Alternative splicing plays a major role in regulating tissue-specific expression of cytoskeletal protein 4.1R isoforms. In particular, expression of the protein's functionally critical spectrin-actin binding domain, essential for maintenance of red cell membrane mechanical properties, is governed by a developmentally regulated splicing switch involving alternative exon 16. Using a model 3-exon 4.1R pre–messenger RNA (pre-mRNA), we explored the sequence requirements for excision of the introns flanking exon 16. These studies revealed that splicing of this alternative exon occurs preferentially in an ordered fashion. The first step is excision of the downstream intron to join exons 16 and 17, followed by excision of the upstream intron. Constructs designed to test the converse pathway were spliced less efficiently and with less fidelity, in part due to activation of a cryptic 5′ splice site in exon 16. This downstream-first model for ordered splicing is consistent with the hypothesis that regulated alternative splicing requires cooperation between multiple exonic and/or intronic regulatory elements whose spatial organization is critical for recruitment of appropriate splicing factors. Our results predict that exon 16 splicing is regulated at the first step—excision of the downstream intron—and that cells unable to catalyze this step will exhibit exon 16 skipping. In cells that include exon 16, adherence to an ordered pathway is important for efficient and accurate production of mature 4.1R mRNA encoding an intact spectrin-actin binding domain.
The gene encoding structural protein 4.1R generates a complex set of tissue-specific alternatively spliced transcripts. Two of these splicing events regulate expression of alternative translation initiation sites in exons 2′ (AUG1) and 4 (AUG2), thereby controlling synthesis of 2 size classes of 4.1R with distinct N-termini.1-4 In addition, regulated splicing of several internal exons leads to tissue-specific expression of diverse protein isoforms with distinct functional properties. The best characterized example involves exon 16, which encodes part of the spectrin-actin binding domain (SAB). Exon 16 is strongly included in late erythroid cells and to a lesser extent in selected nonerythroid cells, including muscle, brain, spleen, and testis.2,5 Importantly for the current study, Xenopus oocytes also exhibit alternative splicing of endogenous 4.1R exon 16, with approximately equal amounts of E16 inclusion and E16 skipping.6 Other 4.1R exons that exhibit tissue- or development-specific splicing patterns include exon 17A (expressed mainly in muscle7,8), exon 17B (expressed in epithelial cells7), and exons 14 and 15 (expressed mainly in brain2 5). Our studies aim to elucidate the molecular mechanisms responsible for regulating the tissue-specific expression of these diverse isoforms of the protein 4.1R family.
During erythroid differentiation, exon 16 exhibits a dramatic alternative splicing switch, being almost entirely skipped in early erythroid progenitors but included with high efficiency in more mature erythroblasts.3,4,9 Because exon 16 encodes a critical part of protein 4.1's SAB domain, this splicing switch results in an important functional change in the protein. A variety of genetic, biochemical, and biophysical data support the notion that expression of an intact SAB domain is essential for assembly of erythroid membranes with the appropriate strength and deformability properties to survive in the circulation without fragmentation.9-14 More recently, it has been shown that exon 16 also encodes part of a nuclear localization signal required for import of selected 4.1 isoforms in nucleated cells.15 16The regulation of exon 16 alternative splicing therefore represents an intriguing model system to use to study a physiologically relevant splicing event.
Alternative exons are frequently flanked by suboptimal splice sites. An increasing body of evidence indicates that regulation of such exons requires, in addition to the constitutive spliceosomal components, participation of enhancer and/or repressor factors that help define the splice sites at the appropriate developmental times. Cis-regulatory sequences that bind to these splicing factors may reside in the regulated exon itself,17 in flanking intron sequences,18,19 or even in an adjacent exon.20Since inclusion of an alternative exon is a multistep process involving removal of both flanking introns, proper coordination among the various regulatory elements might occur in a specific sequential manner. Indeed, in several alternatively spliced pre–messenger RNAs (pre-mRNAs), there is evidence that 1 flanking intron is preferentially removed before the other.21-23
In order to begin exploring the regulatory mechanisms that govern 4.1 exon 16 splicing, we have directly tested the hypothesis that the flanking introns are excised preferentially in a specific order. The substrate for these studies was a model 3-exon 4.1 pre-mRNA derived from the mouse gene. This pre-mRNA exhibited efficient inclusion of exon 16 in microinjected Xenopus oocytes, but exon 16 was predominantly skipped in HeLa nuclear extracts. This difference suggested that specific alternative factors are present in oocytes to mediate exon 16 inclusion. Analysis of the behavior of several 4.1R pre-mRNA constructs led to the conclusion that the splicing pathway for inclusion of exon 16 occurs preferentially in an ordered manner through removal of the downstream intron in the first step, followed by excision of the upstream intron. Constructs designed to test the converse splicing order were spliced inefficiently and/or exhibited activation of a nonphysiological cryptic splice site in exon 16. These results are discussed in the context of current models for regulation of alternative splicing.
Materials and methods
Minigene construction
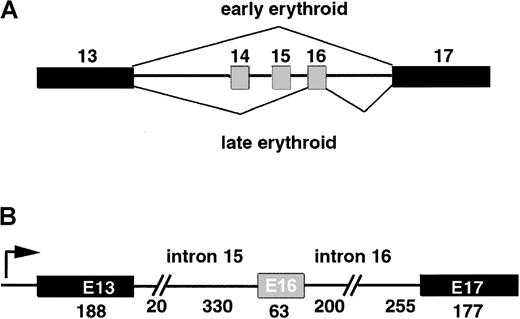
A mouse genomic library, designated 129SV (Stratagene, La Jolla, CA), was screened with the use of a 32P-labeled mouse 4.1 complementary DNA (cDNA). Genomic clones containing exon 16 and 17 were isolated and characterized. Because the introns flanking exon 16 were too large for practical use in in vitro transcription experiments, and because most known splicing regulatory sequences are located near alternative exons, we designed a minigene containing exons 16 and 17 and 200 to 300 bases of flanking intron sequences. Since exons 14 and 15 are alternative exons that are skipped in most tissues, the first exon of the 4.1 minigene was derived from the nearest upstream constitutive exon, exon 13. Exon 13, together with a short 5′ flanking sequence,5 was cloned by polymerase chain reaction (PCR) directly from total mouse genomic DNA. These 3 fragments were assembled in pBluescript (Stratagene, La Jolla, CA), sequenced to confirm the correct structure, and transcribed in vitro to generate synthetic pre-mRNA using T7 polymerase. The 3-exon minigene, having the structure 13i16i17 (where “i” designates intron), was designated pBS4.1.
Truncated derivatives of pBS4.1 were constructed to test the sequence requirements for excision of the individual flanking introns. Sense strand (S) and antisense strand (AS) primers used in construction of these substrates are as follows (location of each primer indicated in parentheses): S1 (exon 13), 5′-TAATACGACTCACTATAGAGCCATTGCTCAGAGTCAGG-3′ (underlined sequence represents an added T7 RNA polymerase site); S2 (exon 16), 5′-AAAAAGAGAGAGAGACTAGA-3′; S3 (upstream intron), 5′-TAATACGACTCACTATAGGGCATTT-GCTGCATCGCACAC-3′; AS1 (downstream intron), 5′-ACGTTTACCACCATGCAAAAG-3′; AS2 (exon 16), 5′-CTCCAACATTAAATTGCTAT-3′; AS3 (exon 17), 5′-GAATTCTCCATCTCCAGTAGGGAC-3′ (underlined sequence represents an EcoRI site added for cloning purposes. Construct 13i16i was derived from pBS4.1 by PCR with the use of primers S1 and AS1. Construct 13i16/17 was derived by splice overlap extension24 of 2 fragments: a 5′ piece generated by amplification of pBS4.1 with primers S1 and AS2, and a 3′ piece produced from cloned 4.1R cDNA by amplification with primers S2 and AS3. Construct i16i17 was made by PCR of pBS4.1 with the use of primers S3 and AS3. Finally, construct 13/16i17 was synthesized by splice overlap extension of 2 fragments: a 5′ piece generated by amplification of cloned 4.1R cDNA with the use of primers S1 and AS2, and a 3′ piece produced from pBS4.1 by amplification with primers S2 and AS3.
Synthesis of model pre-mRNAs and microinjection into oocytes
pBS4.1 was linearized with EcoRI. SP6-HB6 (a gift of Drs A. Mayeda and A. Krainer) was linearized with BamHI. Synthesis of capped RNA transcripts was done according to manufacturer's recommendation (Ambion, Inc, Austin, TX) in the presence of m7G5′)ppp5′)G. Transcripts were either diluted and microinjected directly, or purified by using RNeasy columns with reagents and protocols supplied by the manufacturer (Qiagen, Valencia, CA).
Stage IV and VI oocytes25 were defolliculated manually. A 20 nL solution of pre-mRNA or sterile water was injected into the oocyte nucleus. Typically, a set of 3 oocytes was injected for each sample and incubated in modified Ringer solution (100 mmo/L NaCl, 1.8 mM KCl, 2 mmo/L CaCl2, 1 mmo/L MgCl2, 4 mmo/L sodium bicarbonate, 7.05 mmo/L HEPES at pH7.2, 0.5mg/mL bovine serum albumin, and 50μg/mL gentamicin). Following incubation, oocytes were frozen on dry ice, then stored at −80°C until RNA extraction. RNA was extracted from oocytes with the use of RNeasy columns as described above.
Splicing assays in nuclear extract
HeLa nuclear extract was prepared by the method of Mayeda and Krainer.26 A typical 25μL reaction contained 6.25 fmol of RNA substrate in 40% HeLa nuclear extract, 3.2 mmo/L MgCl2, 1 mmo/L adenosine triphosphate, 20 mmo/L creatine phosphate, 3.1% polyvinyl alcohol, and 40 units RNasin (Promega, Madison, WI). The splicing reaction was incubated at 30°C for 2 hours, unless otherwise indicated, and terminated by the addition of 25 μL of stop solution (0.3 mol/L Tris-HCl/pH7.4, 0.3 mol/L NaOAc, 0.5% SDS, 2 mmo/L EDTA, and 3 μg/mL transfer RNA). RNA was then purified with the use of RNeasy columns (Qiagen, Inc, Valencia, CA) according to the manufacturer's protocols.
RT/PCR analysis of spliced pre-mRNA
RT/PCR analysis has been previously used to analyze regulation of alternative pre-mRNA splicing.27 The conditions outlined below were designed to amplify products representing exon inclusion (13/16/17) and exon skipping (13/17) in one reaction, so that each band serves as an internal control for the other. Functional effects of cis-sequence alterations were judged by changes in the relative efficiency of E16 inclusion, measured by densitometry of the products displayed after polyacrylamide gel electrophoresis, and calculated as (inclusion products/inclusion plus skipping products).
Total RNA from 0.6 oocyte-equivalents was transcribed into single-stranded cDNA in the presence of antisense primers using the first strand cDNA synthesis kit. Mouse erythroleukemia (MEL) cDNA was prepared as previously described.2 Two μL of cDNA was amplified in a 25 μL PCR containing Taq polymerase buffer, 50 pmol each of sense and antisense primers, 0.2 mmol/L dNTPs, and 0.625 units of Taq polymerase (Perkin-Elmer). In some reactions, Tfl polymerase was used according to the manufacturer's specifications (Epicentre Technologies, Madison, WI) with similar results. Thirty-five cycles of amplification were performed with the use of an automated Perkin-Elmer Cetus 2400 thermal cycler under the following conditions: denaturation for 20 seconds at 94°C; annealing for 20 seconds at 60°C; extension for 40 seconds at 72°C. DNA fragments were analyzed by 5% polyacrylamide gel electrophoresis. The identity of all major PCR products discussed in this paper was confirmed by subcloning the fragments into pBluescript and performing DNA sequence analysis. Primers used for RT/PCR analysis of 4.1 spliced products were the following: sense-strand primer (in exon 13), 5′-TAATACGACTCACTATAGAGCCATTGCTCAGAGTCAGG-3′; antisense primer (in exon 17), 5′-CACTGATGCTGGCATGGTGC-3′. In some reactions in which splicing of a single intron was tested, primers S1 plus AS2 (for the upstream intron) or S2 plus AS3 (for the downstream intron) were employed. Primers for β-globin were the following: sense, 5′-ACATTTGCTTCTGACACAAC-3′; antisense, 5′-GTGCAGCTTGTCACAGTGCA-3′.
Extensive analysis of the oocyte splicing system revealed that reproducibility of results within a given preparation of oocytes processed under identical conditions was quite high; ie, efficiency of E16 inclusion as measured by densitometry showed little variability in parallel samples. Significant batch-to-batch variation in the absolute efficiency of E16 inclusion was observed among oocytes prepared and assayed on different days; however, the same relative order of splicing efficiency was always observed in comparison of different substrates.
Determination of E16 flanking sequences in the frog 4.1R gene
A Xenopus genomic library, a gift of Dr. R. Harland, was screened with 32P-labeled Xenopus 4.1 cDNA. A 5-kilobase (kb) DNA fragment hybridizing to exon 16 was subcloned, and flanking introns were sequenced with the use of Xenopus-specific primers located within exon 16.
Results
Figure 1A illustrates the E16 splicing switch activated during the erythroid differentiation program. E16 is skipped in early erythroid progenitors but included at later stages. In order to analyze the contribution of various sequence elements in regulation of exon 16 splicing, we constructed a 3-exon minigene containing exon 16 and its nearest constitutive exons, 13 and 17, each flanked by a portion of the native intron sequence (Figure 1B). In vitro transcription of this minigene yielded a model pre-mRNA that became the substrate for our splicing studies. Initial attempts to splice this pre-mRNA substrate in vitro with the use of HeLa nuclear extracts yielded predominantly exon 16 skipping, eg, direct splicing of exon 13 to 17. As an alternative strategy, we explored whether the frog oocyte splicing system might offer a suitable approach for analyzing sequence elements important for the splicing of exon 16. This strategy was based on the earlier observations (1) that oocytes are capable of accurately splicing exogenously microinjected pre-mRNA28and (2) that endogenous oocyte 4.1 mRNA is alternatively spliced with regard to E16 inclusion. The experiment shown in Figure2 demonstrates that oocytes can splice a control β-globin pre-mRNA (Figure 2A, lane 2) to its mature product (lane 3). Moreover, a model 3-exon 4.1 pre-mRNA (lane 4) also was accurately and efficiently spliced in oocytes into 2 mature products (lane 5). The larger product represented a 13/16/17 splice, which included exon 16, while the smaller band represented a 13/17 splice, an exon-skipping product. These 2 products comigrated exactly with the authentic PCR fragments derived from 4.1 mRNA in differentiating MEL cells (lane 6), and DNA sequence analysis verified that these products possessed the correct splice junctions. Mock-injected oocytes did not yield any PCR products, indicating that the mouse-specific oligonucleotides did not amplify endogenous frog 4.1 mRNA sequences (lane 7). Thus, the splicing machinery in frog oocytes can accurately recognize splicing regulatory sequences in the synthetic 4.1 pre-mRNA.
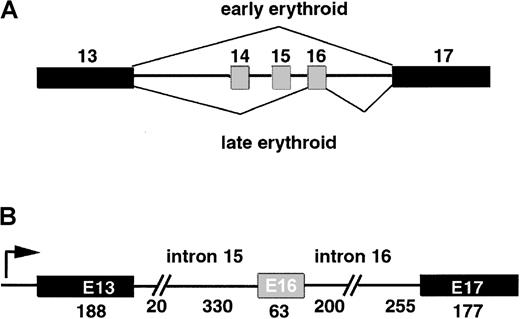
Alternative splicing of erythroid specific exon 16.
(A) Model of the splicing switch that accompanies erythroid differentiation. Constitutive exons are filled in in black; alternatively spliced E16 is filled in in gray. Early erythroid progenitors skip E16 almost entirely, while later progenitors include E16 with high efficiency. (B) Structure of the 1.2-kb minigene that serves as template for transcription of pre-mRNA splicing substrates. Arrow indicates start site for in vitro transcription.
Alternative splicing of erythroid specific exon 16.
(A) Model of the splicing switch that accompanies erythroid differentiation. Constitutive exons are filled in in black; alternatively spliced E16 is filled in in gray. Early erythroid progenitors skip E16 almost entirely, while later progenitors include E16 with high efficiency. (B) Structure of the 1.2-kb minigene that serves as template for transcription of pre-mRNA splicing substrates. Arrow indicates start site for in vitro transcription.
Parameters of splicing in the oocyte system.
(A) Gel analysis of a typical splicing experiment. Diagrams at sides indicate deduced structures of mRNAs from which the PCR products derive. Lane 1 shows size standards (1353, 1078, 872, 603, 310, 281, 271, 234, 194, 118, and 72 base pairs [bp], respectively). Lane 2 depicts control showing amplified β-globin pre-mRNA; lane 3, oocytes injected with β-globin pre-mRNA, showing E1/E2 spliced product; lane 4, control showing amplified 4.1 minigene pre-mRNA; lane 5, positive control showing 13/16/17 and 13/17 authentic 4.1 spliced products from differentiating MEL cells; lane 6, oocytes injected with 4.1 pre-mRNA, showing alternatively spliced 4.1 products 13/16/17 and 13/17; lane 7, oocytes mock-injected as negative control. (B) Reproducibility of the assay. Three groups of oocytes were injected with 4.1 pre-mRNA and processed in parallel. The relative efficiency of E16 inclusion among the triplicate samples, determined by densitometry and indicated above each lane, was nearly identical. (C) Time course of splicing in oocytes. Left: 50 pg of pre-mRNA was microinjected per oocyte and harvested at the indicated times for analysis. Total oocyte RNA recovery from each sample was similar (not shown). Substantial amounts of 13/16/17 and 13/17 splicing products were detected 2 hours after injection. Total spliced products remained fairly stable up to 16 hours postinjection. Right: RT/PCR products derived from endogenous frog 4.1 mRNA isoforms, representing E16 inclusion (upper band) and skipping (lower band) products as a control to demonstrate recovery of intact, amplifiable RNA from all time points. (D) Concentration dependence of splicing. Oocytes were injected with pre-mRNA as follows: Lane 1, 6 pg; lane 2, 25 pg; lane 3, 100 pg; lane 4, 400 pg; lane 5, 1.6 ng. All samples were incubated under identical conditions and harvested after 16 hours for analysis. The relative efficiency of E16 splicing given is calculated as: (inclusion products) / (inclusion plus skipping products). The efficiency varied dramatically from about 80% inclusion at low concentrations of injected substrate, to only less than 20% inclusion at high concentrations. (E) A consensus 5′ splice site mutation promotes better splicing of E16. Protein 4.1 pre-mRNA bearing the natural weak 5′ splice site yielded 55% inclusion of exon 16 (E16wt), while pre-mRNA with a strong consensus 5′ splice site exhibited 85% inclusion of exon 16 (E16↑). (Experiment performed in 2E utilized a different oocyte preparation than the remainder of this figure, thus explaining the different baseline level of wild type E16 inclusion.)
Parameters of splicing in the oocyte system.
(A) Gel analysis of a typical splicing experiment. Diagrams at sides indicate deduced structures of mRNAs from which the PCR products derive. Lane 1 shows size standards (1353, 1078, 872, 603, 310, 281, 271, 234, 194, 118, and 72 base pairs [bp], respectively). Lane 2 depicts control showing amplified β-globin pre-mRNA; lane 3, oocytes injected with β-globin pre-mRNA, showing E1/E2 spliced product; lane 4, control showing amplified 4.1 minigene pre-mRNA; lane 5, positive control showing 13/16/17 and 13/17 authentic 4.1 spliced products from differentiating MEL cells; lane 6, oocytes injected with 4.1 pre-mRNA, showing alternatively spliced 4.1 products 13/16/17 and 13/17; lane 7, oocytes mock-injected as negative control. (B) Reproducibility of the assay. Three groups of oocytes were injected with 4.1 pre-mRNA and processed in parallel. The relative efficiency of E16 inclusion among the triplicate samples, determined by densitometry and indicated above each lane, was nearly identical. (C) Time course of splicing in oocytes. Left: 50 pg of pre-mRNA was microinjected per oocyte and harvested at the indicated times for analysis. Total oocyte RNA recovery from each sample was similar (not shown). Substantial amounts of 13/16/17 and 13/17 splicing products were detected 2 hours after injection. Total spliced products remained fairly stable up to 16 hours postinjection. Right: RT/PCR products derived from endogenous frog 4.1 mRNA isoforms, representing E16 inclusion (upper band) and skipping (lower band) products as a control to demonstrate recovery of intact, amplifiable RNA from all time points. (D) Concentration dependence of splicing. Oocytes were injected with pre-mRNA as follows: Lane 1, 6 pg; lane 2, 25 pg; lane 3, 100 pg; lane 4, 400 pg; lane 5, 1.6 ng. All samples were incubated under identical conditions and harvested after 16 hours for analysis. The relative efficiency of E16 splicing given is calculated as: (inclusion products) / (inclusion plus skipping products). The efficiency varied dramatically from about 80% inclusion at low concentrations of injected substrate, to only less than 20% inclusion at high concentrations. (E) A consensus 5′ splice site mutation promotes better splicing of E16. Protein 4.1 pre-mRNA bearing the natural weak 5′ splice site yielded 55% inclusion of exon 16 (E16wt), while pre-mRNA with a strong consensus 5′ splice site exhibited 85% inclusion of exon 16 (E16↑). (Experiment performed in 2E utilized a different oocyte preparation than the remainder of this figure, thus explaining the different baseline level of wild type E16 inclusion.)
The basic parameters of 4.1 pre-mRNA splicing in oocytes are explored in Figure 2. First, to demonstrate the reproducibility of the technique, 3 identical sets of oocytes were injected with pre-mRNA and processed in parallel. The efficiency of E16 inclusion among these samples was essentially identical (Figure 2B). Extensive additional experimentation confirmed that samples processed in parallel from the same batch of oocytes yielded very reproducible results. Therefore, each experiment was always performed with a single oocyte preparation (ie, no mixed batches of oocytes).
The kinetics of splicing within oocytes is demonstrated in Figure 2C (left panel). Spliced products were first apparent after a lag of approximately 1 hour, presumably representing the time required for exon recognition and spliceosome assembly on the injected pre-mRNA, and products continued to accumulate until about 4 hours postinjection. Spliced products were then stable until at least 16 hours postinjection. As a control to demonstrate that RNA of equivalent quality was extracted from each time point, we amplified in parallel the endogenous frog 4.1 mRNA sequences using frog-specific primers (Figure 2C, right panel). This experiment also illustrated the alternative splicing of exon 16 in the endogenous frog 4.1 RNA.
Finally, the concentration dependence of alternative splicing was tested by microinjecting serial dilutions of 4.1 pre-mRNA. Figure 2D shows that the relative efficiency of E16 inclusion was inversely related to the concentration injected. Below a threshold of approximately 100 pg per oocyte, the efficiency of inclusion was quite high; 70% to 80% of total spliced products contained exon 16. The amount of E16-inclusion products dropped dramatically, relative to E13/17-skipping products, when larger quantities of pre-mRNA were injected (eg, < 20% inclusion at 1.6 ng/oocyte). This concentration effect was quite reproducible and suggests that limited quantities of splicing factor(s) required for E16 inclusion are present in oocytes. For practical purposes, the experiments in this paper were therefore performed with pre-mRNA concentrations well below this threshold, in order to minimize this source of potential variation.
A major purpose for developing the oocyte splicing assay was to facilitate functional analysis of putative cis-regulatory elements around exon 16. To demonstrate that the oocyte splicing machinery is responsive to changes in cis-regulatory elements, we performed a simple functional test of E16 splicing as a consequence of sequence alterations in the 5′ splice donor site. Two 4.1 pre-mRNAs were analyzed: 4.1E16wt contained the natural weak 5′ site AG/gtttgt, with nonconsensus pyrimidines at intron positions 3 and 4; 4.1E16↑ represented a consensus 5′ splice site AG/gtaagt constructed by substitution of consensus purine nucleotides at these positions. When these substrates were assayed in parallel, pre-mRNA 4.1E16↑ (Figure 2E, lane 2) exhibited significantly more efficient E16 inclusion than did pre-mRNA with the nonconsensus 5′ splice site (lane 1). This supports the general proposal that oocytes can be used to analyze cis-regulatory elements in pre-mRNA, as well as the important result that a weak 5′ splice site in 4.1 pre-mRNA contributes to the E16-skipping phenotype in early erythroid cells as well as many nonerythroid cells.
Ordered removal of introns flanking exon 16
The next series of experiments was designed to test the hypothesis that a regulated splicing event, such as that involving exon 16, would be mediated by means of a stepwise ordered process. A priori, it was possible that the splicing of exon 16 into mature 4.1 mRNA could proceed through either of 2 ordered pathways, whereby 1 of the flanking introns is preferentially excised before the other (Figure3A). Another possibility is that splicing of exon 16 is not intrinsically an ordered process and that either pathway would generate authentic mature product.
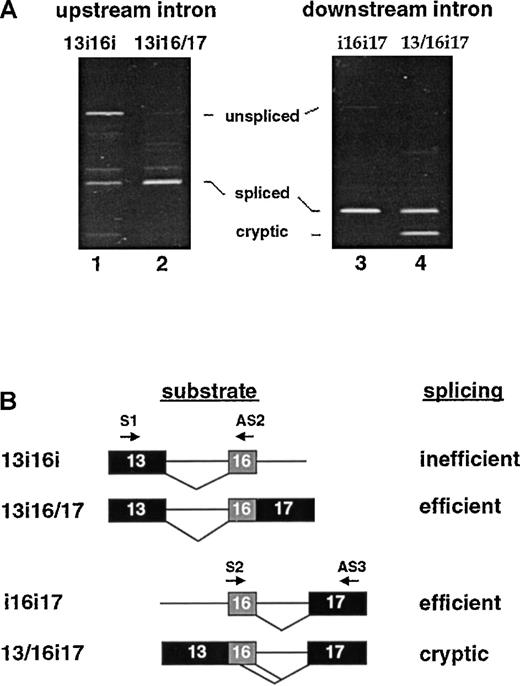
Ordered splicing pathways for inclusion of exon 16.
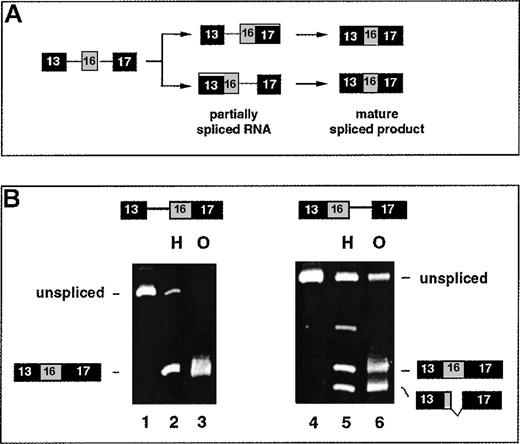
(A) The 2 possible ordered splicing pathways by which exon 16 could be included and the structure of the partially spliced RNAs unique to each pathway. (B) The ability of the partially spliced RNAs to be chased into mature products in either HeLa nuclear extract (H) or oocytes (O). Lanes 1-3 show downstream-first RNA 13i16/17 before splicing (lane 1), or after splicing in HeLa nuclear extract (lane 2) or in oocytes (lane 3). Lanes 4-6 show upstream-first RNA 13/16i17 before splicing (lane 4), or after splicing in HeLa nuclear extract (lane 5) or oocytes (lane 6). Lower band corresponds to splicing at a cryptic 5′ splice site in exon 16.
Ordered splicing pathways for inclusion of exon 16.
(A) The 2 possible ordered splicing pathways by which exon 16 could be included and the structure of the partially spliced RNAs unique to each pathway. (B) The ability of the partially spliced RNAs to be chased into mature products in either HeLa nuclear extract (H) or oocytes (O). Lanes 1-3 show downstream-first RNA 13i16/17 before splicing (lane 1), or after splicing in HeLa nuclear extract (lane 2) or in oocytes (lane 3). Lanes 4-6 show upstream-first RNA 13/16i17 before splicing (lane 4), or after splicing in HeLa nuclear extract (lane 5) or oocytes (lane 6). Lower band corresponds to splicing at a cryptic 5′ splice site in exon 16.
These models make very different predictions about the structure of the partially spliced RNAs produced in the first step of the exon 16–inclusion pathway, and the sequence requirements for removal of the upstream and downstream introns. As a first approach toward characterizing the order of intron removal around E16, the reaction products derived from splicing of the full 3-exon pre-mRNA were examined for evidence of partially spliced RNAs from which only 1 intron had been removed. No significant accumulation of such RNAs was detected in oocytes at any time up to 16 hours postinjection. In any event, it would be difficult to interpret whether partially spliced RNA species detected in this manner represented authentic intermediates in the splicing pathway or aberrant products incapable of yielding the correct mature mRNA.
To directly test whether either of the hypothetical partially spliced RNAs can be spliced into mature product, pre-mRNAs corresponding to those structures were constructed in vitro and assayed in the oocyte system. Microinjection of synthetic pre-mRNA 13i16/17 resulted in almost complete conversion into the correct mature product (Figure3B, lane 3); 13i16/17 was also spliced into mature product when incubated in HeLa nuclear extract (lane 2). In contrast, synthetic RNA 13/16i17 was partially aberrantly spliced in both assay systems (lanes 5, 6). Approximately half of the products derived from this substrate yielded the correct mature product 13/16/17, with the remainder corresponding to an aberrant smaller product spliced at a cryptic site within exon 16 (see below). Together these results are most consistent with an ordered pathway in which the downstream intron is removed first to generate 13i16/17 and the upstream intron is removed subsequently. The alternative pathway appears less favorable because it proceeds through RNA 13/16i17 and results in aberrantly spliced products.
As a complementary strategy, we constructed a series of truncated 4.1 pre-mRNAs designed to analyze each intron-splicing event separately, and asked whether the intron could be excised efficiently and accurately as a first step (ie, in the presence of the opposing intron) or as a second step (ie, if the opposing intron is already replaced by the appropriate flanking exon sequence). We reasoned that an ordered splicing process might require a particular arrangement of nearby enhancer or repressor elements to promote splice site recognition in the proper temporal sequence. Therefore, synthetic substrates corresponding to authentic partially spliced RNAs would be properly spliced into mature 13/16/17 products, while altered substrates that represent nonphysiological structures might be spliced poorly or aberrantly.
The sequence requirements for excision of the upstream intron were explored in Figure 4A (left panel). Construct 13i16i, which retained the downstream intron sequence, was designed to test the feasibility of splicing the upstream intron as the first step of an ordered process. Even in the absence of competing acceptor sites, much of the product recovered after incubation of this substrate in oocytes corresponded to the unspliced RNA, with only a modest amount of 13/16 product observed (lane 1). As shown above, the same upstream intron was very efficiently spliced using construct 13i16/17 in which exons 16 and 17 were already juxtaposed (lane 2). In essence, these results indicate that splicing of the upstream intron would not be favorable as the first step, especially when a larger pre-mRNA has competing splice sites. However, excision of the upstream intron would be very favorable as the second step of an ordered splicing process.
Sequence requirements for splicing of introns flanking exon 16.
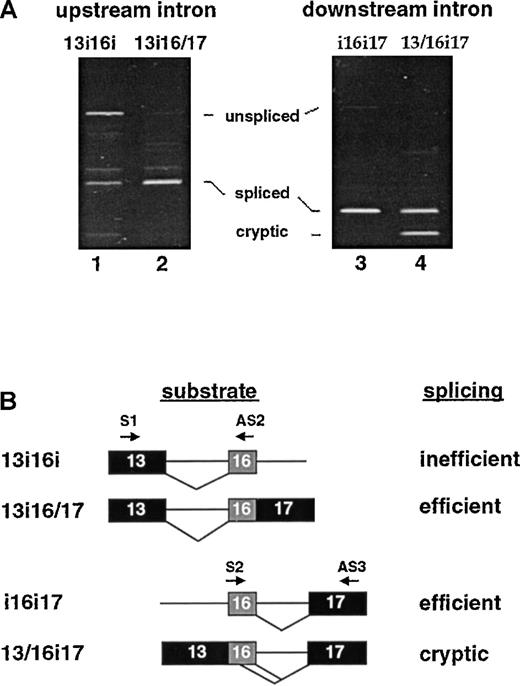
(A) Oocyte splicing assays. Left: excision of the upstream intron in the presence of downstream intron sequences (lane 1) or in the presence of juxtaposed exon 17 sequences (lane 2). Removal of the upstream intron was assayed by PCR with the use of primers in E13 (oligo S1) and E16 (AS2). Right: excision of the downstream intron in the presence of upstream intron sequences (lane 3) or in the presence of juxtaposed exon 13 sequences (lane 4). Removal of the downstream intron was assayed by PCR with the use of primers in E16 (oligo S2) and E17 (AS3). (B) Summary of results. Most importantly, excision of the downstream intron was most accurate if it occurred in conditions consistent with first step removal, ie, in the presence of the upstream intron (construct i16i17). Excision of the upstream intron appeared most efficient as a second step reaction (construct 13i16/17). The product slightly larger than the authentic spliced band represents a potential cryptic splicing event that was predominantly formed when splicing of the upstream intron was attempted as a first step (lane 1).
Sequence requirements for splicing of introns flanking exon 16.
(A) Oocyte splicing assays. Left: excision of the upstream intron in the presence of downstream intron sequences (lane 1) or in the presence of juxtaposed exon 17 sequences (lane 2). Removal of the upstream intron was assayed by PCR with the use of primers in E13 (oligo S1) and E16 (AS2). Right: excision of the downstream intron in the presence of upstream intron sequences (lane 3) or in the presence of juxtaposed exon 13 sequences (lane 4). Removal of the downstream intron was assayed by PCR with the use of primers in E16 (oligo S2) and E17 (AS3). (B) Summary of results. Most importantly, excision of the downstream intron was most accurate if it occurred in conditions consistent with first step removal, ie, in the presence of the upstream intron (construct i16i17). Excision of the upstream intron appeared most efficient as a second step reaction (construct 13i16/17). The product slightly larger than the authentic spliced band represents a potential cryptic splicing event that was predominantly formed when splicing of the upstream intron was attempted as a first step (lane 1).
The converse experiment was also performed to test whether sequences upstream of exon 16 can influence excision of the downstream intron. Splicing substrates with identical core elements (16i17) but different upstream sequences (upstream intron or exon 13) were assayed in the oocyte system. Figure 4A (right panel) shows that the downstream intron was efficiently excised if the upstream intron was still present and there were no competing 5′ splice sites (lane 3). Under the same conditions, the identical downstream intron was partially aberrantly spliced with the use of substrate 13/16i17 (lane 4). Only about half of the products derived from this substrate yielded the correct mature product 13/16/17. The remainder corresponded to an aberrant smaller product that arose from a cryptic splice site within exon 16 (see below). With respect to the ordered splicing hypothesis, these results strongly argue that removal of the downstream intron is more favorable as a first step reaction, resulting in the joining of exon 16 to 17; the splicing of exon 13 to 16 appears more favorable as the second step of the pathway. The alternative pathway would occur less efficiently (owing to the poor splicing of the upstream intron) and with lower fidelity (due to the cryptic splice site activation in exon 16) (Figure 4B).
Interestingly, the cryptic product appeared stable under the conditions of the oocyte splicing assays and could therefore be used as an indirect indicator of which ordered pathway is followed during splicing of the more complex 3-exon substrate. Extensive splicing assays performed with 3-exon substrates failed to detect significant quantities of this cryptic product (see, eg, experiments in Figure 2). Similarly, this cryptic product has not been detected upon amplification of authentic endogenous 4.1 mRNAs. These results provide circumstantial evidence in favor of the ordered model in which the downstream intron is preferentially removed first, although it is possible that the alternative pathway also occurs at much lower efficiency.
The cryptic 5′ splice site in exon 16 does not regulate usage of the authentic downstream 5′ splice site
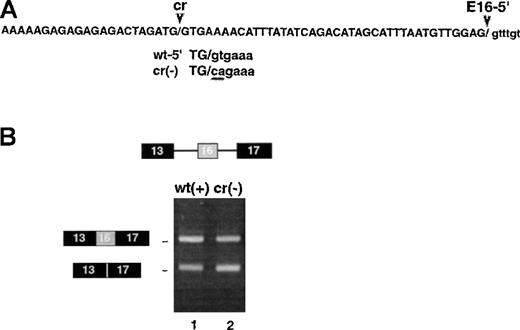
Sequence analysis of the aberrant short splicing product derived from 13/16i17 revealed it arose by activation of a cryptic 5′ splice site within exon 16 (Figure 5A). Splicing at the cryptic site led to inclusion of only 22 nucleotides of exon 16 in the final spliced product, rather than the normal 63 nucleotides. Subsequent splicing to E17, if it occurred, would generate a translational frameshift and premature translation termination. This cryptic product presumably represents a nonphysiological aberrant RNA, since it has never been reported in bona fide 4.1 cDNAs isolated from several cDNA libraries.
Effect of a pseudo-5′ splice site in exon 16.
Presence of a pseudo-5′ splice site in exon 16 does not negatively regulate splicing at the authentic 5′ splice site. (A) Sequence of the cryptic splice site in exon 16. The similarity to a consensus 5′ splice site and its ability to be activated in construct 13/16i17 suggest that U1 snRNP can bind here. Also shown is a gt-> ca mutation introduced to block potential U1 binding. (B) Splicing of 3-exon constructs containing the cryptic splice site (lane 1) or its mutated variant (lane 2). The failure of the mutation to activate exon 16 inclusion argues against a model in which U1 binding at the cryptic site represses exon 16 splicing by inhibiting recognition of the authentic 5′ splice site.
Effect of a pseudo-5′ splice site in exon 16.
Presence of a pseudo-5′ splice site in exon 16 does not negatively regulate splicing at the authentic 5′ splice site. (A) Sequence of the cryptic splice site in exon 16. The similarity to a consensus 5′ splice site and its ability to be activated in construct 13/16i17 suggest that U1 snRNP can bind here. Also shown is a gt-> ca mutation introduced to block potential U1 binding. (B) Splicing of 3-exon constructs containing the cryptic splice site (lane 1) or its mutated variant (lane 2). The failure of the mutation to activate exon 16 inclusion argues against a model in which U1 binding at the cryptic site represses exon 16 splicing by inhibiting recognition of the authentic 5′ splice site.
The cryptic splice site sequence in exon 16, g/gtgaaa, possesses partial sequence homology to the 5′ end of U1 small nuclear RNA (snRNA) and indeed resembles a weak 5′ splice site found at the exon/intron boundaries of some alternative exons. Use of this splice site in selected artificial 4.1 pre-mRNAs provides strong circumstantial evidence that U1 snRNP can bind at this cryptic site and could potentially act as a negative regulator of exon 16 splicing by interfering with recognition of the authentic downstream 5′ splice site. Such a model would predict that mutating the cryptic 5′ splice site would relieve the inhibition and thereby improve the efficiency of splicing, as has been demonstrated in the case of the Drosophila P element intron 3.29 We therefore generated a pre-mRNA in which the cryptic 5′ splice site g/gtgaaa was mutated to g/cagaaa; this mutation eliminated the essential “gt” dinucleotide and thus should have strongly inactivated the cryptic site. When this mutation was introduced into the 3-exon minigene, inclusion of exon 16 was not improved and in fact appeared to be slightly less efficient (Figure 5B, compare lanes 1 and 2). The failure to observe de-repression of exon 16 splicing upon cryptic site mutation is most consistent with the idea that negative regulation of exon 16 splicing is not mediated by U1 binding to the cryptic site.
Discussion
This study represents a first step toward understanding the regulation of alternative pre-mRNA splicing for exon 16 of the protein 4.1R gene. We have constructed a 3-exon model pre-mRNA that exhibits accurate and efficient inclusion of alternative exon 16. Most importantly, our results suggest that the splicing pathway for E16 inclusion occurs preferentially via an ordered process initiated by excision of the downstream intron. A key element of this downstream-first model appears to operate similarly in both the oocyte and the in vitro nuclear extract splicing system: neither splicing machinery was able to splice the upstream intron with maximum efficiency unless (downstream) intron 16 sequences were replaced with exon 17. The model strongly predicts that the differential regulation of exon 16 splicing among various cell types is determined by the ability to catalyze the first step, excision of the downstream intron. Once this first step is accomplished, removal of the upstream intron is efficient even in HeLa extracts, which normally exhibit very low inclusion of E16. It is important to note that such an ordered splicing model would allow regulation at a single rate-limiting step, after which the remaining steps of E16 splicing would proceed efficiently. Therefore, cell types capable of activating downstream intron splicing would include exon 16, while cells that cannot do so would exhibit an exon 16–skipping phenotype.
The hypothesis that splicing of exon 16 occurs by means of an ordered process was supported by analysis of the splicing phenotype of several model 4.1R pre-mRNA constructs, each designed to test individual steps of that pathway. All the results were consistent with preferential splicing by means of a downstream-first model in which formation of the correctly spliced 13/16/17 product proceeds via initial excision of the downstream intron to join exons 16 and 17. In contrast, first-step excision of the upstream intron appeared intrinsically less efficient and less productive in generating mature 4.1R mRNA, in part owing to activation of a cryptic splice site within exon 16. While we cannot entirely exclude the possibility that cryptic splicing arises as an artifact of incorrectly folded synthetic pre-mRNA, we note that splicing at the cryptic site did not occur in the majority of constructs 13i16i17, i16i17, or 13i16/17. The activation of cryptic splicing only in 13/16i17 suggests that premature removal of the upstream intron is necessary for induction of any potential misfolding. RNA spliced at this cryptic site would exhibit a translational frameshift and in vivo would likely be subject to premature translation termination and nonsense-mediated decay.30 It is interesting to speculate that such degradation might function as part of a proofreading mechanism designed to eliminate 4.1 pre-mRNAs spliced inappropriately through a nonregulated pathway. Together these observations indicate that adherence to the ordered pathway illustrated in Figure 6 is important in efficient production of properly spliced, translatable 4.1 mRNA.
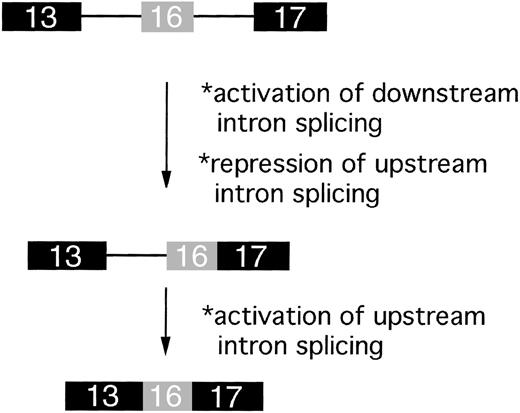
Downstream-first model for ordered splicing of introns flanking exon 16.
The first step of the pathway is excision of the downstream intron, generating a partially spliced RNA with exons 16 and 17 joined. This step is hypothesized to remove repressor elements in intron 16 and/or juxtapose enhancer elements in exon 17, thus activating splicing of the upstream intron in the second step of the reaction.
Downstream-first model for ordered splicing of introns flanking exon 16.
The first step of the pathway is excision of the downstream intron, generating a partially spliced RNA with exons 16 and 17 joined. This step is hypothesized to remove repressor elements in intron 16 and/or juxtapose enhancer elements in exon 17, thus activating splicing of the upstream intron in the second step of the reaction.
The proposed model for ordered splicing of exon 16 is consistent with the general hypothesis that regulated alternative splicing requires cooperation between multiple exonic and/or intronic regulatory elements whose spatial organization is critical to recruitment of appropriate RNA binding proteins/splicing factors. During exon recognition and the subsequent removal of upstream and downstream introns, dramatic rearrangements in overall RNA structure can lead to excision of some key regulatory sequences and novel juxtaposition of others. It is likely that for many alternative exons, removal of flanking intron sequences will necessarily occur in an ordered fashion to maintain the appropriate geometry among a cooperating array of weak interaction sites that together define the proper splice sites.31However, while the concept of ordered splicing probably has wide acceptance, there are only a few examples for which direct evidence is available to support such a model. Specific cases include the pre-mRNAs for preprotachykinin (alternative exon 4),22β-tropomyosin (alternative exon 6),21 src (alternative exon N1).23 In each of these model pre-mRNAs, the intron upstream of the regulated exon is removed poorly or not at all in the presence of the downstream intron, but quite efficiently when the downstream intron is replaced by the appropriate downstream exon. The mechanistic details of these regulated splicing events are not well understood and may vary considerably. Excision of the downstream intron may release an inhibitory block to upstream intron splicing by removal of intronic inhibitory sequences,23 and/or the juxtaposition of enhancer elements in the adjoined downstream exon21 or its associated 5′ splice site sequence.22
The ordered splicing hypothesis for protein 4.1 exon 16 makes testable predictions regarding the key regulatory steps (Figure 6). According to this model, initiation of E16 splicing involves activation of downstream intron splicing under conditions where upstream splicing is repressed. This step might require the function of splicing enhancer factor(s) to overcome the inherently weak splicing of E16 that is due at least in part to a suboptimal 5′ splice site (Figure 2E). Candidate enhancer factors that might promote E16 splicing include members of the SR protein family, a group of nuclear RNA binding proteins that can interact with regulatory sequences in alternative exons and promote spliceosomal assembly.32-34 SR proteins often bind purine-rich sequence elements in regulated exons32-35; therefore, the 15-nucleotide purine stretch at the 5′ end of exon 16 needs to be examined as a potential splicing enhancer.
Another prediction of the model is that there must be some mechanism for repressing upstream intron splicing until after the downstream intron is excised. One might therefore expect that E16 could contain an exonic splicing silencer, a class of regulatory elements recently identified in several other alternative exons.36-39Splicing silencer elements have been shown to inhibit splicing at nearby splice sites, in some cases by binding to members of the hnRNP A/B family.36,38 Finally, there must also be a mechanism to activate upstream intron splicing following the ligation of exons 16 and 17. It is tempting to speculate that downstream splicing could eliminate inhibitory sequences in intron 16 and/or juxtapose enhancer elements in exon 17, either of which might de-repress upstream intron splicing. Future analysis of the model 4.1 pre-mRNA will be directed toward identifying the putative regulatory sequence elements that mediate ordered splicing of exon 16. In that regard, the highly conserved 3′ end of exon 16, which is identical in human, mouse, dog, and frog,6 40 is an obvious candidate regulatory element.
Finally, it is important to note that alternative splicing of protein 4.1R exon 16 is conserved in several vertebrate orders, including mammals, birds, and amphibians. Moreover, E16 inclusion is not exclusively an erythroid phenomenon, as partial inclusion of E16 is also observed in selected other cell types, such as muscle5and oocytes.6 Therefore, although our long-range goal is to understand the molecular mechanism(s) responsible for alternative splicing of E16 during erythroid development, the experiments reported here provide us with an important general model of E16 regulation. Given the high degree of evolutionary conservation that exists among splicing mechanisms in widely divergent species, it seems likely that many of the key features of E16-ordered splicing will be conserved in erythroid cells.
Supported by grant HL45182 from the National Institutes of Health and grant DE-AC03-76SF0098 from the Office of Biological and Environmental Research, Department of Energy.
Reprints:John G. Conboy, Lawrence Berkeley National Laboratory, Life Sciences Division, Mailstop 74-157, 1 Cyclotron Road, Berkeley, CA 94720; e-mail: jgconboy@lbl.gov.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 2. Parameters of splicing in the oocyte system. / (A) Gel analysis of a typical splicing experiment. Diagrams at sides indicate deduced structures of mRNAs from which the PCR products derive. Lane 1 shows size standards (1353, 1078, 872, 603, 310, 281, 271, 234, 194, 118, and 72 base pairs [bp], respectively). Lane 2 depicts control showing amplified β-globin pre-mRNA; lane 3, oocytes injected with β-globin pre-mRNA, showing E1/E2 spliced product; lane 4, control showing amplified 4.1 minigene pre-mRNA; lane 5, positive control showing 13/16/17 and 13/17 authentic 4.1 spliced products from differentiating MEL cells; lane 6, oocytes injected with 4.1 pre-mRNA, showing alternatively spliced 4.1 products 13/16/17 and 13/17; lane 7, oocytes mock-injected as negative control. (B) Reproducibility of the assay. Three groups of oocytes were injected with 4.1 pre-mRNA and processed in parallel. The relative efficiency of E16 inclusion among the triplicate samples, determined by densitometry and indicated above each lane, was nearly identical. (C) Time course of splicing in oocytes. Left: 50 pg of pre-mRNA was microinjected per oocyte and harvested at the indicated times for analysis. Total oocyte RNA recovery from each sample was similar (not shown). Substantial amounts of 13/16/17 and 13/17 splicing products were detected 2 hours after injection. Total spliced products remained fairly stable up to 16 hours postinjection. Right: RT/PCR products derived from endogenous frog 4.1 mRNA isoforms, representing E16 inclusion (upper band) and skipping (lower band) products as a control to demonstrate recovery of intact, amplifiable RNA from all time points. (D) Concentration dependence of splicing. Oocytes were injected with pre-mRNA as follows: Lane 1, 6 pg; lane 2, 25 pg; lane 3, 100 pg; lane 4, 400 pg; lane 5, 1.6 ng. All samples were incubated under identical conditions and harvested after 16 hours for analysis. The relative efficiency of E16 splicing given is calculated as: (inclusion products) / (inclusion plus skipping products). The efficiency varied dramatically from about 80% inclusion at low concentrations of injected substrate, to only less than 20% inclusion at high concentrations. (E) A consensus 5′ splice site mutation promotes better splicing of E16. Protein 4.1 pre-mRNA bearing the natural weak 5′ splice site yielded 55% inclusion of exon 16 (E16wt), while pre-mRNA with a strong consensus 5′ splice site exhibited 85% inclusion of exon 16 (E16↑). (Experiment performed in 2E utilized a different oocyte preparation than the remainder of this figure, thus explaining the different baseline level of wild type E16 inclusion.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/2/10.1182_blood.v95.2.692/6/m_bloo00218002w.jpeg?Expires=1767899634&Signature=Ez9ES0k4VYVb5W6H9S4hbKeJiVcYmh8TP0qrN2dCDgU3~jd3RVdCIgA3YaCVkT5pDi6HbdyZVPxiQhvjFfZ8g9vRFfEfZXEZjdMzLjZan0HKkFI1S0qTWoXiClSzTwdALmcsoKNSYpm9hizitlCMzEcBkXaGrwE4jkdX5-M3dQnNiBQ-Bj95woBeMX3bCEc~DU46Y0VSiJZ99C7A3c8Ftl8kr6yOYq6WZl5D4G~vi-ka0KFN-hWYnnmLo99CWWOb3lE3PhFJmmTwjdHrgBSr9LAHVmblt4GJMjvtJXj5QsoM7CmQpPqygkBZhYQ6PqTpFHSSxdTD9rtS1sxjvgEveA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Fig. 2. Parameters of splicing in the oocyte system. / (A) Gel analysis of a typical splicing experiment. Diagrams at sides indicate deduced structures of mRNAs from which the PCR products derive. Lane 1 shows size standards (1353, 1078, 872, 603, 310, 281, 271, 234, 194, 118, and 72 base pairs [bp], respectively). Lane 2 depicts control showing amplified β-globin pre-mRNA; lane 3, oocytes injected with β-globin pre-mRNA, showing E1/E2 spliced product; lane 4, control showing amplified 4.1 minigene pre-mRNA; lane 5, positive control showing 13/16/17 and 13/17 authentic 4.1 spliced products from differentiating MEL cells; lane 6, oocytes injected with 4.1 pre-mRNA, showing alternatively spliced 4.1 products 13/16/17 and 13/17; lane 7, oocytes mock-injected as negative control. (B) Reproducibility of the assay. Three groups of oocytes were injected with 4.1 pre-mRNA and processed in parallel. The relative efficiency of E16 inclusion among the triplicate samples, determined by densitometry and indicated above each lane, was nearly identical. (C) Time course of splicing in oocytes. Left: 50 pg of pre-mRNA was microinjected per oocyte and harvested at the indicated times for analysis. Total oocyte RNA recovery from each sample was similar (not shown). Substantial amounts of 13/16/17 and 13/17 splicing products were detected 2 hours after injection. Total spliced products remained fairly stable up to 16 hours postinjection. Right: RT/PCR products derived from endogenous frog 4.1 mRNA isoforms, representing E16 inclusion (upper band) and skipping (lower band) products as a control to demonstrate recovery of intact, amplifiable RNA from all time points. (D) Concentration dependence of splicing. Oocytes were injected with pre-mRNA as follows: Lane 1, 6 pg; lane 2, 25 pg; lane 3, 100 pg; lane 4, 400 pg; lane 5, 1.6 ng. All samples were incubated under identical conditions and harvested after 16 hours for analysis. The relative efficiency of E16 splicing given is calculated as: (inclusion products) / (inclusion plus skipping products). The efficiency varied dramatically from about 80% inclusion at low concentrations of injected substrate, to only less than 20% inclusion at high concentrations. (E) A consensus 5′ splice site mutation promotes better splicing of E16. Protein 4.1 pre-mRNA bearing the natural weak 5′ splice site yielded 55% inclusion of exon 16 (E16wt), while pre-mRNA with a strong consensus 5′ splice site exhibited 85% inclusion of exon 16 (E16↑). (Experiment performed in 2E utilized a different oocyte preparation than the remainder of this figure, thus explaining the different baseline level of wild type E16 inclusion.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/2/10.1182_blood.v95.2.692/6/m_bloo00218002w.jpeg?Expires=1768269068&Signature=xn-WhhuAsR-3ElkrH5s7KOmfwguxGiAwehRZzVGRjquTjVWPD4Qjwj9J~fiGreOBDAllhdcrJveJltEosPjorS-lCFzXFpPxw70UlXmpFRI0P4D0~HNkpKiHrIquRfPW3zFhy9zoRLw3oskeH7w8dxKZXUrEnqymlM0ydvFpn~vdNe7yfrV7PIQplchLvFQyMudOg7Aumx9H~iNytVY4HOxM4Mxrkw4z8hNNq3ktsbr0PH8-rWS4px6LAvWGx6VZmdxMcV0GQEsPJMVaCcxnKtQVnSdBJdQB7ecX6yCqtRqw1RAhhOLVJ72fNJ1gmmgLsnugxbhdvMN8TF2YUhg3BQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)