using MRP1 antibody (diluted 1:100) and antigoat IgG (whole molecule) FITC conjugate (Sigma, St Louis, MO).
Results
STI571 inhibits tyrosine phosphorylation in Ba/F3.p210 and K562 cells
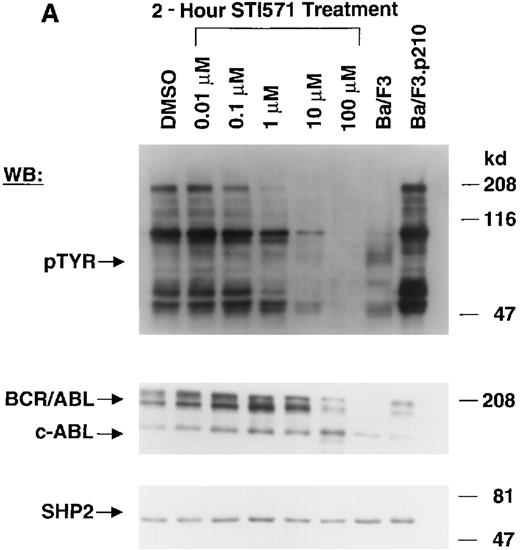
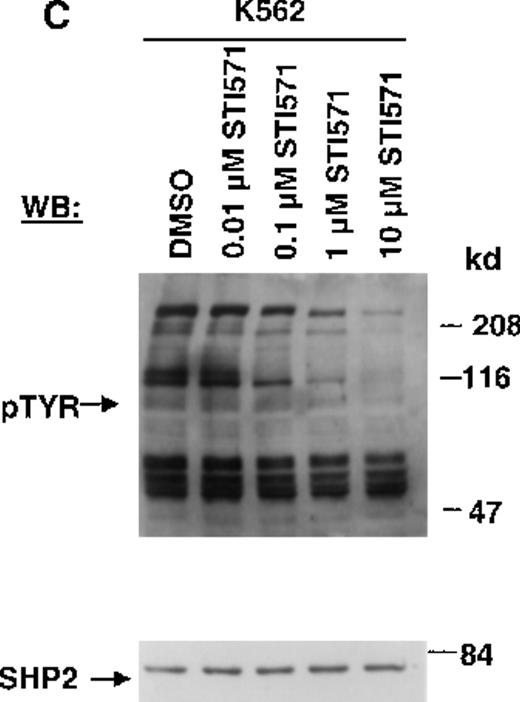
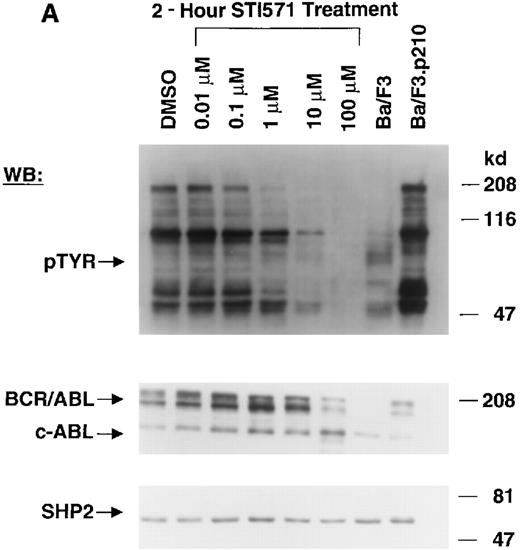
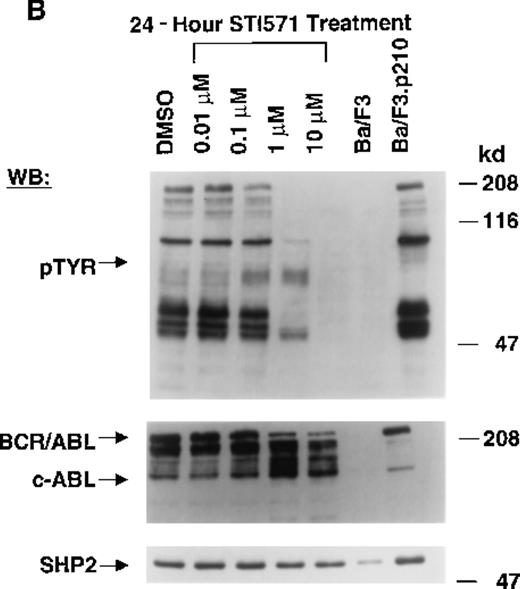
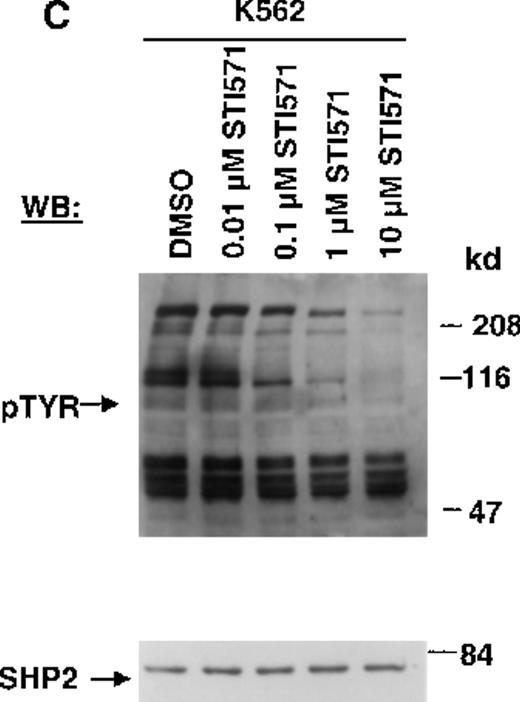
The Abl tyrosine kinase inhibitor STI571 has previously been shown to inhibit the kinase activity of BCR/ABL.11 12 The concentration of STI571 that inhibits tyrosine phosphorylation by at least 50% in the BCR/ABL–transformed hematopoietic cell line, Ba/F3.p210, was determined by measuring tyrosine phosphorylation of cellular substrates of BCR/ABL by immunoblotting. Figure1A shows that at 2 hours, levels of tyrosine phosphorylation began to decrease at 1 μmol/L STI571 and were barely detectable at 10 μmol/L STI571. Figure 1B shows that after 24 hours, 1 μmol/L STI571 decreased cellular tyrosine phosphorylation by more than 50%, as compared with DMSO-treated and untreated Ba/F3.p210 cells. STI571 did not appear to affect expression levels of BCR/ABL and c-Abl after 2 hours, although treatment for 24 hours with, respectively, 1 and 10 μmol/L STI571 resulted in a small reduction in the level of BCR/ABL. As with Ba/F3.p210 cells, tyrosine phosphorylation in K562 cells was potently inhibited by culturing cells in the presence of STI571 for 2 hours (Figure 1C).
Inhibition of tyrosine phosphorylation in STI571-treated Ba/F3.p210 cells.
(A) Ba/F3.p210 cells were incubated in the presence of increasing concentrations of STI571 (0.01 to 100 μmol/L) for 2 hours. Immunoblotting of the lysates from these cells was performed with anti-pTyr (upper panel), anti-ABL (3F12) (middle panel), and anti-SHP2 as a control for protein loading (lower panel). (B) Ba/F3.p210 cells were incubated in the presence of increasing concentrations of STI571 (0.01 to 10 μmol/L) for 24 hours. Immunoblotting of the lysates from these cells was performed as in (A). (C) K562 cells were incubated in the presence of increasing concentrations of STI571 (0.01 to 10 μmol/L) for 2 hours. Immunoblotting of the lysates from these cells was performed as in (A).
Inhibition of tyrosine phosphorylation in STI571-treated Ba/F3.p210 cells.
(A) Ba/F3.p210 cells were incubated in the presence of increasing concentrations of STI571 (0.01 to 100 μmol/L) for 2 hours. Immunoblotting of the lysates from these cells was performed with anti-pTyr (upper panel), anti-ABL (3F12) (middle panel), and anti-SHP2 as a control for protein loading (lower panel). (B) Ba/F3.p210 cells were incubated in the presence of increasing concentrations of STI571 (0.01 to 10 μmol/L) for 24 hours. Immunoblotting of the lysates from these cells was performed as in (A). (C) K562 cells were incubated in the presence of increasing concentrations of STI571 (0.01 to 10 μmol/L) for 2 hours. Immunoblotting of the lysates from these cells was performed as in (A).
Development of STI571-resistant Ba/F3.p210 cells
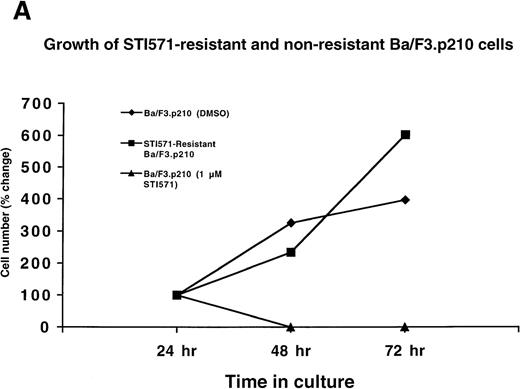
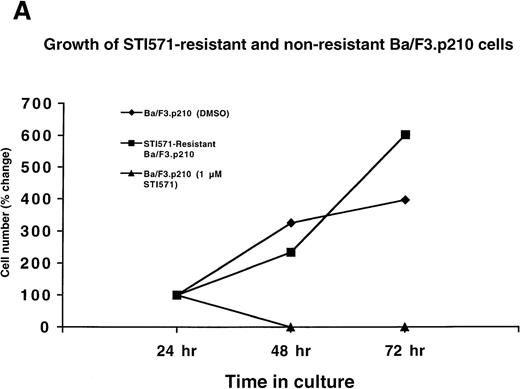
Ba/F3.p210 cells were cultured in the presence of gradually increasing concentrations (0.01 μmol/L to 0.2 μmol/L) of STI571 over a period of 56 days. A subpopulation of these cells was placed in the presence of 1 μmol/L STI571, and the concentration of STI571 increased to 2.5 μmol/L in 4 steps over a period of 31 days. Protein, RNA, and DNA analyses were performed on cells resistant to 2 μmol/L STI571. Interestingly, although the cells achieved complete resistance to 2.5 μmol/L STI571, they were rapidly killed by 3 μmol/L STI571, even following several months of culturing in the presence of 2.5 μmol/L STI571 (data not shown). Whereas nonresistant Ba/F3.p210 cells rapidly die within 2 days of culture in the presence of 1 μmol/L STI571, drug-resistant Ba/F3.p210 cells grow well in 1 μmol/L STI571 (Figure 2A).
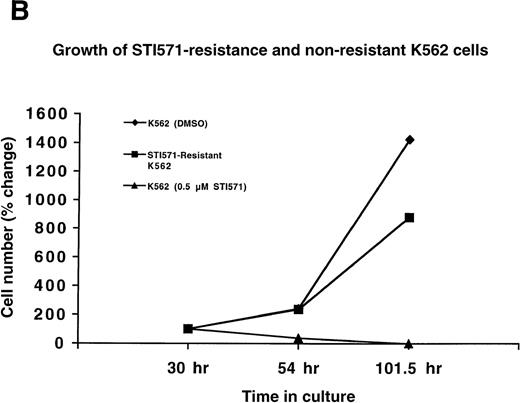
Growth of STI571-resistant and nonresistant cells.
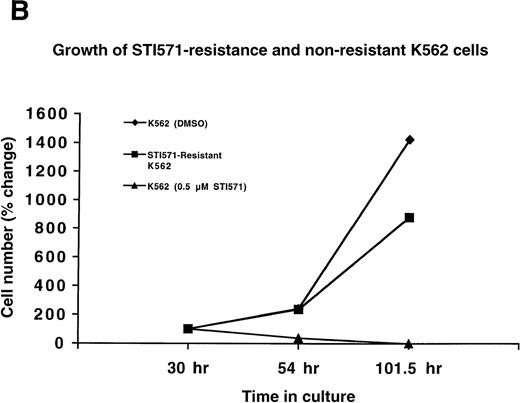
(A) Growth of STI571-resistant and nonresistant Ba/F3.p210 cells. Diamonds represent nonresistant Ba/F3.p210 cells cultured in the presence of DMSO and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Squares represent STI571-resistant Ba/F3.p210 cells cultured in the presence of 1 μmol/L STI571 and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Triangles represent nonresistant Ba/F3.p210 cells cultured in the presence of 1 μmol/L STI571 and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Cell number was determined by counting viable cells. (B) Growth of STI571-resistant and nonresistant K562 cells. Diamonds represent nonresistant K562 cells cultured in the presence of DMSO and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Squares represent STI571-resistant K562 cells cultured in the presence of 0.5 μmol/L STI571 and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Triangles represent nonresistant K562 cells cultured in the presence of 0.5 μmol/L STI571 and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Cell number was determined by counting viable cells.
Growth of STI571-resistant and nonresistant cells.
(A) Growth of STI571-resistant and nonresistant Ba/F3.p210 cells. Diamonds represent nonresistant Ba/F3.p210 cells cultured in the presence of DMSO and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Squares represent STI571-resistant Ba/F3.p210 cells cultured in the presence of 1 μmol/L STI571 and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Triangles represent nonresistant Ba/F3.p210 cells cultured in the presence of 1 μmol/L STI571 and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Cell number was determined by counting viable cells. (B) Growth of STI571-resistant and nonresistant K562 cells. Diamonds represent nonresistant K562 cells cultured in the presence of DMSO and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Squares represent STI571-resistant K562 cells cultured in the presence of 0.5 μmol/L STI571 and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Triangles represent nonresistant K562 cells cultured in the presence of 0.5 μmol/L STI571 and harvested and counted by trypan blue exclusion with the use of a hemacytometer at the designated time points. Cell number was determined by counting viable cells.
BCR/ABL protein expression and cellular tyrosine phosphorylation in STI571-resistant Ba/F3.p210 cells
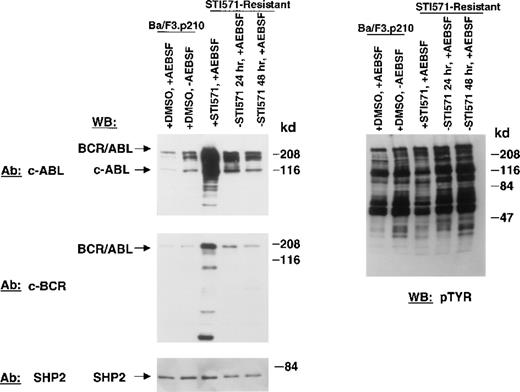
Protein lysates were prepared from STI571-resistant cells with the use of lysis buffer with a standard cocktail of protease inhibitors, including aprotinin, phenyl methyl sulfonyl fluoride, and leupeptin. BCR/ABL was then detected by immunoblotting with a monoclonal antibody directed against the SH2 domain of c-Abl (3F12). There was an increase in BCR/ABL protein in STI571-resistant Ba/F3.p210 cells, but multiple proteolytic cleavage products were observed (data not shown). STI571-resistant cells were then treated with a variety of proteasome and protease inhibitors while being continuously cultured in the presence of STI571. MG-132 (20 μmol/L, 4 hours), a reversible proteasome inhibitor that diminishes 26S complex–mediated degradation of ubiquitin-conjugated proteins, and the 20S proteasome inhibitors, Proteasome Inhibitor I (30 μmol/L to 100 μmol/L, 4 hours) and clasto-Lactacystin β-lactone (30 μmol/L, 4 hours) did not reduce proteolytic cleavage. Calpain Inhibitor I, an inhibitor of calpain I, calpain II, cathepsin B, cathepsin L, and neutral cysteine proteases (20 μmol/L and 50 μmol/L, 6 hours) had a modest effect. However, AEBSF, a specific, cell-permeable, irreversible inhibitor of serine proteases, such as trypsin, chymotrypsin, plasmin, thrombin, and kallikrein, used at a concentration of 1 mmol/L for 20 minutes effectively reduced accumulation of truncated BCR/ABL–related products and led to the reappearance of apparently full-length p210 BCR/ABL. Therefore, all subsequent experiments included AEBSF as indicated to reduce proteolytic cleavage of the amplified BCR/ABL proteins occurring primarily during cell lysis.
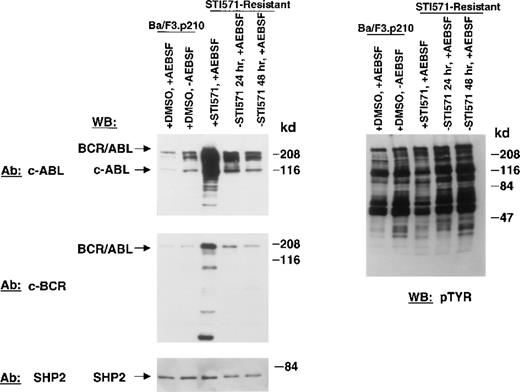
Figure 3 shows BCR/ABL protein expression in nonresistant and STI571-resistant Ba/F3.p210 cells. An anti-Abl antibody and an anti-Bcr antibody were used individually to detect the BCR/ABL protein in these cells. Compared with nonresistant cells, BCR/ABL is significantly overexpressed in STI571-resistant cells. Pretreatment of nonresistant Ba/F3.p210 cells with 1 mmol/L AEBSF did not significantly affect BCR/ABL protein expression in these cells. Removal of STI571 from STI571-resistant Ba/F3.p210 cells for 24 or 48 hours substantially reduced levels of BCR/ABL protein in the drug-resistant cells, although BCR/ABL protein levels were still elevated as compared with nonresistant cells. This suggests that the continuous presence of the kinase inhibitor may be required to maintain BCR/ABL overexpression. No significant changes in expression levels of SHP2 were seen for all samples, suggesting that differences observed in BCR/ABL protein expression are not due to differences in protein loading. Elevated BCR/ABL protein levels in STI571-resistant Ba/F3.p210 cells were not associated with an increase in BCLXL protein expression (data not shown).
BCR/ABL protein expression in STI571-resistant Ba/F3.p210 cells.
Nonresistant Ba/F3.p210 cells cultured in the presence of DMSO were treated for 20 minutes with 1 mmol/L AEBSF or water, as vehicle. STI571-resistant Ba/F3.p210 cells were cultured in the continuous presence of 1 μmol/L STI571 and then treated with 1 mmol/L AEBSF for 20 minutes. STI571-resistant Ba/F3.p210 cells were deprived of STI571 for 24 and 48 hours, respectively, and then treated with 1 mmol/L AEBSF for 20 minutes. Immunoblotting of the lysates from these cells was performed with anti-ABL (upper left-hand panel), anti-BCR (middle left-hand panel), and anti-SHP2 as a control for loading (lower left-hand panel), and anti-pTyr (upper right-hand panel).
BCR/ABL protein expression in STI571-resistant Ba/F3.p210 cells.
Nonresistant Ba/F3.p210 cells cultured in the presence of DMSO were treated for 20 minutes with 1 mmol/L AEBSF or water, as vehicle. STI571-resistant Ba/F3.p210 cells were cultured in the continuous presence of 1 μmol/L STI571 and then treated with 1 mmol/L AEBSF for 20 minutes. STI571-resistant Ba/F3.p210 cells were deprived of STI571 for 24 and 48 hours, respectively, and then treated with 1 mmol/L AEBSF for 20 minutes. Immunoblotting of the lysates from these cells was performed with anti-ABL (upper left-hand panel), anti-BCR (middle left-hand panel), and anti-SHP2 as a control for loading (lower left-hand panel), and anti-pTyr (upper right-hand panel).
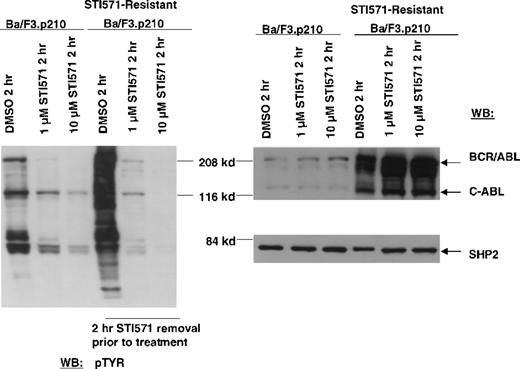
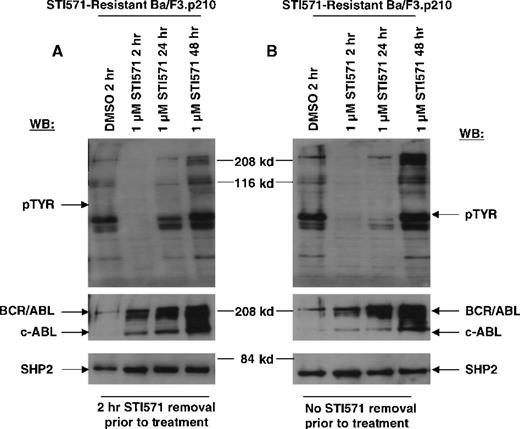
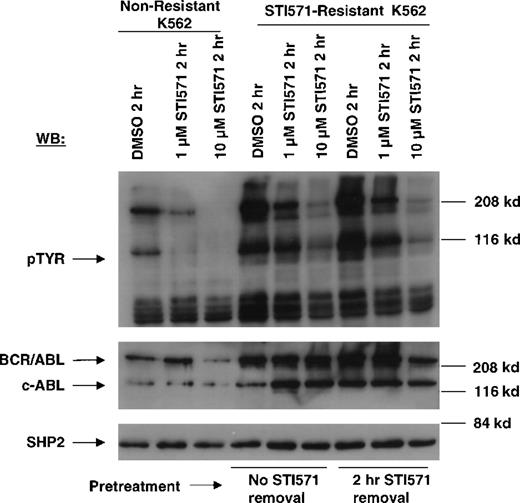
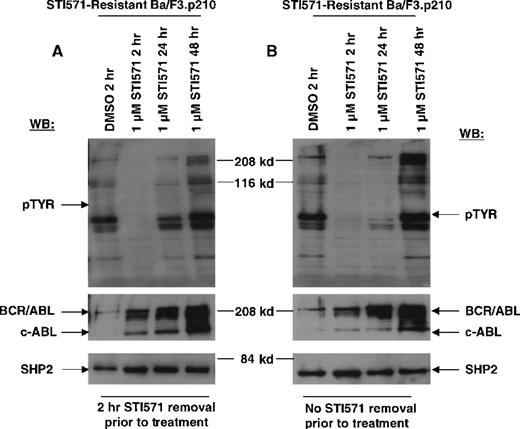
In contrast to drug-sensitive cells, STI571-resistant Ba/F3.p210 cells cultured in the continuous presence (media changed every 2 to 3 days) of STI571 maintained high levels of cellular protein tyrosine phosphorylation (Figure 3). Interestingly, when fresh drug was added to these cells, tyrosine phosphorylation of cellular proteins declined rapidly (Figure 4), but transiently, returning to steady state levels in less than 24 hours, and in the absence of cell death (Figure 5). These results were obtained whether the media containing STI571 were removed and cells underwent short-term (2-hour) drug deprivation before undergoing culturing in fresh STI571, or whether cells were removed from “spent” media (normally changed every 2 to 3 days) and placed immediately in fresh media containing fresh STI571 (Figure 5). Also of interest is that significant changes were observed in the level of BCR/ABL protein that depended on the presence of drug. Removal of STI571 resulted in lower levels of BCR/ABL while addition of STI571 increased BCR/ABL protein levels (Figure 5). These results suggest that BCR/ABL protein levels in drug-resistant cells may be in some way modulated directly by the concentration of drug in the medium and that the BCR/ABL kinase activity remains highly sensitive to the addition of fresh drug to the cells. However, in contrast to nonresistant cells, STI571 treatment of drug-resistant cells fails to cause apoptosis, and the inhibition of tyrosine phosphorylation is quite transient.
Inhibition of cellular tyrosine phosphorylation in nonresistant Ba/F3.p210 cells and STI571-resistant Ba/F3.p210 cells by STI571.
Nonresistant Ba/F3.p210 cells were treated for 2 hours with DMSO, 1 μmol/L STI571, or 10 μmol/L STI571. STI571-resistant Ba/F3.p210 cells were treated for 2 hours with DMSO, 1 μmol/L STI571, or 10 μmol/L STI571, following a 2-hour STI571 deprivation. Immunoblotting of the lysates from these cells was performed with anti-pTyr (left-hand panel), anti-ABL (upper right-hand panel), and anti-SHP2 as a control for loading (lower right-hand panel).
Inhibition of cellular tyrosine phosphorylation in nonresistant Ba/F3.p210 cells and STI571-resistant Ba/F3.p210 cells by STI571.
Nonresistant Ba/F3.p210 cells were treated for 2 hours with DMSO, 1 μmol/L STI571, or 10 μmol/L STI571. STI571-resistant Ba/F3.p210 cells were treated for 2 hours with DMSO, 1 μmol/L STI571, or 10 μmol/L STI571, following a 2-hour STI571 deprivation. Immunoblotting of the lysates from these cells was performed with anti-pTyr (left-hand panel), anti-ABL (upper right-hand panel), and anti-SHP2 as a control for loading (lower right-hand panel).
Transient inhibition of cellular tyrosine phosphorylation by STI571 in STI571-resistant Ba/F3.p210 cells.
(A) STI571-resistant Ba/F3.p210 cells treated with 1 μmol/L STI571 for 2 to 48 hours, following a 2-hour STI571 deprivation. Immunoblotting of the lysates from these cells was performed with anti-pTyr (upper left-hand panel), anti-ABL (middle left-hand panel), and anti-SHP2 as a control for loading (lower left-hand panel). (B) STI571-resistant Ba/F3.p210 cells treated with 1 μmol/L STI571 for 2 to 48 hours, with no STI571 deprivation prior to treatments. Immunoblotting of the lysates from these cells was performed as in (A).
Transient inhibition of cellular tyrosine phosphorylation by STI571 in STI571-resistant Ba/F3.p210 cells.
(A) STI571-resistant Ba/F3.p210 cells treated with 1 μmol/L STI571 for 2 to 48 hours, following a 2-hour STI571 deprivation. Immunoblotting of the lysates from these cells was performed with anti-pTyr (upper left-hand panel), anti-ABL (middle left-hand panel), and anti-SHP2 as a control for loading (lower left-hand panel). (B) STI571-resistant Ba/F3.p210 cells treated with 1 μmol/L STI571 for 2 to 48 hours, with no STI571 deprivation prior to treatments. Immunoblotting of the lysates from these cells was performed as in (A).
Expression of BCR/ABL RNA in STI571-resistant and nonresistant Ba/F3.p210 cells
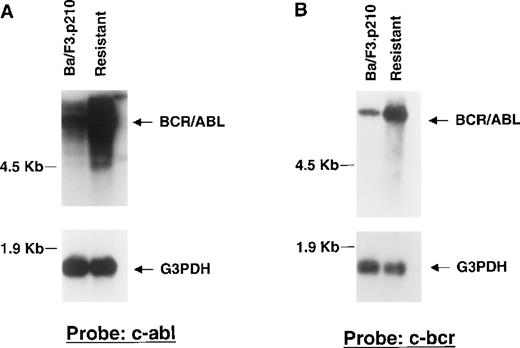
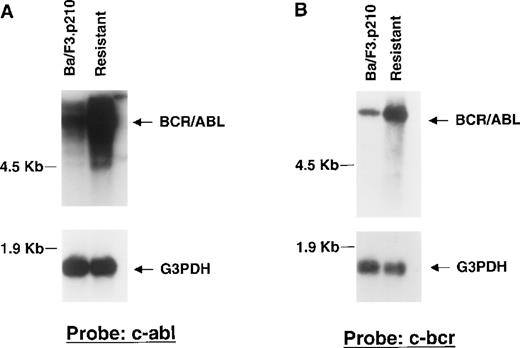
To determine if drug resistance was associated with alteration of Bcr/Abl RNA expression, Northern analysis was performed on RNA extracted from DMSO-treated nonresistant and STI571-resistant Ba/F3.p210 cells, with the use of cDNA probes corresponding to c-Bcr and the C-terminus of c-Abl, respectively. A higher level of Bcr/Abl mRNA in resistant cells was observed to hybridize to both c-Bcr and c-Abl probes, as compared with nonresistant cells (Figure6A, 6B). Re-probing the blots with a G3PDH cDNA probe confirmed equal loading of mRNA in all lanes. Northern results shown here are representative of 2 independent experiments.
Bcr/Abl RNA expression in STI571-resistant Ba/F3.p210 cells.
Northern blotting was performed on mRNA extracted from nonresistant and STI571-resistant Ba/F3.p210 cells. (A) Northern blot using a cDNA probe from the C-terminus of c-Abl. (B) Northern blot using a cDNA probe from c-Bcr. See “Materials and methods” for details of each probe. Each Northern filter was stripped and re-probed with human G3PDH, as a control for loading.
Bcr/Abl RNA expression in STI571-resistant Ba/F3.p210 cells.
Northern blotting was performed on mRNA extracted from nonresistant and STI571-resistant Ba/F3.p210 cells. (A) Northern blot using a cDNA probe from the C-terminus of c-Abl. (B) Northern blot using a cDNA probe from c-Bcr. See “Materials and methods” for details of each probe. Each Northern filter was stripped and re-probed with human G3PDH, as a control for loading.
A quantitative PCR assay was used to estimate the increase in Bcr/Abl RNA present in the resistant cell lines. Quantitative PCR was performed on total RNA extracted from STI571-resistant and DMSO-treated nonresistant Ba/F3.p210 cells. The number of Bcr/Abl mRNA copies/G3PDH mRNA copies in STI571-resistant Ba/F3.p210 cells was determined to be 2.35 × 106/4.45 × 106 (or 0.528). The number of Bcr/Abl mRNA copies/G3PDH mRNA copies in nonresistant Ba/F3.p210 cells was determined to be 2.5 × 105/9.55 × 106 (or 0.026). There is thus an approximately 20-fold increase in Bcr/Abl mRNA copy number in resistant versus nonresistant cells. Quantitative PCR results are representative of 2 independent experiments.
Analysis of BCR/ABL gene in STI571-resistant and nonresistant Ba/F3.p210 cells
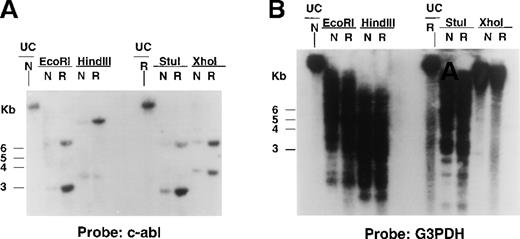
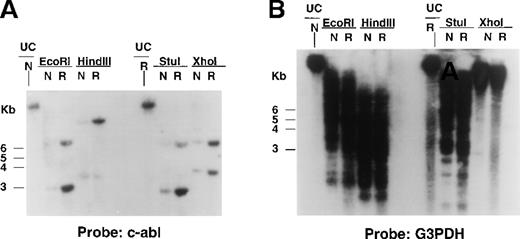
To determine if the increased Bcr/Abl RNA observed in STI571-resistant cells was associated with alterations in the Bcr/Abl transgene, Southern analysis was performed on digested genomic DNA isolated from DMSO-treated Ba/F3.p210 cells and STI571-resistant cells with the use of a cDNA probe corresponding to the C-terminus of c-Abl. HindIII cleaves the 6935-bp Bcr/Abl transgene at bp 2729 and yields a single (8- to 10-kb) molecular weight DNA band that hybridized to the c-Abl probe and was amplified in STI571-resistant Ba/F3.p210 as compared with nonresistant Ba/F3.p210 (Figure7A). Similar results were obtained with genomic DNA isolated from nonresistant and resistant Ba/F3.p210 cells that was digested with Xho I, which cleaves BCR/ABL at bp 779, and EcoRI and StuI, which cleave the transgene at or near the beginning and end of the Bcr/Abl coding region, respectively (Figure 7A). A human G3PDH probe was used to demonstrate equal loading of DNA in all lanes (Figure 7B). Southern results shown here are representative of 2 independent experiments. Stripping and re-probing of Southern filters with a c-Bcr cDNA probe also resulted in hybridization of the probe to an amplified level of genomic DNA in resistant versus nonresistant cells (data not shown).
Bcr/Abl gene expression in STI571-resistant Ba/F3.p210 cells.
Southern blotting was performed on EcoRI-, HindIII-, StuI-, and XhoI-digested genomic DNA prepared from nonresistant and STI571-resistant Ba/F3.p210 cells. (A) Southern blot using a cDNA probe from the C-terminus of c-Abl. (B) Southern blot shown in (A) after stripping and re-probing with human G3PDH, a control for loading.
Bcr/Abl gene expression in STI571-resistant Ba/F3.p210 cells.
Southern blotting was performed on EcoRI-, HindIII-, StuI-, and XhoI-digested genomic DNA prepared from nonresistant and STI571-resistant Ba/F3.p210 cells. (A) Southern blot using a cDNA probe from the C-terminus of c-Abl. (B) Southern blot shown in (A) after stripping and re-probing with human G3PDH, a control for loading.
Development of STI571-resistant K562 cells
Since Ba/F3.p210 cells express BCR/ABL from a transgene introduced by transfection, we also generated STI571 resistance in the human Ph-chromosome–positive cell line K562. K562 cells were cultured in the presence of 0.2 μmol/L STI571 for a total of 4 months, and the drug concentration was gradually increased to 0.5 μmol/L STI571 over a period of 1 and a half months. Whereas nonresistant K562 cells die at 0.5 μmol/L STI571, STI571-resistant K562 cells proliferate well in the presence of 0.5 μmol/L STI571 (Figure 2B).
BCR/ABL protein expression and cellular tyrosine phosphorylation in STI571-resistant K562 cells
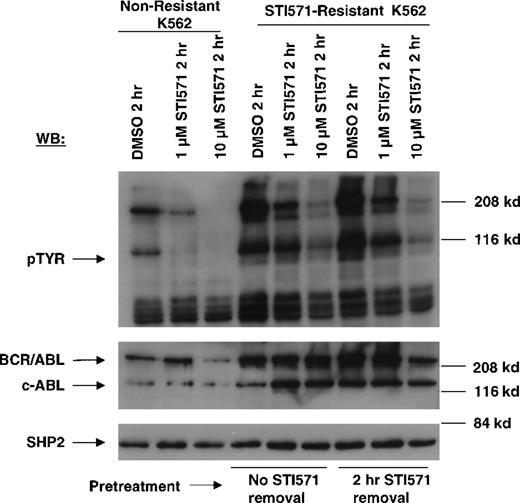
Immunoblotting studies performed with an anti–c-ABL antibody reproducibly demonstrated an increase in levels of BCR/ABL protein in STI571-resistant cells, as compared with nonresistant K562 cells (Figure 8). Results shown here are representative of 7 independent experiments. Cellular protein tyrosine phoshorylation is elevated, consistent with increased BCR/ABL protein expression. Addition of fresh STI571 again transiently decreased cellular protein phosphorylation, as was the case with drug-resistant Ba/F3.p210 cells, but to a lesser extent.
Inhibition of cellular tyrosine phosphorylation by STI571 in nonresistant and STI571-resistant K562 cells.
Nonresistant K562 cells were treated for 2 hours with DMSO, 1 μmol/L STI571, and 10 μmol/L STI571 (lanes 1-3). STI571-resistant K562 cells were treated with DMSO, 1 μmol/L, and 10 μmol/L STI571 following continuous culturing in the presence of 0.5 μmol/L STI571 prior to treatments (lanes 4-6), and following STI571 deprivation for 2 hours prior to treatments (lanes 7-9). Immunoblotting of the lysates from these cells was performed with anti-pTyr (upper panel), anti-ABL (middle panel), and anti-SHP2 as a loading control for protein (bottom panel).
Inhibition of cellular tyrosine phosphorylation by STI571 in nonresistant and STI571-resistant K562 cells.
Nonresistant K562 cells were treated for 2 hours with DMSO, 1 μmol/L STI571, and 10 μmol/L STI571 (lanes 1-3). STI571-resistant K562 cells were treated with DMSO, 1 μmol/L, and 10 μmol/L STI571 following continuous culturing in the presence of 0.5 μmol/L STI571 prior to treatments (lanes 4-6), and following STI571 deprivation for 2 hours prior to treatments (lanes 7-9). Immunoblotting of the lysates from these cells was performed with anti-pTyr (upper panel), anti-ABL (middle panel), and anti-SHP2 as a loading control for protein (bottom panel).
Expression of BCR/ABL RNA in STI571-resistant and nonresistant K562 cells
To determine if drug resistance in K562 cells was associated with changes in BCR/ABL RNA expression, Northern analysis was performed on mRNA extracted from DMSO-treated nonresistant and STI571-resistant K562 cells, with the use of a cDNA probe corresponding to the C-terminus of c-Abl. No detectable difference in BCR/ABL mRNA could be seen between resistant cells and nonresistant cells (data not shown). Quantitative PCR performed with the use of total RNA extracted from nonresistant and STI571-resistant K562 cells confirmed Northern results (data not shown).
Analysis of BCR/ABL gene in STI571-resistant and nonresistant K562 cells
The BCR/ABL gene was investigated for amplification/alteration in STI571-resistant K562 cells. Southern analysis was performed on HindIII- and EcoRI-digested genomic DNA isolated from DMSO-treated nonresistant K562 cells and STI571-resistant K562 cells as described above. No differences were observed between nonresistant and STI571-resistant K562 cells in either DNA-banding pattern or signal intensity (data not shown).
Mdr-1 and MRP1 protein expression in STI571-resistant K562 cells
Flow cytometry was performed to determine if drug-resistant K562 cells had altered p-glycoprotein (Pgp) expression, as compared with nonresistant cells. No significant difference in levels of Mdr-1 or MRP1 was observed between the 2 cell populations (data not shown).
Discussion
STI571 inhibits c-Abl tyrosine kinase activity by acting as a competitor for ATP.11 As a result of this inhibition, the proliferation of cells transformed by v-Abl, p190Bcr/Abl, p210Bcr/Abl, and TEL-ABL is suppressed.11-13,17 STI571 also inhibits colony formation and tumorigenicity of BCR/ABL– and Ph+ cells.11-13 STI571 induces apoptosis in BCR/ABL–positive cells14-16 without causing cell differentiation14; STI571-induced programmed cell death is accompanied by caspase activation.15
Here we report the generation of 2 BCR/ABL–positive cell lines that have developed partial resistance to STI571. BCR/ABL–transformed Ba/F3 hematopoietic cells were made resistant to apoptotic death in the presence of up to 2.5 μmol/L STI571. Resistant cells were found to have an increase in Bcr/Abl mRNA, amplification of the Bcr/Abl transgene by over 5-fold, and increased levels of Bcr/Abl protein. The Southern blots using either Abl or Bcr-derived probes did not show any major rearrangements of the transgene in STI571-resistant Ba/F3 cells. Similarly, the existence of 1 major mRNA transcript of the same size in resistant and nonresistant cells, as observed in Northern blotting experiments, also suggests that the entire transgene was amplified without major rearrangements. In addition, a region of C-ABL spanning the ATP-binding site (amino acid sequence LGGGQYGEVYE) was amplified from nonresistant and ST1571-resistant Ba/F3.p210 and K562 cells by RT-PCR. No difference in sequence was observed between resistant and nonresistant cells (data not shown).
In contrast to Ba/F3.p210 cells, drug-resistant K562 cells did not undergo detectable amplification of the BCR/ABL gene, despite the fact they displayed a several-fold increase in p210BCR/ABL protein. However, although both cell lines displayed increased levels of BCR/ABL protein and kinase activity, we hypothesize that the mechanism of resistance in both cell lines is likely to involve additional mechanisms. The addition of fresh drug to cultures still resulted in rapid, but transient, inhibition of protein tyrosine phosphorylation, but with preservation of cell viability. Several mechanisms could explain these results. First, the cell lines may have developed methods to increase degradation or metabolism of the drug, which cannot currently be assayed. Second, increased drug efflux may be present, and further studies are ongoing in our laboratory to identify such mechanisms. We did not detect an increase in Mdr-1 or MRP1 protein expression by flow cytometry. Finally, the cells may have accumulated compensating mutations in other genes, such as loss of tyrosine phosphatase activity, loss of apoptosis genes, or gain-of-function mutations in viability genes. One prominent BCR/ABL–associated viability gene, Bcl-X, was studied here, and no changes were seen. Overall, our results suggest that the mechanism of resistance in these lines may be multifactorial and interesting from a biochemical standpoint, with the common end result of an increase in the expression of the target protein and a reduction in the apoptosis effects of the drug.
Gene amplification as a mechanism of drug resistance is also commonly observed where the drug inhibits an enzyme within a critical biosynthetic pathway. For example, methotrexate resistance is associated with amplification of the dihydrofolate reductase (DHFR) gene22; thymidylate synthase (TS) can be amplified in response to a variety of TS inhibitors23; resistance to hydroxyurea is associated with amplification of the M2 subunit of dihydrofolate reductase24; and PALA (N-phosphonoacetyl-L-aspartate) resistance is correlated with amplification of the carbamyl phosphate synthetase-aspartate transcarbamylase-dihydroorotase gene.25 Thus, amplification of genes encoding drug targets or enzymes that can bypass a drug effect is a frequent mechanism of drug resistance. However, amplification of a tyrosine kinase in response to a tyrosine kinase inhibitor has not previously been reported.
The mechanisms of gene amplification are not well understood. Genes may be amplified in situ at intrachromosomal locations through a mechanism that may involve repeated cycles of breakage-fusion-bridge cycles where chromosome breakage is initiated at fragile sites.26,27Amplification of the DHFR gene has also been reported to occur owing to unequal sister chromatid exchange.28 Intrachromosomal gene amplification is typically associated with “homogeneously staining regions” (HSR) in specific chromosomes, and HSR has been used to localize genes involved in drug resistance.28-30 In other situations, gene amplification may be associated with extrachromosomal accumulation of genetic material in the form of double minute (DM) chromosomes.31 DMs are acentric and atelomeric chromatin composed of circular DNA and are seen in a substantial proportion of human cancers. In most cases, however, the genes being amplified by the accumulation of DM are still unknown. Spontaneous amplification of Bcr/Abl has been described during the transition from chronic phase to blast phase, either in the form of duplication of the Ph chromosome or as isolated gene amplification.32,33 Similarly, Ph+ cell lines have been reported to show amplification of the Bcr/Abl gene during tissue culture.34
Although we observed increased expression of Bcr/Abl in both resistant Ba/F3.p210 and K562 cells, overall our results do not suggest that the increase in the level of target protein, by itself, is sufficient to account for the high level of drug resistance we observed. Altered drug metabolism or efflux deserves further study and could account for some of the resistance seen in this report. Although we did not observe overexpression of Mdr-1 in the resistant K562 cells studied here, overexpression of Mdr-1 or other “drug pumps” is commonly thought to occur in drug-resistant cells.35,36 Other genes conferring multidrug resistance, such as MRP and LRP, have also been shown to be amplified in cell lines and patient specimens.29
In the case of both K562 and Ba/F3.p210 cell lines, we were unable to derive lines that were resistant to drug levels of more than 0.5 μmol/L and more than 2.5 μmol/L STI571, respectively, despite several attempts and many months of drug exposure. Overall, the results suggest that resistance to STI571 may be relative, and it may be worthwhile to treat patients who become resistant to standard doses of STI571 with increased doses of the drug.
Acknowledgments
We are grateful to Dr Jerry Ritz and Antoinette Chillemi from the Dana Farber Cancer Institute for technical assistance with quantitative PCR.
Reprints: James D. Griffin, Dana Farber Cancer Institute, Department of Adult Oncology, 44 Binney St, Boston, MA 02115; e-mail: james_griffin@dfci.harvard.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.