The Rh blood group system is one of the most polymorphic and immunogenic systems known in humans. In the past decade, intense investigation has yielded considerable knowledge of the molecular background of this system. The genes encoding 2 distinct Rh proteins that carry C or c together with either E or e antigens, and the D antigen, have been cloned, and the molecular bases of many of the antigens and of the phenotypes have been determined. A related protein, the Rh glycoprotein is essential for assembly of the Rh protein complex in the erythrocyte membrane and for expression of Rh antigens. The purpose of this review is to provide an overview of several aspects of the Rh blood group system, including the confusing terminology, progress in molecular understanding, and how this developing knowledge can be used in the clinical setting. Extensive documentation is provided to enable the interested reader to obtain further information.
The Rh blood group system was first described 60 years ago. A woman had a severe transfusion reaction when she was transfused with blood from her husband following delivery of a stillborn child with erythroblastosis fetalis. Her serum agglutinated red blood cells (RBCs) from her husband and from 80% of Caucasian ABO-compatible donors.1 The following year, Landsteiner and Wiener2 found that sera from rabbits (and later guinea pigs) immunized with RBCs from Macaca mulatta (Macacus rhesus in the original paper) agglutinated 85% of human RBC samples. Initially, it was thought that the animal and human antibodies identified a common factor, Rh, on the surface of rhesus and human RBCs. It was soon realized that this was not the case.3 Therefore, the original terms (Rh factor and anti-Rh) coined by Landsteiner and Wiener, although being misnomers, have continued in common usage. The heteroantibody was renamed anti-LW (after Landsteiner and Wiener), and the human alloantibody was renamed anti-D.4
The Rh blood group system is the most polymorphic of the human blood groups, consisting of at least 45 independent antigens and, next to ABO, is the most clinically significant in transfusion medicine. The ability to clone complementary DNA (cDNA) and sequence genes encoding the Rh proteins has led to an understanding of the molecular bases associated with some of the Rh antigens. Serologic detection of polymorphic blood group antigens and of phenotypes provides a valuable source of appropriate blood samples for study at the molecular level. This review summarizes our present understanding of the complexities of Rh blood group expression and how this knowledge impacts on clinical situations that arise through Rh blood group incompatibility.
Terminology
Several nomenclatures have been used to describe antigens, proteins, and genes in the Rh system. Throughout this review, we will use traditional terminology recommended by the International Society of Blood Transfusion (ISBT) committee for terminology of blood group antigens.5 The numeric portion of the ISBT terminology for Rh antigens is based on the nomenclature described by Rosenfield et al.6-9RH30 and RH50 have been used to describe genes encoding Rh proteins (Rh30) and Rh glycoprotein (Rh50), respectively, where the numbers relate to the apparent molecular mass of the proteins on a SDS-polyacrylamide gel. Because Rh30 and Rh50 also relate to Goa and FPTT antigens, respectively, we will useRH as a generic term for genes encoding either the RhD protein or the RhCcEe (also known as RhCE) protein and use RHAG for the gene encoding the Rh-associated glycoprotein (RhAG). The common Rh antigens: D, C or c, and E or e, were originally written in alphabetical order (CDE) but later, when it was recognized that C and E antigens are inherited en bloc, the order was changed to DCE. Although d antigen, which was thought to be antithetical to D, does not exist, the letter “d” is used to indicate the D-negative phenotype. The most frequently occurring forms of RHCE and RHD encode 8 haplotypes: Dce, dce, DCe, dCe, DcE, dcE, DCE, and dCE, known in short, respectively, as R0, r, R1, r′, R2, r″, Rz, and ry. The uppercase “R” is used when the D antigen is expressed, lowercase “r” when it is not. This notation has practical value in transfusion medicine as a means to communicate the Rh phenotype of a patient or donor. Rare deletion phentoypes use dashes in the notation to indicate a lack of antithetical antigens; eg, Dc−. RBCs lack E and e antigens, and D−− RBCs lack C, c, E, and e antigens. RBCs with the Rhnullphenotype do not express any of the Rh antigens.
The Rh complex
Biochemical studies, protein purification, and amino acid sequencing of Rh and RhAG are beyond the scope of this article but have been reviewed elsewhere.10-16
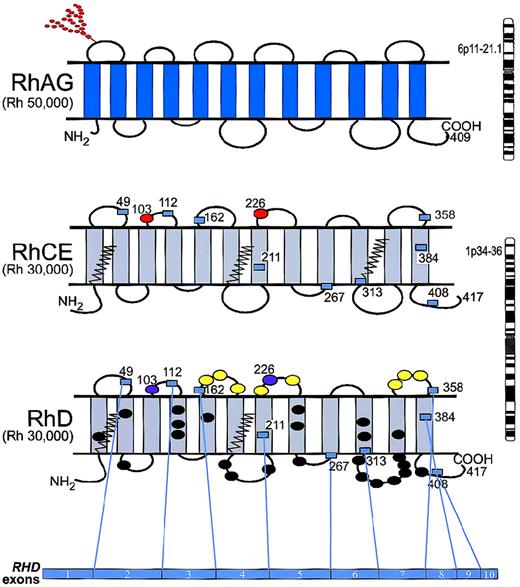
The Rh proteins carry Rh antigens but are only expressed on the erythrocyte surface if RhAG is also present. The amino acid sequence homology (approximately 40%) of the Rh and RhAG proteins indicates an ancestral relationship, and collectively they are referred to as the “Rh protein family.” Hydrophobicity profiles, immunochemical analyses, and data obtained through site-directed mutagenesis imply that Rh and RhAG proteins have 12 transmembrane spans with both the N-terminus and C-terminus oriented to the cytoplasm (Figure1).17-24
Model of topology for RhAG, RhCE, and RhD.
RhAG (Mr 50 000) consists of 409 amino acids and is encoded by RHAG on chromosome 6p11-p21.1. RhCE and RhD (Mr 30 000) are predicted to have a similar topology and are encoded by RHCE and RHD, which are adjacent on chromosome 1p34-p36. The domain of the RhD protein encoded by each exon is depicted by numbered boxes, which represent the start and finish of each exon. Of the D-specific amino acids, 8 are on the exofacial surface (yellow ovals), and 24 are predicted to reside in the transmembrane and cytoplasmic domains (black ovals). Red ovals represent amino acids that are critical for C/c (Ser103Pro) and E/e (Pro226Ala) antigens; purple ovals represent Ser103 and Ala226 on RhD. The zigzag lines represent the Cys-Leu-Pro motifs that are probably involved in the palmitoylation sites. The N-glycan on the first loop of RhAG is indicated by the branched structure of red circles.
Model of topology for RhAG, RhCE, and RhD.
RhAG (Mr 50 000) consists of 409 amino acids and is encoded by RHAG on chromosome 6p11-p21.1. RhCE and RhD (Mr 30 000) are predicted to have a similar topology and are encoded by RHCE and RHD, which are adjacent on chromosome 1p34-p36. The domain of the RhD protein encoded by each exon is depicted by numbered boxes, which represent the start and finish of each exon. Of the D-specific amino acids, 8 are on the exofacial surface (yellow ovals), and 24 are predicted to reside in the transmembrane and cytoplasmic domains (black ovals). Red ovals represent amino acids that are critical for C/c (Ser103Pro) and E/e (Pro226Ala) antigens; purple ovals represent Ser103 and Ala226 on RhD. The zigzag lines represent the Cys-Leu-Pro motifs that are probably involved in the palmitoylation sites. The N-glycan on the first loop of RhAG is indicated by the branched structure of red circles.
“Rh accessory proteins” is a collective term for other glycoproteins that are associated with the Rh protein family as defined by their absence or deficiency from Rhnull RBCs (see below and Table 1).25 Together, the association of the Rh protein family and the Rh accessory proteins is called the “Rh complex.”
Rh protein family
The complex of the Rh protein family is estimated by density ultracentrifugation to be 170 000 daltons26 and to consist of a tetramer with 2 RhAG molecules and 2 RhCcEe or RhD protein molecules stabilized by both N-terminal and C-terminal domain associations.18,19,26 27 The mode of association of this core complex with Rh-accessory proteins, some of which interact directly with the membrane skeleton, remains undefined.
RhD and RhCcEe proteins.
The RhD protein expresses the D antigen, while the RhCcEe protein carries either C or c antigens (involving the second extracellular loop) together with E or e antigens (involving the fourth extracellular loop) on the same protein.19,28-30 Characteristics of the RhD protein (synonyms: Rh30, Rh30B, Rh30D, D30, Rh30 polypeptide [30 kd], RhXIII, Rh13) and of the RhCcEe protein (synonyms: Rh30, Rh30A, Rh30C [RhCE], Rh30 polypeptide [32 kd], RhIXb cDNA, [RhcE], Rh21 cDNA [RhcE], R6A32, Rhce, RhCe, RhcE, RhCE, CcEe) are summarized in Table 1 and depicted in Figure 1. Analysis of the primary amino acid sequences (inferred from cDNAs) shows that the first 41 N-terminal amino acids of RhD and RhCE/e are identical.20,31-33 and that RhD differs from the common forms of RhCE by only 30 to 35 amino acids along the entire protein.20,28,31-33,35 36 Despite the high degree of homology, the various RhCcEe proteins do not express any D epitopes, and RhD protein does not express C or e antigens.
The Rh proteins are thought to interact with the membrane bilayer by palmitoylation,26,37 where acylated palmitic acid residues are attached to cysteine side chains. These cysteine residues are predicted to be at the boundary of the cytosol and lipid bilayer (Figure 1). Cys-Leu-Pro motifs, flanked by charged amino acids (2 are on RhD and 3 are on RhCcEe) are likely candidates although 2 other cysteine residues (315 and 316) may be alternative sites.20,26 This interaction may explain why alteration of membrane cholesterol concentration affects the accessibility of the D antigen.38 The ability to label Rh proteins with3H-palmitate26 37 indicates that a reversible coenzyme A and adenosine triphosphate (ATP)-dependent acylation-deacylation cycle occurs in mature RBC membranes, which is of unknown significance.
Rh-associated glycoprotein.
The characteristics of RhAG (synonyms: Rh50, Rh glycoprotein Rh50A, D50, MB-2D10 protein, R6A45, GP50, GP50A) are summarized in Table 1 and depicted in Figure 1. One of the 2 potential N-glycan sites is glycosylated. A third site is predicted to be cytoplasmic and, therefore, not accessible for glycosylation.17,39 The N-glycan carries ABH antigens,12 but RhAG is not known to possess a protein-based blood group polymorphism. Based on the predicted amino acid sequence, RhAG shares 39.2% and 38.5% amino acid sequence identity with, respectively, the Rhce and RhD proteins.17,20 31-33
Expression of Rh proteins and RhAG during erythropoiesis.
Rh antigens appear early during erythropoietic differentiation. Anti-D binds to approximately 3% of BFU-E (burst-forming unit, erythroid), 68% of CFU-E (colony-forming unit, erythrocyte), and to all of the more mature erythroid cells. However, the binding of anti-D to proerythroblasts, basophilic erythroblasts, polychromatophilic erythroblasts, and normoblasts was, respectively, 25%, 50%, 66%, and 75% compared with mature RBCs.33 RhAG protein is detectable on CD34 progenitors isolated from cord blood, after culture for 3 to 5 days, while RhCcEe appears after 5 to 7 days, and RhD appears after 9 to 11 days of culture.40 In the fetus, Rh antigens are expressed on RBCs from the 6-week conceptus.41
Possible function of Rh protein family.
The function of the Rh complex remains unclear. Rh proteins have approximately 20% homology to the methylamine permease (Mep) transporters and ammonium transporters (Amt) in yeast, bacteria, and simple plants.42 This family of transporters are uniporters that have evolved to concentrate ammonium salts from the surrounding environment. Higher animals use more complex nitrogen sources, and they eliminate toxic ammonium via the urea cycle and transport it in the form of glutamine and alanine. The role of the Rh complex as a dedicated ammonium transporter is unlikely, but the complex could cotransport ammonium with other cations; however, further study is needed. Matassi et al43 report that RHAG shares greatest homology to MEP2, which behaves as an ammonium sensor and transporter in yeast.44 Furthermore, the presence of RhAG homologs in Caenorhabditis elegans and Geodia cydonium infers they have roles that are not confined to RBCs.
Rh accessory proteins
The blood group antigens associated with the Rh family of proteins, the gene location, their molecular mass, number of copies per RBC, and selected accession numbers are summarized in Table1.
LW glycoprotein.
The LW glycoprotein (synonym: ICAM-4) is a single pass (type I) membrane protein with homology to intercellular adhesion molecules (ICAMs), which are ligands for β2 integrins. LW has been reported to be a ligand for the integrin LFA-1 (synonyms: αLβ2, CD11a/CD18).45
While the LW glycoprotein is absent from RBCs of LW(a−b−) and Rhnull individuals, expression of Rh antigens is normal on LW(a−b−) RBCs. LW antigens are more abundant on D-positive RBCs than on D-negative RBCs from adults, which led to the initial interpretation that anti-D and anti-LW were the same.46,47 It is possible that the LW glycoprotein interacts preferentially with RhD as compared with RhCcEe; however, the nature of such an interaction awaits definition. Interestingly, LW antigens are expressed equally well on D-positive and D-negative RBCs from fetuses and newborns and more strongly than on RBCs from adults.48 49
Integrin-associated protein.
Isoform 2 of integrin-associated protein (IAP; synonyms: CD47, BRIC 125 glycoprotein, AgOAB, 1D8) is present in the RBC membrane, where it is predicted to pass through the RBC membrane 5 times and have 6 potential N-glycan motifs.50,51 IAP carries ABH antigens but no known protein-based blood group antigen. IAP occurs as different isoforms in various tissues where it binds to β3 integrins.50,52 The IAP isoform in RBCs does not bind integrins but does bind thrombospondin53 and may be involved in calcium transport, possibly as a gated channel.54 While the amount of IAP is reduced in RBC membranes from Rhnull and D−− people, it is present in normal levels in lymphoblastoid cell lines from these people.55-57
Glycophorin B.
Glycophorin B (GPB; synonyms: Ss sialoglycoprotein [SGP], δ-SGP, PAS-3) is a type I membrane glycoprotein that has several O-glycans but no N-glycan. The Rh complex appears to aid, but is not essential for, the correct insertion of GPB in RBC membranes. In S−s−U− RBCs that lack GPB, the Rh proteins are apparently normal, but RhAG has increased glycosylation, suggesting a slower migration through the endoplasmic reticulum and Golgi apparatus.39 An interaction of GPB and RhAG may be required for full expression of the U antigen58 59 and, to a lesser extent, S and s antigens (Table 2). Further, the known ability of GPB to form heterodimers with glycophorin A (GPA) may bridge the Rh complex with the band 3/GPA complex, forming a large unit in the RBC membrane.
Fy glycoprotein.
A possible association between the Fy glycoprotein (synonyms: Duffy, DARC) and the Rh complex is indicated by the Fy5 antigen, which is absent from Fy(a−b−) and RhnullRBCs.60 However, Rhnull RBCs have normal Fya, Fyb, Fy3, and Fy6 antigens, and Fy(a−b−) RBCs have normal Rh antigens. The specific requirements for expression of the Fy5 antigen remain unknown.
Band 3.
Band 3 (synonyms: AE1, anion exchanger, solute carrier family 4 anion exchanger member 1) is a glycosylated protein that is predicted to pass through the RBC membrane 12 or 14 times and is the major anion transporter.61,62 Unlike the proteins described above, band 3 is apparently normal in Rhnull RBCs; however, based on hemagglutination studies, antigens on Rh proteins and on band 3 are decreased in South-East Asian ovalocytic RBCs.63 The molecular defect associated with South-East Asian ovalocytic RBCs results from a deletion of a segment of DNA encoding 9 amino acids located at the boundary of the cytoplasmic N-terminal domain and membrane domain of band 3.64-67 Recent evidence that the expression of endogenous and retrovirally expressed Rh antigens were enhanced following transduction of K562 cells with band 3 suggests that band 3 and Rh proteins associate in erythroid cells.68
Structure of RH and RHAG genes
The genes encoding RhD and RhCcEe are highly homologous, while the gene encoding RhAG is almost 40% homologous. The 3 genes are each composed of 10 exons; RHCE and RHD in tandem encompass 69 kilobases (kb) of DNA (Figure 2), whileRHAG encompasses 32 kb. The RhD protein is encoded byRHD (synonyms: RH30, RH30B, RH30D, RHXIII, RH13); the RhCcEe protein is encoded by RHCE (synonyms: RH30, RH30A, RH30C (RHCE),RHIXB, RH21); and the RHAG glycoprotein is encoded by RHAG (synonyms: RH50, RH50A).
RHCE-RHD gene organization.
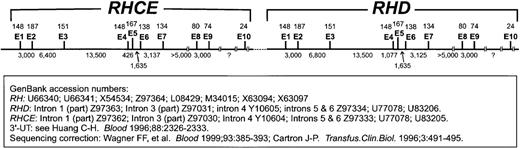
Organization of exons (E) and introns of RHCE and RHDis shown. Exon sizes are indicated above the line as number of nucleotides, and intron sizes are indicated below the line. A c-specific short tandem repeat (STR) is located in intron 2 ofRHCE and another in intron 6 of RHD. The information used to compile this figure came from the database accession numbers given in the figure.
RHCE-RHD gene organization.
Organization of exons (E) and introns of RHCE and RHDis shown. Exon sizes are indicated above the line as number of nucleotides, and intron sizes are indicated below the line. A c-specific short tandem repeat (STR) is located in intron 2 ofRHCE and another in intron 6 of RHD. The information used to compile this figure came from the database accession numbers given in the figure.
The intron-exon boundaries of the RHCE gene21 and the complete nucleotide sequences of some RHCE and RHDintrons have been described.69-74 Selected GenBank accession numbers for cDNA are listed in Table 1, and those for introns are given in Figure 2. The intron-exon structure of the RHAGgene also has been defined and is remarkably similar to RHCEand RHD.22 75-77 Several mutations in RHAGhave been described that cause the regulator type of Rh deficiency syndrome (see below).
Evolution of the RH gene family
It was thought that Rh proteins were erythroid-specific and confined to higher vertebrates. However, the discovery of sequence-relatedRHAG homologs in invertebrates suggests otherwise. These homologs have been found as 2 different RHAG-like genes inCaenorhabditis elegans (a nematode; GenBank accession U64 847 and Z74 026)78 and as at least 1 in Geodia cydonium (a marine sponge; GenBank accession Y12 397).79 These genes are predicted to encode proteins with remarkably high (respectively, 46%, 39%, and 47%) amino acid identity to human RhAG. The highest homology is within the transmembrane domains, suggesting a conserved functional role for the RhAG protein family. Recent work has also demonstrated the presence ofRHAG counterparts in mouse (GenBank accession AF065 395; AF057 524-27, AF012 430), macaque (AF058 917) and RHorthologs in chimpanzee (L37 048-50), gorilla (L37 052, L37 053), orangutan (AF012 425), gibbon (L37 051), baboon (AF012 426), macaque (L37 054 570 343), New World monkeys (AF012 427-9, AF021 845) and cow (U59 270).77,80 81
As the invertebrate homologs more strongly resemble human RHAGthan human RH, it is likely that an ancient gene duplication event, estimated to have occurred 250 million to 346 million years ago, caused divergence of RH from RHAG.75Subsequent to the gene duplication, RH and RHAGunderwent different evolutionary pathways.43 A second gene duplication event, being the origin of the human RHCE andRHD genes, occurred much later in a primate ancestor 5 million to 12 million years ago. Based on the evolutionary rates ofRHAG and RH genes in different species, it appears thatRHAG evolved some 2.6 times slower than RH,suggesting that RhAG has a more important functional role than Rh proteins.22,73,77 82
The order of the Rh genes on chromosome 1 is probablyRHCE-RHD.83 (After submission of this manuscript, a paper was published that questions the order of the Rh genes on chromosome 1. Sequencing the intergenic region of the two RH genes suggests that the order may in fact beRHD-RHCE.211) The primordial human Rh haplotype is believed to be Dce, and the other 7 common Rh haplotypes most likely each arose from this gene complex by a single genetic event. The predominant Caucasian D-negative haplotype (dce) probably arose by a deletion of the RHD gene84 from theRHce/RHD gene complex, whereas the DCe haplotype (the most common D-positive haplotype in Caucasians) arose by gene conversion with exon 1 and 2 from RHD replacing the same exons of RHce. The remaining haplotypes arose through point mutations (eg, the E/e polymorphism) or rare recombination events of the various haplotypes.83
Molecular basis of Rh antigens
Since the first descriptions of Rh cDNAs,20,31-33 much effort has been expended in differentiating the molecular bases underlying the antigens of the Rh system. The different genetic mechanisms that give rise to the major clinically relevant Rh antigens are described within this section. These include gene deletion (D-negative phenotype); gene conversion (C/c polymorphism); antithetical missense mutations (E/e); and other missense mutations (VS and V). The RH genes appear to be a source of massive diversity, and combinations of these different genetic rearrangements abound among all racial groups. We have selected examples of Rh polymorphisms that are of clinical significance and have been defined at the molecular level. Figures 3-6 detail the molecular basis of published examples of Rh variants. Enthusiastic readers requiring more data regarding Rh variants should consult references.16,25,85 86
D antigen
The D antigen is a collection of conformation-dependent epitopes along the entire RhD protein. While in most D-negative Caucasians there is a deletion of RHD, in other populations (notably Japanese and African blacks) the D-negative phenotype is associated with a grossly normal RHD, and the reason for the lack of expression of the D antigen is not known (except in Africans; see later). Figure 3 depicts the molecular basis of some D-negative phenotypes.70,74,84 87-89
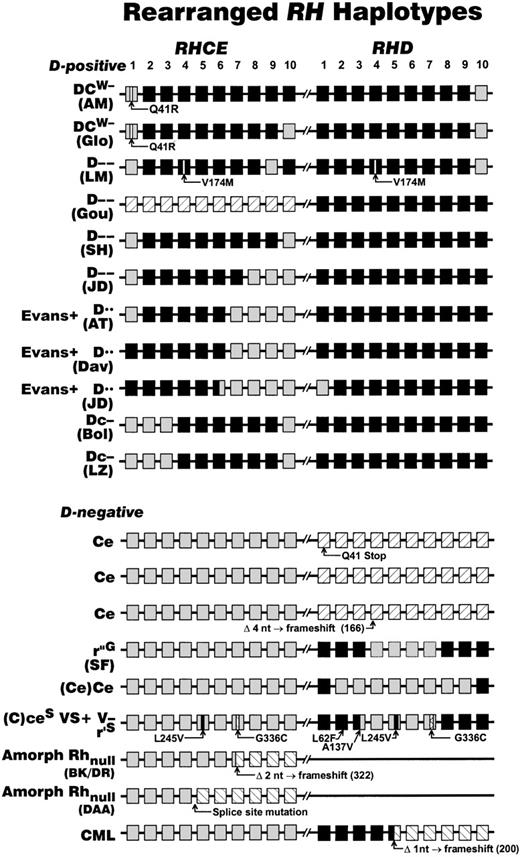
Rearrangements at the Rh locus giving rise to D-negative and Rh deletion haplotypes.
The structures of the RH locus (located at 1p34-36) that has been defined in various D-negative phenotypes and rare Rh antigen deletion phenotypes are depicted. Each RH gene is represented as 10 boxes, each box representing an exon, where RHCE is shown as gray, RHD as black. Crosshatched boxes depict silent RHDalleles (eg, RHD Q41 X ). The positions of microinsertions or deletions of DNA that cause or are indicative of D-negative phenotypes are shown as triangles. Because exon 8 ofRHCE and RHD are of identical sequence and their origins are not possible to define, they are shaded according to the gene loci position. The significance of these rearrangements, and their impact in particular on molecular genotyping, is discussed within the text. Sources for the information in this figure: DCW − (AM)184; DCW − (Glo)185; D−− (LM)186; D−− (Gou)186; D−− (SH)72; D−− and Evans+ D•• (JD)187; Evans+ D•• (AT)69; Evans+ D•• (Dav)186; Dc− (Bol)186; Dc− (LZ)188; Ce70,74,149; r”G(SF)105,140; (Ce)Ce184; (C)ceS VS+ (Donor 1077) V−r'S 29;98;104; Amorph Rhnull (BK/DR)176,189; Amorph Rhnull (DAA)177; CML.179
Rearrangements at the Rh locus giving rise to D-negative and Rh deletion haplotypes.
The structures of the RH locus (located at 1p34-36) that has been defined in various D-negative phenotypes and rare Rh antigen deletion phenotypes are depicted. Each RH gene is represented as 10 boxes, each box representing an exon, where RHCE is shown as gray, RHD as black. Crosshatched boxes depict silent RHDalleles (eg, RHD Q41 X ). The positions of microinsertions or deletions of DNA that cause or are indicative of D-negative phenotypes are shown as triangles. Because exon 8 ofRHCE and RHD are of identical sequence and their origins are not possible to define, they are shaded according to the gene loci position. The significance of these rearrangements, and their impact in particular on molecular genotyping, is discussed within the text. Sources for the information in this figure: DCW − (AM)184; DCW − (Glo)185; D−− (LM)186; D−− (Gou)186; D−− (SH)72; D−− and Evans+ D•• (JD)187; Evans+ D•• (AT)69; Evans+ D•• (Dav)186; Dc− (Bol)186; Dc− (LZ)188; Ce70,74,149; r”G(SF)105,140; (Ce)Ce184; (C)ceS VS+ (Donor 1077) V−r'S 29;98;104; Amorph Rhnull (BK/DR)176,189; Amorph Rhnull (DAA)177; CML.179
People whose RBCs have an altered form of RhD protein (partial D) may make alloanti-D. Such RBCs, depending on which D epitopes are altered, are agglutinated by a proportion of anti-D reagents. Figure4 summarizes the molecular changes that are associated with partial D antigens.
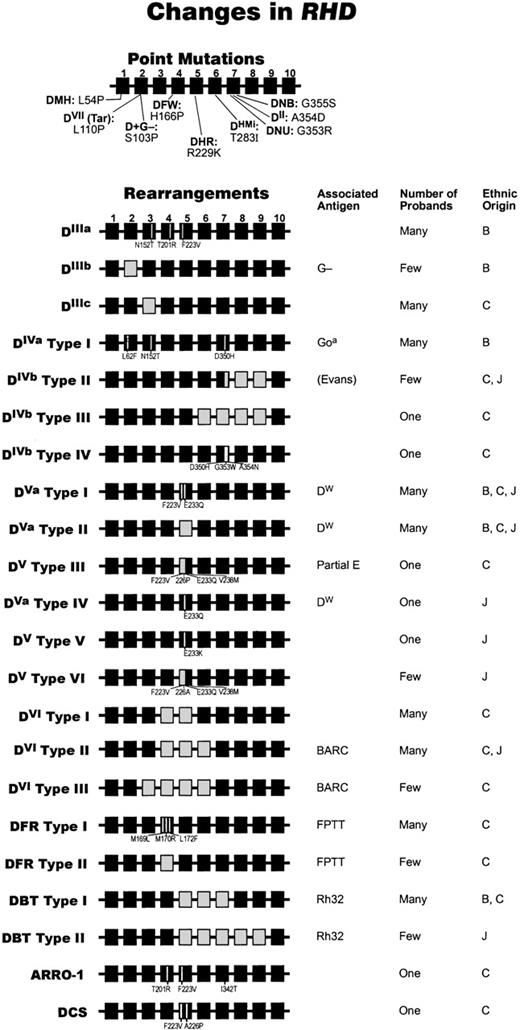
Molecular bases of partial D phenotypes.
The different alleles of RHD that cause partial D phenotypes are depicted here graphically. The genetic structure of each partial DRHD 10-exon gene is shown, as are associated low-incidence antigen(s) and the estimated gene frequency. RHD (ie, wild type) exons are shown as black boxes; where they have been replaced byRHCE equivalents is shown as white boxes. Missense mutations are indicated within the exon where they occur. We have used the original Roman numeral notation (ie, DII to DVII) and the more recent 3-letter notation (eg, DFR, DBT) for the different D categories. Where partial D phenotypes have identical (or very similar) serologic profiles but different genetic backgrounds, we have adapted the classification originally described by Mouro et al190 to describe different DVIphenotypes (types I and II). Thus, we depict DIV types I to IV, DV types I to VI; DVI types I to III, and DFR types I and II. We use DVa to indicate the presence of the DW antigen and DV to represent samples that have a similar molecular background but that either do not express the DW antigen or have not been tested for this antigen.Few = 1 to 10 examples. Many = 11 or more examples as indicated by serological testing. DVII is common (1 in 900) in the German population.191 Under “Ethnic Origin,” B = black, C = Caucasian, and J = Japanese. The information used for the point mutations used in this figure are as follows: D+G−106; DNU and DII192; DHMi92; DVII193; DVa71,194 DFW195; DHR.196 The information used for the rearrangements in this figure was obtained from the following: DIIIa197; DIIIb106; DIIIc 198; DIVa type I 194; DIVbtype II194; DIVb type III92; DIVb type IV195; DVa type I194; DVa type II194; DV type III102; DVa type IV156; DV type V156; DVtype VI156; DVI type I199,200; DVI type II190; DVI type III71; DFR type I194; DFR type II201; DBT type I202; DBT type II203; ARRO-1204; DCS205.
Molecular bases of partial D phenotypes.
The different alleles of RHD that cause partial D phenotypes are depicted here graphically. The genetic structure of each partial DRHD 10-exon gene is shown, as are associated low-incidence antigen(s) and the estimated gene frequency. RHD (ie, wild type) exons are shown as black boxes; where they have been replaced byRHCE equivalents is shown as white boxes. Missense mutations are indicated within the exon where they occur. We have used the original Roman numeral notation (ie, DII to DVII) and the more recent 3-letter notation (eg, DFR, DBT) for the different D categories. Where partial D phenotypes have identical (or very similar) serologic profiles but different genetic backgrounds, we have adapted the classification originally described by Mouro et al190 to describe different DVIphenotypes (types I and II). Thus, we depict DIV types I to IV, DV types I to VI; DVI types I to III, and DFR types I and II. We use DVa to indicate the presence of the DW antigen and DV to represent samples that have a similar molecular background but that either do not express the DW antigen or have not been tested for this antigen.Few = 1 to 10 examples. Many = 11 or more examples as indicated by serological testing. DVII is common (1 in 900) in the German population.191 Under “Ethnic Origin,” B = black, C = Caucasian, and J = Japanese. The information used for the point mutations used in this figure are as follows: D+G−106; DNU and DII192; DHMi92; DVII193; DVa71,194 DFW195; DHR.196 The information used for the rearrangements in this figure was obtained from the following: DIIIa197; DIIIb106; DIIIc 198; DIVa type I 194; DIVbtype II194; DIVb type III92; DIVb type IV195; DVa type I194; DVa type II194; DV type III102; DVa type IV156; DV type V156; DVtype VI156; DVI type I199,200; DVI type II190; DVI type III71; DFR type I194; DFR type II201; DBT type I202; DBT type II203; ARRO-1204; DCS205.
Analysis of genes encoding the weak D phenotype (previously known as DU) showed a normal RHD sequence but a severely reduced expression of RHD messenger RNA (mRNA), suggesting a defect at the level of transcription or pre-mRNA processing.70,90 More recently, RHD transcripts from people whose RBCs express a weak form of the D antigen were found to have missense mutation(s) within the predicted transmembrane or cytoplasmic domains of RhD (Figure5).91 92 RBCs with some weak D antigens may not be agglutinated by all monoclonal anti-D. People whose RBCs express this type of weak D antigen do not make anti-D.
Molecular basis of weak D phenotypes.
This figure depicts missense mutations in the RHD gene associated with weak D phenotypes.92 153 The locations of these mutations on the predicted topology of the RhD protein are depicted as checkered ovals; the D-specific amino acids are shown as open ovals. Most of the missense mutations are located within nonconserved membrane spans (gray) and cytoplasmic regions. Regions of conserved Rh protein family sequence are indicated as black rectangles.
Molecular basis of weak D phenotypes.
This figure depicts missense mutations in the RHD gene associated with weak D phenotypes.92 153 The locations of these mutations on the predicted topology of the RhD protein are depicted as checkered ovals; the D-specific amino acids are shown as open ovals. Most of the missense mutations are located within nonconserved membrane spans (gray) and cytoplasmic regions. Regions of conserved Rh protein family sequence are indicated as black rectangles.
CcEe antigens
The RhC/c and RhE/e polymorphisms are caused by nucleotide substitutions in RHCE.28,93 While 6 nucleotide substitutions causing 4 amino acid changes (Cys16Trp; Ile60Leu; Ser68Asn; Ser103Pro) are associated with the C to c polymorphism (Figure 6), only the Ser103Pro polymorphism strictly correlates with C/c antigenicity.94 However, Pro102 appears to be a critical part of the c antigen.95,96The presence of 2 adjacent proline residues (102 and 103) would be expected to form a relatively rigid structure that is resistant to changes in nearby amino acid residues and may explain the relatively low number of c variants as compared with other Rh antigens. It has been generally accepted that a single nucleotide substitution is sufficient for expression of the E to e polymorphism (Pro226Ala). However, variants of the e antigen have been described,97showing that the requirements for expression of the e antigen are not fully understood. For example, the presence of Val at residue 245 instead of Leu,29,98,99 a deletion of Arg at amino acid residue 229,100 or the presence of Cys (instead of Trp) at amino acid residue 16 101 affects the expression of the e antigen. The molecular basis of partial E antigens (categories I, II, and III, and DV type III) has been determined and are shown in Figures 4 and 6.102 103
Changes in RHCE.
Amino acids encoded by RHCE are shown by gray boxes, and those encoded by exons from RHD are shown by black boxes. The amino acids associated with E/e and C/c antigens28,93 are shown at the top, and single amino acid changes associated with variant forms of RhCE are shown in the middle. The bottom portion of the figure shows rearrangements of the RHCE and associated antigens. Polymorphism that does not have a 100% correlation with expression of c and C antigens. The information depicted in this figure was obtained from the following sources: Point Mutations35,98,99,104,185,206; Rearrangements DHAR 207; rG208;N90; E Cat II and III185; and Variant e100 101.
Changes in RHCE.
Amino acids encoded by RHCE are shown by gray boxes, and those encoded by exons from RHD are shown by black boxes. The amino acids associated with E/e and C/c antigens28,93 are shown at the top, and single amino acid changes associated with variant forms of RhCE are shown in the middle. The bottom portion of the figure shows rearrangements of the RHCE and associated antigens. Polymorphism that does not have a 100% correlation with expression of c and C antigens. The information depicted in this figure was obtained from the following sources: Point Mutations35,98,99,104,185,206; Rearrangements DHAR 207; rG208;N90; E Cat II and III185; and Variant e100 101.
VS and V antigens
The imultaneous presence of 2 low-incidence antigens (VS and V) occurs with a single amino acid substitution (Leu245Val) that is predicted to be within a transmembrane domain (Figure6).104 The V antigen (in the presence of VS) is not expressed when another transmembrane amino acid substitution is present at residue 336 (Gly→Cys) (Figure 3).98 104 The membrane location of residues 245 and 336 illustrate that Rh antigen expression is affected significantly by nonexofacial amino acids and suggests that the prediction of some Rh epitope expression cannot be based solely on externalized residues.
G antigen
RhD and RhC proteins carry the G antigen, which is associated with residues in the second extracellular loop encoded by exon 2.105,106 In DVIcE (DVI type I) RBCs, which are predicted to have a hybrid RhD (exons 1-3)–RHCE (exons 4 and 5)–RhD (exons 6-10) protein, the G antigen was not detected by 1 of 2 monoclonal anti-G.107 Thus, it would appear that the G antigen is conformation-dependent and not solely dependent on the second external domain of RhC(e/E) or RhD proteins.
Rh variants
Rh-variant phenotypes arise through at least 4 mechanisms: (1) rearrangements of the tandemly arrangedRHCE and/or RHD (Figures 3, 4, and 6); (2) point mutation(s) in either gene causing amino acid change(s), with subsequent loss of some epitopes and/or expression of a low-incidence antigen; (3) nonsense mutations, and (4) deletion of nucleotides causing a frameshift and premature stop codon. There is some evidence that there are recombination hot spots due to Alu IV elements in the RH genes.72 108
Rearranged RHCE genes, associated with D−− and D••, ablate expression of C, c, E, and e antigens, while the D antigen expression is exalted to the extent that immunoglobulin (Ig) G anti-D can agglutinate the RBCs in saline.109 It is now clear that this increased expression is due to a large insert ofRHD into RHCE in tandem with a RHD gene (Figure3). In DCW− and Dc− phenotypes, the region of the RHCE gene encoding the E/e antigen is replaced by an RhD equivalent with loss of E/e antigenicity (Figure 3). While these appear as RHCE deletion phenotypes at the protein level, they are encoded by rearranged RHCE and thus are RHCE-depleted.
Low-incidence antigens associated with partial D antigens.
Low-incidence antigens associated with some partial D phenotypes are due to novel structures on the RBC surface and are useful markers for the identification of the partial D (Figure 4).110 A few low-incidence antigens are associated with more than 1 molecular background, eg, the FPTT (Rh50) antigen is expressed on DFR, RoHar, and DIVa(C)− phenotype RBCs; the Rh32 antigen is expressed on DBT andThe Evans antigen is expressed on D••, and a weak form of Evans is present on DIVb RBCs. RBCs expressing Rh23 or Rh32 possess an antigen (Rh23/32) present on both phenotypes.111In these cases, it is likely that external surfaces of the altered proteins have localized similarities.
RhD epitope mapping
Partial D antigens were classically identified by testing the RBCs with well-characterized polyclonal anti-D made by other people with partial D phenotypes and, also, by testing the patient's anti-D against RBCs with known partial D antigens. Human monoclonal antibodies are now being used to classify partial D antigens in terms of expressed epitopes. The original model consisted of 8- and 9-epitope D (epD)112,113 but has been expanded to consist of 16,110 30,114 and 37 epitopes.115 When using monoclonal anti-D to define D epitopes, it is important to perform the testing at the correct pH, temperature, ionic strength, and antibody concentration; to use RBCs that have been stored appropriately; and to include controls.110,114 Most D epitopes are conformation-dependent and may be influenced by other proteins and lipids in the RBC membrane. Indeed, only 1 monoclonal anti-D has been described that reacts strongly by immunoblotting, implying that the epitope it recognizes may be linear.116
Predictions as to the location of various D epitopes have been based on which epitopes are absent from RBCs with a partial D for which the molecular basis is known.117,118 However, the absence of a D epitope may not always be a direct result of the change in molecular structure, and the presence of Rh proteins encoded by cis andtrans genes can effect the binding of certain monoclonal anti-D. For example, R0Har and DVado not have any RHD exons in common, but they have overlapping reactivity with monoclonal antibody anti-D, demonstrating the difficulty of correctly defining the molecular basis of D epitopes. A model proposed by Chang and Siegel119 suggests that anti-D are essentially similar in that they react with the basic footprint of the D protein. In this model, a change in the footprint, induced by an amino acid substitution or a hybrid protein, is predicted to interfere with binding of anti-D. The involvement of certain residues for binding of monoclonal anti-D has been investigated by site-directed mutagenesis (SDM), which showed that incorporation of 3 D-specific amino acids (Asp350, Gly353, and Ala354) into an RhcE construct generated some epD3 and epD9 expression,120,121 and incorporation of 9 exofacial D residues generated epitopes that were recognized by 40 of 50 monoclonal anti-D.122 These data argue that at least some D epitopes are spatially distinct. However, SDM studies have not yet addressed the impact of amino acids located within the lipid bilayer or on the cytoplasmic side of the RBC membrane. Accurate determination of the contact points of interaction(s) between antigen and antibody awaits crystallographic data.
Clinical aspects
Clinical complications result from RBC destruction due to the interaction of an alloantibody with RBCs carrying the corresponding antigen. The D antigen is highly immunogenic and induces an immune response in 80% of D-negative persons when transfused with 200 mL of D-positive blood.123 For this reason, in most countries D typing is performed routinely on every blood donor and transfusion recipient so that D-negative patients receive D-negative RBC products. Consequently, clinical complications due to mismatched transfusions are infrequent. In contrast, despite the use of immunosuppressive therapy with anti-D immunoglobulin prophylaxis, D alloimmunization in pregnancy still occurs.
Alloantibodies
Alloantibodies that recognize Rh antigens are usually IgG and react by the indirect antiglobulin test. This is a test in which RBCs are incubated in serum, washed to remove free immunoglobulin, and then exposed to an antiglobulin reagent that is formulated to detect the cell-bound IgG. The end point of the test is hemagglutination. Alloantibodies in the Rh blood group system can cause destruction of transfused RBCs and of fetal RBCs in hemolytic disease of the newborn (HDN). People whose RBCs have a rare deleted Rh phenotype (Rhnull, D−−) readily make alloantibodies. People with the Rhnull phenotype of amorph or regulator type can make anti-Rh29 (an antibody to “total” Rh), anti-Rh17 (an antibody to the RhCc/Ee protein), anti-D, anti-C, or a mixture of specificities. Transfusion of a patient with anti-Rh29 is a problem because only Rhnull RBCs will be compatible: People with the Rhnull phenotype are not only rare, but they have a compensated hemolytic anemia and are therefore unlikely to meet predonation criteria.124 People with either the D−−, D••, DCW−, or Dc− phenotype make anti-Rh17. A patient with anti-Rh17 also represents a transfusion conundrum because only RBCs with a deleted phenotype will be compatible.
Autoantibodies
An autoantibody is one that reacts with an antigen on the antibody maker's own RBCs. Autoantibodies that react optimally at 37°C are present in the serum of about 80% of patients with warm autoimmune hemolytic anemia.125 Although most of these autoantibodies appear to be “nonspecific,” many have specificity to an Rh antigen, notably to e. Rarely is the specificity clear-cut, but the autoantibody commonly reacts more weakly with antigen-negative RBCs than with antigen-positive RBCs; however, in these cases, transfused antigen-negative RBCs only rarely survive better than antigen-positive RBCs.123 Autoantibodies in serum from patients with warm autoimmune hemolytic anemia may be nonreactive only with Rhnull and D−− RBCs (autoanti-Rh17), or only with Rhnull RBCs (autoanti-Rh29). In such cases, antigen-negative blood will not be available, and transfusion with antigen-positive RBCs should not be withheld if the patient has life-threatening anemia.125,126 In most cases, the autoantibody is equally reactive with all RBCs tested—whether from donors or antibody detection/identification kits. Thus, in the clinical setting, it is important to perform tests to ensure that the patient's serum does not have potentially clinically significant alloantibodies underlying the autoantibodies before transfusing incompatible RBCs. Detection and identification of such antibodies is required to prevent transfusion reactions but is beyond the scope of this review. For more information, see a current textbook on laboratory aspects of transfusion medicine.125-127
Partial and weak D phenotypes
As described earlier, people whose RBCs have a weak D phenotype (quantitative D variant) do not make anti-D, whereas people whose RBCs have a partial D phenotype (qualitative D variant with or without weakening of the D antigen) can make alloanti-D. This presents a different problem depending on whether the person is a donor or a patient. For donors, detection of weak and partial D antigens would eliminate the possibility of immunization should such blood be transfused to a true D-negative patient. However, historical data show that weakly expressed D antigens are most unlikely to be immunogenic. For transfusion recipients and pregnant women, it is common practice to use a procedure that will classify RBCs with a weak D antigen or some partial D antigens as D-negative. Thus, blood donated from such a person should be labeled as D-positive (Rh-positive), but the same person should be listed as D-negative (Rh-negative) when they are recipients in need of transfusion. The transfusion recipient will receive D-negative RBC products, and the pregnant woman will receive prophylactic Rh immunoglobulin, thereby preventing alloimmunization. Although a pregnant woman with the DVI partial phenotype may make alloanti-D, this has rarely caused a clinical problem to a D-positive fetus.128 In the autologous transfusion setting (in which the person is both the donor and patient), the above policy can cause confusion because partial D RBCs may be typed as D-positive at the donor center but D-negative at the hospital. In practice, it is difficult to distinguish RBCs with the DVI phenotype from other weak D; however, this now can be accomplished by immunoblotting with the unique anti-D, LOR-15C9.129
Rh and hemolytic disease of the newborn
HDN is caused by maternal IgG antibody crossing the placenta, binding to the fetal antigen-positive RBCs, and initiating their destruction, thereby causing anemia. Prior to the use of prophylactic Rh immunoglobulin, anti-D frequently caused fetal brain damage due to increased levels of bilirubin (kernicterus) and even death (erythroblastosis fetalis). Despite the widespread use of prophylactic Rh immunoglobulin, a significant number of women still become alloimmunized during pregnancy for a variety of reasons, including nonadministration of Rh immunoglobulin, unrecognized miscarriage, leakage of fetal RBCs into the maternal circulation late in pregnancy, and exposure to maternal D-positive RBCs while in utero (grandmother effect).130
The D antigen accounts for about 50% of cases of maternal alloimmunization; the remainder is due mainly to incompatibility to K, c, C/G, E, and Fya antigens and to low incidence antigens in Rh, MNS, and Diego blood group systems.131-133Therefore, feto-maternal Rh incompatibility still represents the major cause of HDN. Ultraviolet phototherapy and, occasionally, exchange transfusion or even intrauterine transfusion may be required. Invasive procedures are used as a “last option” in monitoring and treating HDN, because they may cause further leakage of fetal RBCs into the maternal circulation. Measures such as determination of the optical density of amniotic fluid and functional assays (ADCC, MMA, chemiluminescence) have been used to monitor at-risk pregnancies and to identify cases requiring treatment (for review, see Zupanska134). With current molecular technology, it is possible to perform analyses on fetally derived DNA to predict the blood type of a fetus.
Interestingly, a fetus that is ABO incompatible with the maternal anti-A/B is less likely to have HDN due to anti-D, presumably due to rapid removal of the ABO-incompatible RBCs by the naturally occurring anti-A/B. Also, because the number of copies of the D antigen per RBC is higher in the R2 haplotype (range, 14 000 to 16 000) than in the R1 haplotype (range, 9000 to 14 600), fetuses whose RBCs are R2 have more severe anemia than their R1counterparts.123 There is also evidence that male fetuses have more severe HDN than female fetuses.135
Rh immunoglobulin prophylaxis in the prevention of HDN.
The immunologic mechanism responsible for preventing production of maternal anti-D following administration of prophylactic Rh immunoglobulin may be due, at least in part, to antigen blocking and central inhibition of the immune response by negative feedback in the spleen (for review, see Bowman130). In some instances, recommendations have been made to administer anti-D to partial D-phenotype mothers (eg, DVI and DBT phenotypes) following the birth of D-positive babies.110 136 In Europe, anti-D reagents are selected to deliberately type DVI mothers as D-negative and, thus, ensure that such mothers would automatically receive prophylactic Rh immunoglobulin therapy following pregnancy.
Prophylactic Rh immunoglobulin preparations for this purpose are usually for intramuscular injection. However, products approved also for intravascular injection are used for the treatment of idiopathic thrombocytopenia.137 138
Legislative restrictions for immunization of D-negative volunteers with accredited D-positive RBCs are partly responsible for the declining source of polyclonal anti-D for prophylaxis. Thus, clinical trials have explored the possibility of using human monoclonal anti-D to prevent anti-D alloimmunization;139 however, the in vivo use of monoclonal antibodies derived from EBV-transformed cells remains controversial. It is possible that recombinant forms of anti-D can be prepared as an injectable prophylactic product.
Prenatal Rh genotyping
When a pregnant woman has a potentially clinically significant alloantibody and the father of the fetus is phenotypically heterozygous for the gene encoding the corresponding antigen (or is unknown), prenatal determination can be considered. The potential benefits of identifying a fetus whose RBCs are predicted to be antigen-negative is enormous in that the need for further invasive techniques is diminished. Fetal DNA can be obtained from amniocytes, chorionic villi, vaginal swabs, and mother's blood (see later). Following cloning and sequencing of RHCE and RHD, many polymerase chain reaction (PCR)-based tests to analyze DNA prepared from amniocytes have been reported (for recent review, see Flegel86). However, the genetic diversity of the Rh genes, particularly among blacks and Japanese, has reduced the clinical utility of this approach because false-negative and false-positive results can occur. Prenatal diagnosis of fetal RHD status exploits structural differences between theRHD and RHCE genes and is based on the assumption that D-negative individuals have a deleted RHD gene. As the knowledge regarding the molecular basis of partial D antigens evolved, use of multiplex,70,86,140,141heteroduplex,142,143 and multiple sequence-specific PCR reactions144,145 have replaced the single exon genotyping assays32,146,147 in an attempt to avoid “false-negative” typing of a fetus with a partial D antigen. However, because HDN in a fetus whose RBCs have a partial D antigen is rare,148 the clinical value of RHD multiplex analysis may only have marginal added value.
All current RhD genotyping assays will mistype people whose RBCs are D-negative and yet carry an intact, nonfunctional RHD. Such people have been described in Caucasians (rare),70 African blacks (common),89,144 and Asians (common).74,144 Molecular genotyping will have limited clinical utility in populations where the presence of nonexpressedRHD is frequent. The molecular backgrounds of these D-negative phenotypes are beginning to emerge: In 2 Caucasians expressing the dCe phenotype, 1 had an in-frame stop codon in exon 1 of the RHDgene70 and the other a deletion of 4 nucleotides in exon 4.149 Very recently, the molecular basis of the major silent RHD allele (namedRHDψ) found in persons of African ancestry has been defined.150RHDψ has a 37–base pair insertion of DNA, being a duplication of the intron 3/exon 4 boundary, and has missense mutations in exon 5 and a nonsense and missense mutation in exon 6. The Del phenotype (ie, D antigen is detectable only by adsorption-elution tests) was thought to have a deletion ofRHD;151,152 however, a deletion of 1013 base pairs encompassing intron 8, exon 9, and intron 9 has been observed (Figure5).153
Clearly, knowledge of the ethnic group of both parents is helpful in the selection of appropriate genotyping tests. Wherever possible, to limit the gene pool, concurrent analyses of maternal and paternal blood group phenotypes and genotypes should be performed. It is worth noting that samples that have been used for clinical automated instruments are often contaminated with blood from previous tests.144 154
As molecular analysis becomes more common, it is worth remembering that some D variants may be more common than previously thought. An example of this is the hybrid gene encoding the DIIIa phenotype, which has recently been shown to be present in 18% of blacks in New York and 28% of blacks from Brazil.155 Furthermore, a similar pattern of reactivity may be obtained with monoclonal anti-D in tests with RBCs from people with different Rh genes. This is illustrated by the large number of molecular events associated with DVa (or DV-like) samples as defined by the pattern of reactivity with monoclonal anti-D.156 Not all of the molecular events give rise to the DW antigen, whose presence on RBCs is required for DVacategorization.110
Although hemolytic disease due to Rh antibodies other than anti-D is less frequent, PCR-based tests have been designed to defineRHCE alleles using fetally derived DNA.157-159 Most of these are relatively straightforward; however, genotyping C in the presence of D is difficult because RHC(E/e) and RHDhave identical sequences in exons 1 and 2. RhC typing is possible by exploitation of a polymorphism in intron 2 of RHCE, which involves a 109-base pair insert of DNA in RHC(E/e) but notRhc(E/e) or RHD.94 159
Noninvasive prenatal Rh genotyping.
It is now possible to obtain fetally derived DNA using noninvasive procedures. Fetally derived RHD has been detected using nested PCR analysis on genomic DNA (gDNA) extracted from maternal peripheral blood or plasma160-163 or from transcervical samples.164-166 An alternative approach uses cDNA templates derived by reverse transcriptase–PCR from maternal peripheral blood and detection of fetal RhD mRNA targets.167 All noninvasive procedures have limited value because there is no suitable way to assess the presence of fetal cells in a given sample and, thus, negative results cannot be interpreted with confidence. Nevertheless, the fact that fetally derived Rh mRNAs and gDNA can be detected in maternal blood indicates that this area of prenatal diagnosis may soon have an impact. However, it is possible that fetal-nucleated RBCs are the most pertinent target cell type for noninvasive diagnosis168,169 because other fetally derived CD34+ cells have been detected in maternal blood for as long as 27 years postpartum169 and thus could interfere with analyses in women who have had multiple pregnancies.
Rh and other disease states
Rhnull disease.
RBCs from people who have the Rhnull phenotype (synonyms: Rhnull syndrome, Rhnull disease) lack Rh proteins and, thus, Rh antigens. This phenotype is rare (approximately 1 in 6 × 106 individuals)170 and most often results from a consanguineous mating. The syndrome is associated with stomatocytosis, spherocytosis, increased osmotic fragility, altered phospholipid asymmetry, altered cell volume, defective cation fluxes, and elevated Na+/K+ ATPase activity.13,14 171-173 Rhnull RBCs may have a shortened in vivo survival, and the person may have a mild compensated hemolytic anemia.
There are 2 types of Rhnull, amorph and regulator, that historically were classified based on their pattern of inheritance. It is now known that the amorph type is the result of a molecular change in RHCE in tandem with a deleted RHD (Figure 3),whereas the regulator type is associated with a molecular defect inRHAG (Figure7).22,76,174-177 Table 2summarizes the characteristics of the 2 types of Rhnullphenotypes and compares them with the Rhmodphenotype, in which the Rh antigens are suppressed. While RhAG is apparently critical for the correct assembly of the Rh proteins in the RBC membrane, RhAG by itself can form stable complexes, albeit in reduced quantity, in the absence of Rh proteins.22 58
Localization of molecular defects on RhAG.
The regulator type of Rhnull is associated with 2 mutantRHAG genes (homozygote or double heterozygote). The mutations include splice site/frameshift alterations and missense mutations (gray circles). The missense changes predominantly occur within conserved Rh protein family domains (black rectangles), within membrane-spanning regions. It is thought that missense mutations affect either RhAG-RhAG associations/RhAG-Rh protein assocations, resulting in an absence of the Rh protein family from mature RBC membranes. The Rhmodphenotype is associated with missense mutations (crosshatched circles), which lead to a marked reduction of the RhAG-Rh protein complex in mature RBC membranes. The initials refer to the probands. The information used in this figure was obtained from the following: SM209; SF, JL174; AL177; YT27,175; VL174; HT 210; TT, AC22; TB174; WO210.
Localization of molecular defects on RhAG.
The regulator type of Rhnull is associated with 2 mutantRHAG genes (homozygote or double heterozygote). The mutations include splice site/frameshift alterations and missense mutations (gray circles). The missense changes predominantly occur within conserved Rh protein family domains (black rectangles), within membrane-spanning regions. It is thought that missense mutations affect either RhAG-RhAG associations/RhAG-Rh protein assocations, resulting in an absence of the Rh protein family from mature RBC membranes. The Rhmodphenotype is associated with missense mutations (crosshatched circles), which lead to a marked reduction of the RhAG-Rh protein complex in mature RBC membranes. The initials refer to the probands. The information used in this figure was obtained from the following: SM209; SF, JL174; AL177; YT27,175; VL174; HT 210; TT, AC22; TB174; WO210.
Myeloid leukemia
Patients with acute or chronic myeloid leukemia, myeloid metaplasia, polycythemia, or myelofibrosis occasionally have 2 populations of RBCs of different Rh type. In some cases, a loss of Rh antigens is associated with chromosome aberrations.178 Recent analysis of blood from a D-positive patient with CML who became D-negative for the 3 years that she was studied revealed a single base deletion in exon 4 of RHD that occurred by somatic mutation.179
Discussion
Considerable progress has been made in our understanding of the molecular basis of Rh and other blood group antigens in the past 10 years. Despite this, our knowledge concerning the function of many of the components in the RBC remains speculative. The Rh protein complex is a prime example of this; it is a major red cell protein of considerable clinical importance, yet our understanding of its functional significance in human RBCs and other animals relies almost entirely on circumstantial evidence.
The Rh blood group system consists of numerous antigens that are located on variant forms of RhD and RhCE proteins. These proteins form a core complex in the erythrocyte membrane with a glycosylated homolog (RhAG) and are only expressed when it is present. RhD, RhCE, and RhAG associate with other membrane proteins (LW, IAP, GPB, Duffy, and band 3) to form a large complex. Although the function(s) of these proteins has not been defined, it is possible that the complex forms a concerted transporter. The genes encoding the Rh proteins (RHCE andRHD) are highly homologous and adjacent on the short arm of chromosome 1, while the gene encoding RhAG (RHAG) is nearly 40% homologous and is located on the short arm of chromosome 6. Although the molecular basis associated with many of the Rh antigens is known, the actual epitopes have not been defined, but it is apparent that most of the Rh antigens are conformation-dependent. The molecular knowledge is increasingly being used in the clinical setting. However, the allelic diversity in this system is a potential problem for reliable genotyping by PCR-based assays. Hemagglutination is still a powerful, practical, and economical test with a specificity and sensitivity that is appropriate for clinical applications. However, the use of hemagglutination in conjunction with molecular techniques undoubtedly will lead to insights that can enhance approaches for the treatment of Rh incompatibility. Further understanding of the immunologic responses to the Rh antigens will be of importance in the treatment of hemolytic disease, and detailed epitope maps involving serologic, molecular, and protein crystallographic studies of the Rh proteins will contribute to this objective.
Acknowledgments
We thank Christine Lomas-Francis, Narla Mohandas, Olga Blumenfeld, Geoff Daniels, Michael J. A. Tanner, Jill Storry, and Karina Yazdanbakhsh for reading the manuscript and giving helpful suggestions. Neil Avent thanks Willy Flegel, Giorgio Matassi, and Tim Kemp for providing manuscripts prior to publication. We also thank Robert Ratner for preparing the manuscript and cataloging the references.
Supported in part by a National Institutes of Health Specialized Center of Research (SCOR) grant in transfusion medicine and biology HL54 459.
Reprints:Marion E. Reid, Immunochemistry Laboratory, New York Blood Center, 310 East 67th St, New York, NY 10021; e-mail:mreid@nybc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.