Abstract
The Fanconi Anemia (FA) Group C complementation group gene (FANCC) encodes a protein, FANCC, with a predicted Mr of 63000 daltons. FANCC is found in both the cytoplasmic and the nuclear compartments and interacts with certain other FA complementation group proteins as well as with non-FA proteins. Despite intensive investigation, the biologic roles of FANCC and of the other cloned FA gene products (FANCA and FANCG) remain unknown. As an approach to understanding FANCC function, we have studied the molecular regulation of FANCC expression. We found that although FANCCmRNA levels are constant throughout the cell cycle, FANCC is expressed in a cell cycle-dependent manner, with the lowest levels seen in cells synchronized at the G1/S boundary and the highest levels in the M-phase. Cell cycle–dependent regulation occurred despite deletion of the 5′ and 3′ FANCC untranslated regions, indicating that information in the FANCC coding sequence is sufficient to mediate cell cycle–dependent regulation. Moreover, inhibitors of proteasome function blocked the observed regulation. We conclude that FANCC expression is controlled by posttranscriptional mechanisms that are proteasome dependent. Recent work has demonstrated that the functional activity of FA proteins requires the physical interaction of at least FANCA, FANCC, and FANCG, and possibly of other FA and non-FA proteins. Our observation of dynamic control of FANCC expression by the proteasome has important implications for understanding the molecular regulation of the multiprotein complex.
Fanconi anemia (FA) is an autosomal recessive disorder characterized by progressive pancytopenia, high risk for malignancies (especially acute myelogenous leukemia),1 and, in some patients, congenital malformations including skin pigmentation abnormalities, skeletal deformities, and renal anomalies.2-5 The cellular hallmark of FA is a unique hypersensitivity to DNA cross-linking agents. Treatment of FA cells with agents such as mitomycin C or diepoxybutane at doses that have little impact on normal cells results in chromosomal instability and cellular death.6-9
Despite clinical and cellular similarities in the FA phenotype, cell–cell fusion techniques have established that there are at least 8 FA complementation groups, FA[A] to FA[H].10 The genes inactivated in complementation groups A (FANCA), C (FANCC), and G (FANCG) have been cloned.11-13 Additionally, the genes mutated in group D and group E FA have been localized to chromosomes 3p22-26 and 6p21-22, respectively.14 15
The FANCC gene was cloned by Strathdee et al11using a functional complementation strategy, and it encodes a protein of 558 amino acids with a predicted molecular mass of 63 kd. FANCC is predominately hydrophobic and has no obvious transmembrane domain, signal sequence, or other functional motifs. The polypeptide sequence contains no significant homologies with other known proteins in genetic databanks, and the biochemical function of FANCC is unknown. A number of groups have found FANCC localized primarily in the cytoplasm, with recent reports also describing the existence of nuclear FANCC.16,17 Low endogenous protein expression has limited all existing studies of FANCC to immunoblot and overexpression immunofluorescent analyses (IFA). Numerous interacting partners for FANCC have been described, including the cyclin-dependent kinase cdc2, FANCA, FANCG, the molecular chaperone GRP94, NADPH cytochrome P450 reductase, and FAZF—a novel transcriptional repressor with homology to the acute promyelocytic zinc finger protein.17-24
We report here further evidence that FANCC expression is regulated during the cell cycle and that this regulation is controlled by a posttranscriptional mechanism dependent on proteasome function. The lowest expression was found in cells synchronized at the G1/S boundary, and the highest expression was found in cells in the M-phase. We infer that the regulation of FANCC polypeptide expression occurs after transcription because we found that levels of FANCC mRNA were constant throughout the cell cycle. Cell cycle–dependent regulation occurred despite deletion of the 5′ and 3′ FANCC untranslated regions, indicating that information in the FANCC coding sequence is sufficient to mediate cell cycle-dependent regulation. Cell cycle regulation was abrogated with the use of inhibitors of proteasome-dependent proteolysis. We conclude that FANCC expression is controlled by proteasome-dependent processes that regulate FANCC mRNA translation, protein degradation, or both.
Materials and methods
Plasmids
pcFANCC.
The plasmid pcFANCC has been described.25 The FANCC cDNA in this plasmid contains only the coding sequence with no FANCC-derived 5′ or 3′ untranslated region sequences.
pGEM-11z FANCC.
The FANCC cDNA was excised from pLFACSN using XhoI andBamHI and was ligated into aXhoI/BamHI-digested pGEM-11z vector (Promega, Madison, WI).26
2HA-L554P.
The construct 2HA-L554P has been described.23 It contains an HA-epitope–tagged cDNA for the L554P mutant cDNA inserted in the backbone of the pCEP4 vector (Invitrogen, Carlsbad, CA).
pcLUC.
A BglII/XhoI fragment containing the firefly luciferase cDNA was excised from pSP-luc+ (Promega) and ligated intoBamHI/XhoI-digested pcDNA3.1(+) vector (Invitrogen).
Cell lines
The parental 293 cell line was obtained from ATCC (Manassas, VA) and maintained in DMEM (GIBCO-BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (defined, heat-inactivated, low endotoxin; HyClone, Logan, UT). As described previously,23 25 293 FANCC, 293 LUC, 293 L544P, and 293 NEO cells were generated. For this study, subclones of 293 LUC, 293 NEO, and 293 L554P were prepared by limiting-dilution cloning. The 293 FANCC E2 subclone was prepared by 2 successive rounds of limiting-dilution cloning.
FANCC antibodies
Monoclonal antibodies specific for wild-type 8F3, 3A11, or L554P FANCC 13F5 polypeptide were generated by immunizing mice with a KLH-peptide conjugated to a peptide containing wild-type FANCC AA 547-558 or with an overlapping peptide containing the inactivating leucine-to-proline substitution at position 554.11 These antibodies will be described in detail elsewhere (Zhi et al, manuscript in preparation). Immunoblots were performed as described previously.25
Reagents
Polyclonal rabbit anti-luciferase antiserum, dimethyl sulfoxide (DMSO), nocodazole, and hydroxyurea were purchased from Sigma (St Louis, MO). Lactacystin and MG132 were purchased from Calbiochem–Novabiochem (San Diego, CA) and were resuspended in DMSO.
Cell synchronization experiments
Cells were synchronized at the G1/S boundary by culturing in 1.3 mmol/L hydroxyurea for 24 hours.27 To release cells from synchronization, the cells were washed twice with phosphate-buffered saline (PBS) and placed back in regular growth medium. Cell cycle progression was monitored by flow cytometric determination of DNA content. At timed intervals after release, cells were processed for protein, DNA, or RNA analysis. To enrich for cells in M-phase, cultures were treated with 2 μmol/L nocodazole for 24 hours before harvest.28
RNA analysis
Biotin-labeled anti-sense probes to luciferase or FANCC were generated by in vitro transcription of HincII linearized pSP-luc+ or NcoI linearized pGEM-11Z FANCC, respectively. A biotin-labeled anti-sense probe to human β-actin was generated by in vitro transcription of the pTRI-β–actin template (Ambion, Austin, TX). Non-isotopic ribonuclease protection assays were performed using a commercially available kit (RPA II and Brightstar Biodetect; Ambion).
Cell cycle analysis
Cells were harvested and stained with propidium iodide as described previously. After staining, the samples were analyzed for DNA content using a Becton Dickinson FACScan flow cytometer (Becton Dickinson, Mountain View, CA). Data were analyzed by the Multicycle software program, which uses the polynomial S-phase algorithm (Phoenix Flow Systems, San Diego, CA).29
Indirect immunofluorescence
Then 293 FANCC E2 and control 293 NEO cells were seeded at a density of 2.5×104 cells/chamber of 2-well Permanox Lab Tek chamber slides (Nunc Nalge; Rochester, NY). Cells were cultured in complete growth media for 48 hours. For cell synchronization studies, slides were treated with fresh medium (asynchronous control) or medium containing hydroxyurea (final concentration, 1.3 mmol/L) for 24 hours, washed twice with PBS, and released into complete media. The cells were fixed and permeabilized for analysis at 0, 4, 6, 8, 10, and 12 hours after release. To enrich for cells in the M-phase, slides were treated with 2 μmol/L nocodazole for 24 hours and then processed for IFA.28
The slides were fixed with 3.7% paraformaldehyde and permeabilized with 0.2% Triton X-100. Cells were blocked with a solution of 3% normal goat serum in PBS. Primary antibody was added (1:100 8F3) and allowed to bind overnight at room temperature with gentle rocking. After 5 washes with PBS at 5 minutes each, Oregon green-conjugated goat antimouse secondary antibody was added (1:100 [Molecular Probes, Eugene, OR]). Primary and secondary antibodies were diluted in PBS containing 3% normal goat serum. Secondary antibody incubation was performed in the dark. The stained cells were mounted in SlowFade (Molecular Probes).
Fluorescence was observed by using a Bio-Rad MRC 1024ES laser scanning confocal imaging system (Bio-Rad, Cambridge, MA) attached to an inverted Nikon eclipse TE330 microscope (Nikon, Tokyo, Japan). The acquisition system (LaserSharp) uses a krypton/argon laser with an excitation line at 488 nm and an 8-bit photomultiplier tube. Settings were optimized using positively stained cells and were maintained during scanning of control cells to retain relative brightness. Images were processed using the LaserSharp postprocessing software and exported as TIFF files into Adobe Photoshop 4.0 (Adobe, San Jose, CA).
Percentage FANCC expression data were gathered with a Leitz Orthoplan 2 fluorescence microscope (Leitz, Stuttgart, Germany) using a green filter (488 nm) and a ×100 oil lens. Two hundred cells were counted from each slide. Cells were scored as positive for FANCC staining based on a comparison with control cells (293 NEO). Standard error was computed from the standard deviation divided by the square root of the count using StatView 5.0 (SAS Institute, Cary, NC).
Results
Immunofluorescent analysis of FANCC expression
In a previous study using 293 FANCC cells, we noted cell-to-cell variation of FANCC expression and variable subcellular localization.25 Because the expression of endogenous FANCC is regulated during the cell cycle, we wondered whether exogenous FANCC was subject to the same type of regulation and whether this phenomenon might partially or completely explain the cell-to-cell variation we observed.25 30
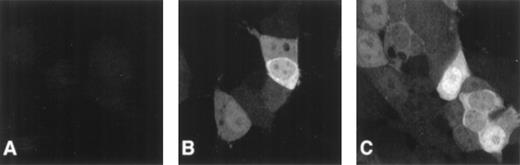
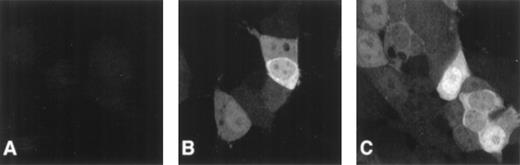
To investigate FANCC expression and subcellular localization during the cell cycle, we transfected 293 cells with pcFANCC and then cloned the resultant cell population twice by limiting dilution to obtain 293/FANCC E2 cells. Control cell lines, 293 NEO and 293 LUC, were created similarly. Using polyclonal and monoclonal anti-FANCC antibodies, we confirmed our previous observation that the cellular expression of exogenous FANCC was variable from cell to cell (Figure1). In contrast, the expression of luciferase was similar in all 293 LUC cells examined. As we reported previously, FANCC was found in the cytoplasm and the nuclei of these cells.23 25 The subcellular localization was highly variable: it ranged from equal amounts in the cytoplasm and the nuclei, to much greater nuclear expression than cytoplasmic expression, to much greater cytoplasmic expression than nuclear expression. Additionally, we observed FANCC nuclear foci in approximately 25% of the cells. Figure 1C depicts a typical range of FANCC expression and localization seen in our experiments.
Heterogeneous cellular expression of FANCC.
Cells were stained with monoclonal anti-FANCC antibody and a secondary Oregon green–labeled goat antimouse antibody. Fluorescence was observed using a confocal imaging system. A single slice through the plane of cells is displayed. (A) 293 NEO cells. No staining of endogenous FANCC is detectable. (B) 293 FANCC E2 cells. In this field, 1 cell has a large amount of cytoplasmic FANCC, whereas an adjacent cell is only weakly positive. (C) 293 FANCE2 cells. Some cells have only cytoplasmic FANCC, others have predominately nuclear FANCC, and some have FANCC in both compartments. Nuclear foci are clearly visible in the top and bottom cells. In addition to the subcellular localization differences, there is marked cell-to-cell variation in total cellular FANCC expression.
Heterogeneous cellular expression of FANCC.
Cells were stained with monoclonal anti-FANCC antibody and a secondary Oregon green–labeled goat antimouse antibody. Fluorescence was observed using a confocal imaging system. A single slice through the plane of cells is displayed. (A) 293 NEO cells. No staining of endogenous FANCC is detectable. (B) 293 FANCC E2 cells. In this field, 1 cell has a large amount of cytoplasmic FANCC, whereas an adjacent cell is only weakly positive. (C) 293 FANCE2 cells. Some cells have only cytoplasmic FANCC, others have predominately nuclear FANCC, and some have FANCC in both compartments. Nuclear foci are clearly visible in the top and bottom cells. In addition to the subcellular localization differences, there is marked cell-to-cell variation in total cellular FANCC expression.
Cell cycle–dependent regulation of FANCC expression
To test the hypothesis that the concentration of exogenous FANCC was regulated in a cell cycle–dependent manner analogous to that of endogenous protein, we used hydroxyurea to synchronize cells at the G1/S boundary.31 32 After synchronization, the cells were washed extensively and allowed to progress through the cell cycle. Cell cycle position was monitored by flow cytometry, and FANCC expression was assessed using immunofluorescence.
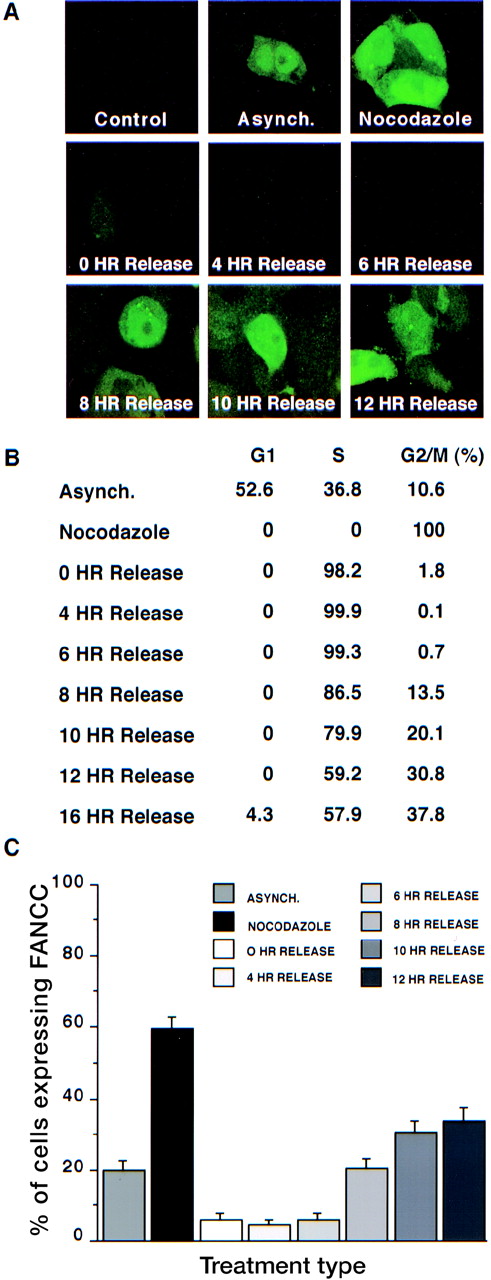
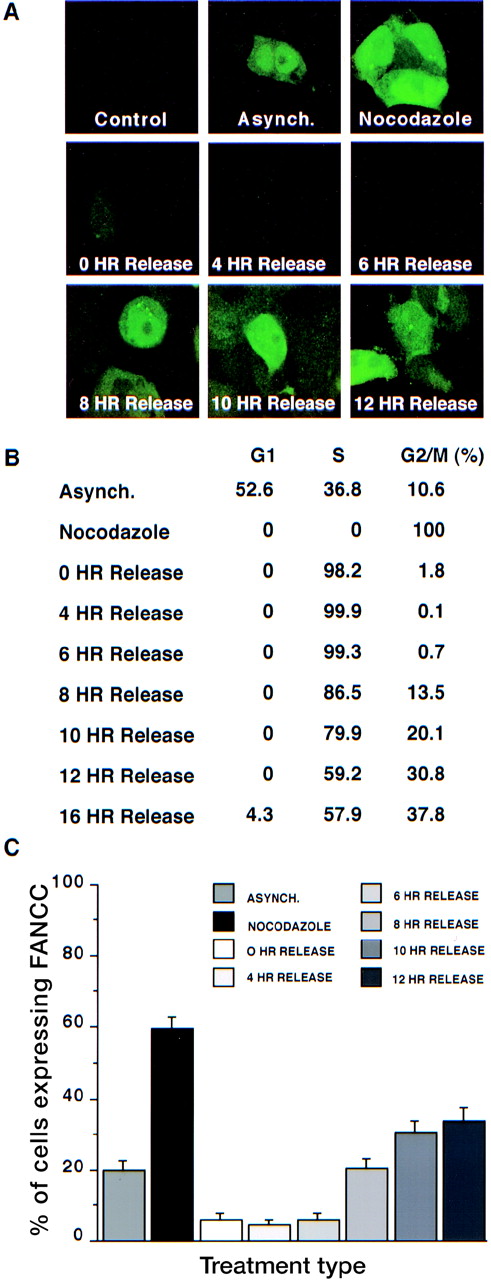
Results of representative fields are shown in Figure2A, and summarized data are depicted in Figures 2B and 2C. Although our impression is that all 293 FANCC E2 cells stain at least weakly positive when using our anti-FANCC mAbs and are brighter than control cells (293 WT, 293 NEO, or 293 LUC), the level of staining was very low in some cells. For the purposes of our experiments, we scored 200 cells as either 0+ (negative/low FANCC expression compared with control cells [“low”]) or 1+ (moderate-to-high FANCC expression compared with control cells [“high”]). In asynchronous cultures approximately 20% of cells have high FANCC expression. The percentage of high FANCC-expressing cells drops to 5% in cells arrested at the G1/S boundary. After release, FANCC expression does not appreciably increase for 8 hours, and it reaches a maximum 10 to 12 hours after release, at which time 30% of the cells express high levels of FANCC. The greatest number of high FANCC-expressing cells (60%) was seen in cultures of nocodazole-treated cells, implying that FANCC expression is highest in the M-phase because nocodazole arrests cells by the disruption of microtubules.28
Cell cycle–dependent regulation of FANCC expression.
293 FANCC E2 cells were synchronized with hydroxyurea for 24 hours and then washed twice and cultured in complete growth medium. Cells were fixed and permeabilized at different time points after release (0 hours, 4 hours, 6 hours, 8 hours, 10 hours, and 12 hours). An asynchronous population of 293 FANCC E2 cells (Asynch.) and 293 Neo cells were also analyzed (Control). Cells were treated with nocodazole for 24 hours to induce M-phase arrest. (A) IFA of cells using monoclonal anti-FANCC antibody. No FANCC staining is detectable in 293 Neo (control cells). FANCC expression is lowest in cells at the time of or immediately after release from hydroxyurea synchronization. Highest FANCC expression is seen in cells 10 to 12 hours after HU release or in cells arrested in the M-phase by nocodazole. (B) Cell cycle distribution of cells shown in panel A. (C) Summary statistics. 200 cells in each condition were scored as negative or positive for FANCC expression. Bars depict mean ± SEM.
Cell cycle–dependent regulation of FANCC expression.
293 FANCC E2 cells were synchronized with hydroxyurea for 24 hours and then washed twice and cultured in complete growth medium. Cells were fixed and permeabilized at different time points after release (0 hours, 4 hours, 6 hours, 8 hours, 10 hours, and 12 hours). An asynchronous population of 293 FANCC E2 cells (Asynch.) and 293 Neo cells were also analyzed (Control). Cells were treated with nocodazole for 24 hours to induce M-phase arrest. (A) IFA of cells using monoclonal anti-FANCC antibody. No FANCC staining is detectable in 293 Neo (control cells). FANCC expression is lowest in cells at the time of or immediately after release from hydroxyurea synchronization. Highest FANCC expression is seen in cells 10 to 12 hours after HU release or in cells arrested in the M-phase by nocodazole. (B) Cell cycle distribution of cells shown in panel A. (C) Summary statistics. 200 cells in each condition were scored as negative or positive for FANCC expression. Bars depict mean ± SEM.
Interestingly, cell cycle position did not account for all the observed cell-to-cell variation in FANCC expression; even in the nocodazole-treated cultures, a full 40% of the cells were scored as having negative or low FANCC expression. Nor did we observe any correlation between cell cycle position and subcellular localization.
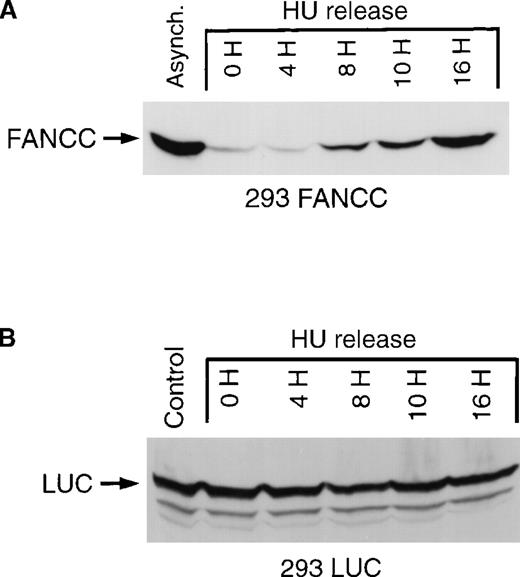
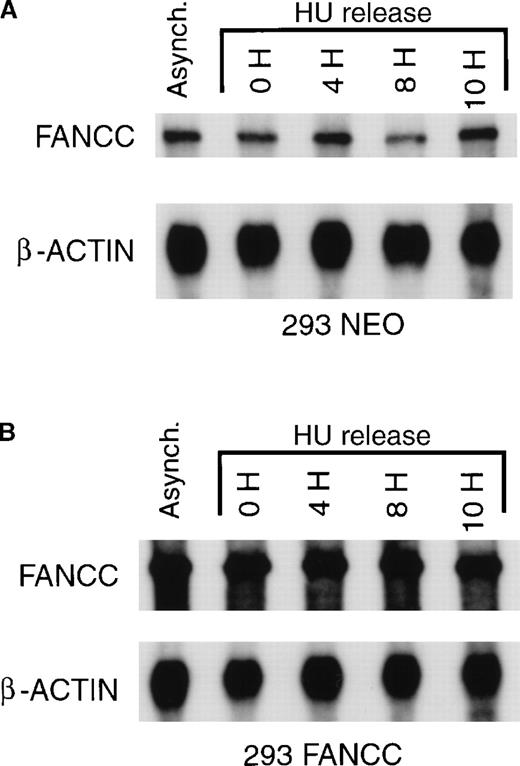
To confirm our IFA results, we used an immunoblot technique to assess FANCC expression. Whole-cell lysates were prepared from asynchronous cells, cells synchronized at the G1/S boundary by hydroxyurea, and populations of cells released from G1/S synchronization (Figure3A) and probed with a murine monoclonal antibody that recognizes a carboxy-terminal FANCC epitope. We detected only a single protein band corresponding to FANCC in 293 FANCC E2 cells. Under these conditions, only exogenous FANCC can be detected. No bands are seen using whole-cell lysates from 293, 293 NEO, or 293 LUC cells (data not shown). Using an immunoblot method, we found excellent agreement in our immunofluorescent results, with FANCC expression increasing from low levels at the G1/S boundary to maximum levels at 16 hours after release.
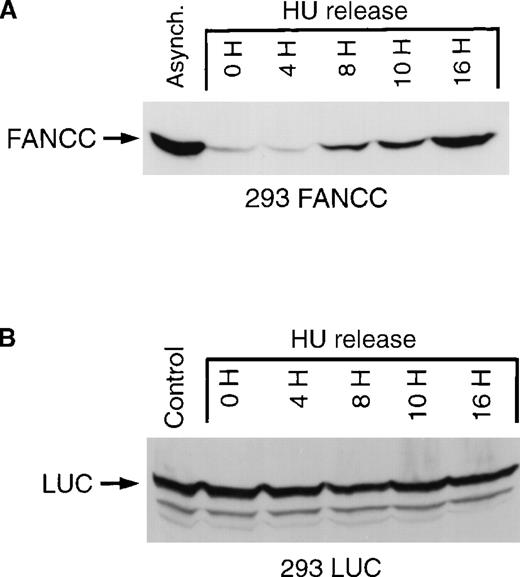
Cell cycle–dependent regulation of exogenous FANCC but not exogenous luciferase expression.
(A) 293 FANCC E2 cells were treated with 1.3 mmol/L HU for 24 hours to synchronize cells at the G1/S boundary. Cells were then washed twice and fed with complete growth medium. Whole-cell lysates were prepared at the indicated times after release. A whole-cell extract was also prepared from a culture of asynchronous FANCC E2 cells (Control). The whole-cell extracts were subjected to Western blotting and probed using a murine anti-FANCC antibody (3A11). Identical results were obtained using a second monoclonal antibody (8F3; data not shown). Cell cycle kinetics in these experiments was identical to that shown in Figure 2B. (B) Whole-cell extracts of 293 LUC cells were prepared under similar experimental conditions. Extracts were analyzed for luciferase expression by Western blotting and probing with a rabbit anti-luciferase antiserum. The top band is a luciferase-specific band (LUC) detected only in 293 LUC cells and not in 293 WT, 293 NEO, 293 FANCC, or 293 L554P cells (data not shown). The lower 2 bands are nonspecific and are seen in 293 WT and derivative cells.
Cell cycle–dependent regulation of exogenous FANCC but not exogenous luciferase expression.
(A) 293 FANCC E2 cells were treated with 1.3 mmol/L HU for 24 hours to synchronize cells at the G1/S boundary. Cells were then washed twice and fed with complete growth medium. Whole-cell lysates were prepared at the indicated times after release. A whole-cell extract was also prepared from a culture of asynchronous FANCC E2 cells (Control). The whole-cell extracts were subjected to Western blotting and probed using a murine anti-FANCC antibody (3A11). Identical results were obtained using a second monoclonal antibody (8F3; data not shown). Cell cycle kinetics in these experiments was identical to that shown in Figure 2B. (B) Whole-cell extracts of 293 LUC cells were prepared under similar experimental conditions. Extracts were analyzed for luciferase expression by Western blotting and probing with a rabbit anti-luciferase antiserum. The top band is a luciferase-specific band (LUC) detected only in 293 LUC cells and not in 293 WT, 293 NEO, 293 FANCC, or 293 L554P cells (data not shown). The lower 2 bands are nonspecific and are seen in 293 WT and derivative cells.
The expression of exogenous FANCC in 293 FANCC cells is controlled by the cytomegalovirus (CMV) immediate early promoter.33 To test whether our results were an artifactual effect of cell cycle position on CMV promoter activity, we used 293 LUC cells in which the expression of the exogenous firefly luciferase gene was dependent on the identical CMV promoter used in our FANCC expression vector. We found that the expression of luciferase protein was constant throughout the cell cycle (Figure 3B). The FANCC cDNA used in our experiments consisted of only the coding sequence with no FANCC-derived 5′ or 3′ UTR sequences. We concluded that the coding sequence of FANCC contained information sufficient for its regulated expression.
Cell cycle–dependent regulation of L544P FANCC expression
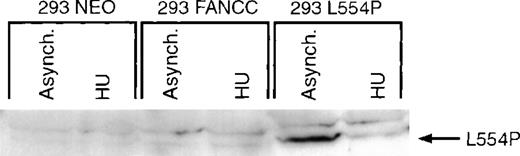
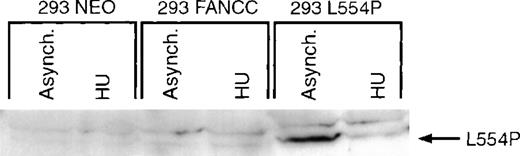
The carboxyl terminus of FANCC is thought to be critical for the proper functioning of FANCC.17 An inactive form of FANCC, L554P, results from a carboxy-terminal mutation identified in FA(C) patients.11 We and others23,25 34 have recently reported that the L554P polypeptide is unable to enter the nucleus, possibly accounting for its failure to function. To test whether the L554P mutation would alter cell cycle–dependent FANCC regulation, we generated an isogenic cell line overexpressing the mutant L554P polypeptide (293 L554P). We have developed and characterized a monoclonal antibody (13F5) that selectively recognizes the epitope present in L554P but absent in wild-type FANCC (Zhi et al, manuscript in preparation). The13F5 monoclonal antibody was used to probe whole-cell lysates prepared from asynchronous or HU-synchronized 293 NEO, 293 FANCC E2, and 293 L554P cells. The 13F5 antibody selectively recognized the L554P but not the wild-type FANCC isoform (Figure4). The expression of L554P was markedly decreased in cells synchronized at the G1/S boundary in a manner identical to that of wild-type FANCC. Thus, the mechanism by which FANCC expression is regulated during the cell cycle is not affected by this particular mutation.
Expression of a patient-derived mutant form of FANCC (L554P) is regulated identically to the wild-type isoform.
293 NEO, 293 FANCC, or 293 L554P cells were grown in the presence (HU 24H) or absence (Asynch.) of 1.3 mmol/L HU for 24 hours. Whole-cell extracts were prepared and analyzed by Western blotting using a monoclonal antibody specific for the L554P FANCC isoform (13F5). The L554P isoform is recognized in asynchronous 293 L554P cells, but no endogenous (293 NEO) or exogenous (293 FANCC) wild-type FANCC is detected using this antibody. Synchronization with HU markedly decreases expression of the mutant isoform in 293 L554P cells.
Expression of a patient-derived mutant form of FANCC (L554P) is regulated identically to the wild-type isoform.
293 NEO, 293 FANCC, or 293 L554P cells were grown in the presence (HU 24H) or absence (Asynch.) of 1.3 mmol/L HU for 24 hours. Whole-cell extracts were prepared and analyzed by Western blotting using a monoclonal antibody specific for the L554P FANCC isoform (13F5). The L554P isoform is recognized in asynchronous 293 L554P cells, but no endogenous (293 NEO) or exogenous (293 FANCC) wild-type FANCC is detected using this antibody. Synchronization with HU markedly decreases expression of the mutant isoform in 293 L554P cells.
Endogenous and exogenous FANCC mRNA levels are constant throughout the cell cycle
To aid in determining potential mechanisms for the cell cycle-dependent regulation of FANCC, we measured FANCC mRNA levels in synchronized 293 FANCC and 293 NEO cells. Cell cycle position did not change the concentration of exogenous FANCC (293 FANCC), endogenous FANCC (293 NEO), or β-actin. Results of a representative experiment are shown in Figure 5. Although slight fluctuations in endogenous FANCC mRNA concentration are seen in Figure5, these changes were not consistently observed. In multiple experiments we found no significant change in either endogenous or exogenous FANCC mRNA levels during cell cycle progression. These results are similar to those reported by Kupfer et al.31Cell cycle position also did not change the concentration of luciferase mRNA in 293 LUC cells (data not shown). We concluded that FANCC expression varied during the cell cycle despite constant levels of FANCC mRNA, suggesting the regulation of FANCC mRNA translation, FANCC protein degradation, or both.
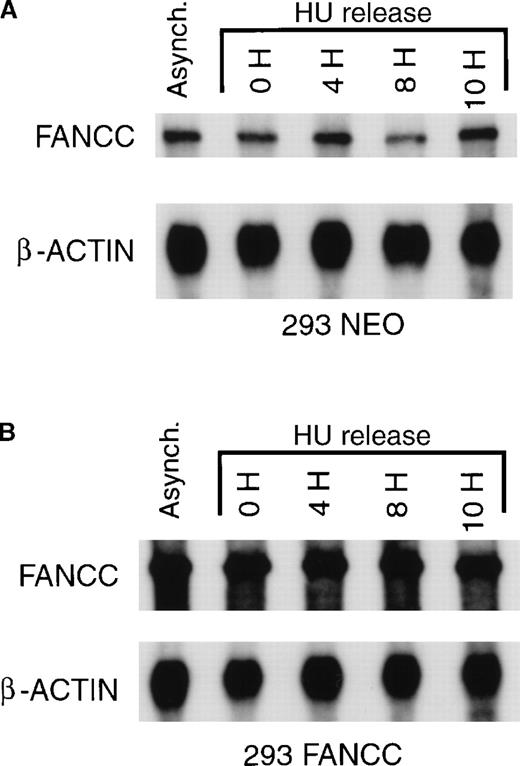
FANCC mRNA expression is constant during cell cycle progression.
(A) 293 NEO cells were synchronized with HU and released from the G1/S boundary as described above. Total cytoplasmic RNA was prepared from synchronized cells or from a culture of asynchronous 293 NEO cells (Asynch.). Endogenous FANCC and β-actin mRNA were analyzed by ribonuclease protection assay. A representative experiment is shown. (B) Identical experiments were performed using 293 FANCC E2 cells. Total FANCC (endogenous plus exogenous) and β-actin mRNA were analyzed by ribonuclease protection assay. A representative experiment is shown. Exposure times were identical for membranes depicted in A and B to demonstrate that 293 FANCC cells express more FANCC mRNA than 293 NEO cells.
FANCC mRNA expression is constant during cell cycle progression.
(A) 293 NEO cells were synchronized with HU and released from the G1/S boundary as described above. Total cytoplasmic RNA was prepared from synchronized cells or from a culture of asynchronous 293 NEO cells (Asynch.). Endogenous FANCC and β-actin mRNA were analyzed by ribonuclease protection assay. A representative experiment is shown. (B) Identical experiments were performed using 293 FANCC E2 cells. Total FANCC (endogenous plus exogenous) and β-actin mRNA were analyzed by ribonuclease protection assay. A representative experiment is shown. Exposure times were identical for membranes depicted in A and B to demonstrate that 293 FANCC cells express more FANCC mRNA than 293 NEO cells.
Inhibitors of proteasome function block cell cycle–dependent regulation of FANCC
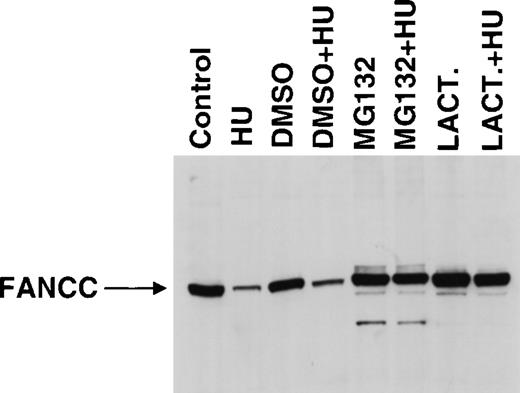
The ubiquitin–proteasome pathway is the best-described system for the regulation of protein degradation. Therefore, we used inhibitors of proteasome function to test whether the regulation of FANCC expression is dependent on proteasome function. For 24 hours, 293 FANCC E2 cells were treated with vehicle only (DMSO) or with proteasome inhibitor (20 μmol/L MG132 or 10 μmol/L lactacystin in DMSO) ± HU (Figure6). Treatment with either inhibitor alone increased FANCC expression. However, the percentage of cells in the G2/M compartment also dramatically increased in inhibitor-treated cells (Table 1), probably because of the inhibition of proteasome-mediated cyclin degradation.35 36Thus, increased FANCC expression could be a direct result of the inhibition of proteasome-dependent degradation of FANCC or an indirect result of the accumulation of cells in the G2/M compartment, where FANCC expression is increased (Figures 2, 3). To help distinguish between these possibilities, we treated cells with both HU and proteasome inhibitor. Combined treatment of cells with HU and MG132 resulted in partial synchronization at the G1/S boundary (70% vs. 97% DMSO + HU). Combined treatment with HU and lactacystin resulted in nearly complete synchronization at the G1/S boundary (93%). However, the synchronization of cells at the G1/S boundary in the presence of either inhibitor did not result in a decrease in FANCC expression when compared with treatment with inhibitor alone. Thus, the inhibition of proteasome function prevented the reduction of FANCC expression associated with partial or complete synchronization at the G1/S boundary, suggesting that this phenomenon was proteasome dependent.
Proteasome inhibitors increase FANCC expression.
293 FANCC E2 cells were treated with media only (Control) or media supplemented with vehicle alone (DMSO), 20 μm MG132 (MG132), or 10 μmol/L lactacystin (LACT.) for 24 hours in the presence or absence of 1.3 mmol/L HU. Whole-cell extracts were analyzed for FANCC expression by Western blotting and probing with monoclonal anti-FANCC antibody (8F3). Ten percent of the cells from each experimental condition were analyzed for DNA content by flow cytometry (Table 1). Arrow indicates full-length FANCC. The apparent increase in size of full-length FANCC in inhibitor-treated cells is an artifact of this particular gel (see also Figure 7).
Proteasome inhibitors increase FANCC expression.
293 FANCC E2 cells were treated with media only (Control) or media supplemented with vehicle alone (DMSO), 20 μm MG132 (MG132), or 10 μmol/L lactacystin (LACT.) for 24 hours in the presence or absence of 1.3 mmol/L HU. Whole-cell extracts were analyzed for FANCC expression by Western blotting and probing with monoclonal anti-FANCC antibody (8F3). Ten percent of the cells from each experimental condition were analyzed for DNA content by flow cytometry (Table 1). Arrow indicates full-length FANCC. The apparent increase in size of full-length FANCC in inhibitor-treated cells is an artifact of this particular gel (see also Figure 7).
To further confirm a direct role of proteasome inhibitor on FANCC expression independent of any secondary effect on cell cycle distribution, we first synchronized the cells at the G1/S boundary for 24 hours using hydroxyurea, and then we added vehicle alone (DMSO), MG132, or lactacystin to our cultures. Treatment of cells with MG132 or lactacystin after prior synchronization at the G1/S boundary did not change the cell cycle distribution compared with treatment with HU alone or HU + DMSO (data not shown). As shown in Figure7, the addition of MG132 or lactacystin to cells synchronized at the G1/S boundary increased FANCC expression. Thus, our results suggested that the observed decrease in FANCC expression at the G1/S boundary was an active process dependent on proteasome function.
Proteasome inhibitors MG132 and lactacystin increase FANCC expression in cells synchronized at the G1/S boundary.
293 FANC E2 cells were treated for 24 hours with complete growth medium (Control) or 1.3 mmol/L HU (HU). HU-synchronized cells were treated with vehicle only (DMSO), 20 μmol/L MG132 (MG132), or 20 μmol/L lactacystin (LACT.) for an additional 6 hours. Whole-cell extracts were analyzed for FANCC expression by Western blotting and probing with a monoclonal anti-FANCC antibody (8F3).
Proteasome inhibitors MG132 and lactacystin increase FANCC expression in cells synchronized at the G1/S boundary.
293 FANC E2 cells were treated for 24 hours with complete growth medium (Control) or 1.3 mmol/L HU (HU). HU-synchronized cells were treated with vehicle only (DMSO), 20 μmol/L MG132 (MG132), or 20 μmol/L lactacystin (LACT.) for an additional 6 hours. Whole-cell extracts were analyzed for FANCC expression by Western blotting and probing with a monoclonal anti-FANCC antibody (8F3).
Discussion
In the past decade, genes for 3 of the 8 known FA complementation groups have been cloned. However, the gene products of the 3 cloned FA genes have no informative homologies to known sequences, nor do the 3 proteins have significant homology to each other.11-13Thus, the FA genes likely regulate a novel biologic pathway. Various hypotheses have been advanced to explain the clinical phenotype, including defects in DNA repair, pre-repair defects, abnormalities in coping with cellular oxidative stress, and abnormal regulation of apoptosis.2,3,37 Additionally, we recently reported that FANCC interacts with and co-localizes in nuclear foci with the novel transcriptional repressor FAZF, suggesting a possible role in chromatin remodeling.23
Understanding the basic features of FA protein fate, including expression and subcellular localization during the cell cycle, is a reasonable first step in confirming and extending notions about FA protein function. In this paper we report that FANCC subcellular localization was surprisingly variable. Under the conditions of our experiments, no obvious correlation between cell cycle transit and subcellular localization was observed. In addition, we report that the level of total cellular FANCC expression was controlled by a proteasome-dependent posttranscriptional mechanism.
We previously reported that FANCC expression varied in a clonal population of cells that had been engineered to overexpress FANCC (293 FANCC).25 The expression vector used in these studies lacked FANCC 5′ and 3′ UTR sequences and contained only the coding sequence under the control of a CMV promoter. Thus, the variable expression we observed suggested that sequence information contained in the FANCC coding region might allow cell cycle–dependent regulation of FANCC expression. Moreover, endogenous FANCC expression was reported by other investigators to change during the cell cycle in HeLa cells, suggesting that exogenous expression of FANCC mirrors the behavior of the endogenous protein in this respect.31 To explore further the basis of FANCC regulation, we measured FANCC mRNA and protein levels in asynchronous and synchronous cell populations of a doubly cloned subline of 293 FANCC (293 FANCC E2). Despite a striking variation in FANCC protein expression during the passage from the G1/S boundary to the G2/M compartment assayed by immunoblot and IFA, we found no corresponding change in FANCC mRNA levels. Regulated expression was not observed for unrelated exogenous cDNA expressed under the same conditions, suggesting that this effect is specific for the FANCC coding sequence. Consistent with a posttranscriptional regulation model, we found that endogenous FANCC mRNA levels remained constant during cellular transition from the G1/S boundary to the G2/M compartment in matched control cells by using a ribonuclease protection assay. We emphasize these similarities between endogenous and overexpressed proteins because a drawback in the FA field is that endogenous FA proteins cannot be detected by IFA because of low expression levels. However, IFA is crucial for investigating which factors regulate subcellular localization. One strength of this work is that endogenous FANCC expression behavior closely matched that observed for overexpressed proteins wherever comparison was possible. We conclude that FANCC expression is regulated at a posttranscriptional level by altering the translation of FANCC mRNA, controlling degradation of FANCC, or both.
An inactive mutant form of FANCC, L554P, results from a mutation identified in FA(C) patients.11 Although wild-type FANCC interacts with FANCA, appears in the nucleus during some parts of the cell cycle, and can form subnuclear foci, the L554P mutation abrogates the interaction with FANCA and the nuclear localization observed for wild-type FANCC.23,25,38 To determine whether this missense mutation alters the cell cycle–dependent regulation of FANCC, we made a cell line overexpressing L554P and examined protein levels during the cell cycle. We found that the expression pattern of L554P was identical to that seen with wild-type FANCC. Thus, the mechanism by which FANCC expression is regulated during the cell cycle is not affected by the L554P mutation. Taken together, this suggests that conditional nuclear localization or interaction with other FA proteins is required for FANCC function. In this regard, Youssoufian39 found that forced nuclear expression of FANCC by attachment of an SV-40 nuclear localization signal (NLS) interferes with FANCC function, leading to the notion that cytoplasmic localization, not nuclear localization, is required for FANCC function. The abundant cytoplasmic expression observed for FANCC and the discovery of cytoplasmic FANCC binding partners are consistent with this view.17-19,21-24 However, now that FANCC expression and subcellular localization seem to be much more complex than originally appreciated, additional interpretations should be considered. For example, attachment of an SV-40 nuclear localization signal may have interfered with appropriate subnuclear localization (eg, in foci). Moreover, experimental evidence suggests that FANCC and FANCA physically interact and that this association is required for the nuclear translocation of FANCA.38,40 Other FA proteins may contribute to the formation of the nuclear FANCA/FANCC complex because cells from certain other FA complementation groups are deficient in this complex.41 42 Our observations suggest that the cell cycle–dependent regulation of FANCC may be an important control point for modulating the assembly or activity of a multiprotein complex of FA and non-FA proteins.
Abundant precedent exists for the posttranscriptional regulation of proteins involved in the control of critical cellular processes. For example, the translation of proopiomelanocortin mRNA is regulated by the interaction of RNA-binding proteins and a stem-loop sequence present in the coding region of proopiomelanocortin mRNA.43 This model is potentially of relevance to our observations of FANCC regulation because the FANCC cDNA used in these experiment contained only the FANCC coding sequence without 5′ or 3′ UTR sequences.
Another possibility for the mechanism of the posttranscriptional regulation of FANCC expression is cellular control of FANCC polypeptide degradation. The proteasome pathway is the best-described mechanism for regulated protein degradation and is involved in the control of many cell cycle-dependent proteins, including some proteins that directly regulate cell cycle progression such as cyclins and cyclin-dependent kinase inhibitors.44,45 Proteasome-dependent degradation of proteins occurs through ubiquitin-dependent (eg, cyclins) and ubiquitin-independent (eg, ornithine decarboxylase) pathways.44,46-48 Interestingly, the expression pattern of FANCC during the cell cycle is similar to that of A- and B-type cyclins. Both cyclin B1 and FANCC are binding partners of cdc2.31,49 50 However, the relationship between FANCC binding to cdc2 and the observed regulation of FANCC expression during cell cycle progression is unknown.
The peptide aldehyde MG132, used in this study, inhibits the proteolytic activities of the proteasome but also inhibits the cysteine proteases calpain and cathepsin B. In contrast, lactacystin is the most selective proteasome inhibitor known, and it inhibits the proteolytic activities of the 20S proteasome without inhibiting other known proteases, including chymotrypsin, trypsin, or papain.51-53Our results with lactacystin strongly suggest a specific role of the 20S proteasome in regulating FANCC expression during the cell cycle.
We do not yet know whether proteasome-dependent proteolysis regulates the translation of FANCC mRNA, controls FANCC degradation, or both. Further studies are needed to discover the precise molecular mechanism(s) by which cell cycle position regulates FANCC expression.
Note added in proof. During the manuscript review process, the cloning and characterization of the FANCF gene was reported by de Winter et al.54
Acknowledgments
The authors thank Michael Moody for his help in preparing the figures and Dr William Skach for his thoughtful review of the manuscript.
Supported by grant HL56045 from the National Institutes of Health (M.E.H.), a grant from the Fanconi Anemia Research Foundation (M.E.H.), and a Merit Review grant from the Department of Veterans Affairs (M.C.H.).
Reprints:Michael Heinrich, R&D-19, Portland Veterans Affairs Medical Center, 3710 SW US Veterans Hospital Road, Portland, OR 97207; e-mail: heinrich@ohsu.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.