STE20-related kinases play significant regulatory roles in a range of cellular responses to environmental stimuli. GCKR (also referred to as KHS1) is a serine/threonine protein kinase that has an STE20-like protein kinase domain and that stimulates the stress-activated protein kinase (SAPK, also referred to as Jun kinase or JNK) pathway. GCKR has a large C-terminal regulatory domain that provides sites for interactions with other proteins. Adaptor proteins mediate the interactions between signaling molecules. In this study we showed that the adaptor proteins Crk and CrkL associated with GCKR. When Crk-I, Crk-II, or CrkL was transiently expressed in HEK 293T cells along with GCKR, each coimmunoprecipitated with GCKR. Furthermore, in the Bcr-Abl transformed cell line, K562 endogenous GCKR and CrkL coimmunoprecipitated, indicating a constitutive association. Detection of the CrkL-GCKR interaction required the SH3 domains of CrkL and 2 regions in GCKR—1 between amino acids 387 and 395 that contains a consensus SH3 binding motif and the other between amino acids 599 and 696. Crk or CrkL overexpression increased GCKR catalytic activity. A dominant negative form of Ras abolished Crk- or CrkL-induced GCKR activation, suggesting a dependence on Ras activation for their activation of GCKR. Finally, we showed impairment of the known ability of CrkL to activate the SAPK pathway by a catalytically inactive form of GCKR or by a GCKR antisense construct. Thus, GCKR associates with other proteins through interactions mediated by SH2/SH3 adaptor proteins, which can lead to GCKR and SAPK activation.
Mammalian cells have several serine/threonine protein kinases related to the StE20 protein kinase in Saccharomyces cerevisiae. Ste20 is of particular interest because it regulates a mitogen-activated protein kinase (MAPK) pathway that controls the mating response of haploid yeast cells.1 Genetic epistasis analysis has positioned STE20 between the heterotrimeric G protein, which is activated by pheromone exposure, and the MAPK cassette. Ste20 associates through its N-terminal regulatory domain with the small GTPase Cdc42 and the scaffolding protein Ste5, interactions that result in the recruitment of STE20 to specific membrane sites.2Among the mammalian proteins related to STE20, the PAK family of protein kinases contains not only a related C-terminal kinase domain but a similar overall protein structure with an N-terminal regulatory domain that also contains a binding site for Cdc42 and for Rac.3 The PAK kinases have also been implicated in activating a MAPK cassette; however, instead of the MAPK pathway, the PAK kinases activate 2 related pathways, the stress-activated protein kinase (SAPK), also referred to as Jun kinase (JNK), and the p38 pathways.3 4
The germinal center kinase (GCK) family is another subfamily of serine/threonine protein kinases with a kinase domain related to that of STE20. Eleven mammalian family members have been identified.5 The GCK-related kinases can be subdivided into 2 broad groups based on their structural and functional properties. The group 1 GCKs are closely related to GCK itself and include GCK, GCK-related (GCKR, KHS), GCK-like kinase (GLK), hematopoietic progenitor kinase-1 (HPK1), and Nck-interacting kinase (NIK).6-12 These enzymes selectively activate SAPKs. The group 1 GCK family members also have an approximately 350-A region that can be divided into a leucine-rich domain and a 150-AA carboxyl terminal (CT) region. Studies of GCK and GCKR indicate that the CT region is required for binding to tumor necrosis factor (TNF) receptor-associated factor-2 (TRAF2).13,14 GCK, GCKR, and GLK are activated in vivo by TNF, a potent activator of the SAPK pathway.7,9,13 Furthermore, TNF stimulation results in the recruitment of GCKR to TRAF2.14
The carboxyl terminal region of the group 1 GCKs also includes at least 2 PEST motifs and a minimum of 2 polyproline-rich regions for binding proteins that contain SH3 domains. The regulatory domain of HPK1 has 4 proline-rich motifs that have been designated P1 to P4. Three of these, P1, P2, and P4, mediate Grb2 binding.10,15 GCKR shares the P2 and P4 proline-rich motifs with HPK1. GCK also shares 2 proline-rich motifs with HPK1, P3 and P4. Recently, HPK1 has been shown to interact with the first SH3 domain of the adaptor proteins Crk and CrkL.16 A peptide that spans the P2 region of HPK1 effectively blocks the binding of CrkL to a known CrkL SH3 ligand. Similar to HPK1, GCKR also bound Crk and CrkL.16
The adaptor protein Crk was originally discovered as an avian retrovirus encoding an oncogene product v-crk.17 The mammalian homologs of v-crk were subsequently identified as Crk-I and Crk-II, alternatively spliced forms of the same gene. Crk-II has an N-terminal SH2 domain followed by 2 SH3 domains, whereas Crk-1 has a single SH3 domain. In addition, a closely related gene, CrkL, was isolated and found to be constitutively phosphorylated in Bcr-Abl–transformed cells.18 The first SH3 domains of Crk and CrkL have similar binding specificities and are known to bind the guanine nucleotide exchange proteins C3G and SOS, the tryosine kinase Bcr-Abl, and DOCK18 0.19,20 A conserved lysine present in the defined binding motif PPxLPxK contributes significantly to the binding affinity and specificity of the first Crk SH3 domain. No binding specificities of the second SH3 domains of Crk-II and CrkL are known. The SH2 domain of Crk binds tyrosine-phosphorylated YxxP motifs present in such proteins as p130Cas, Cas-L, and paxillin.17,18 Transient expression of v-Crk, Crk-I, or Crk-II activated the SAPK pathway in several different cell types.21-24 In this study, we have explored the potential role of GCKR in Crk-induced SAPK activation.
Material and methods
Cell lines and cell culture
HEK 293 and HEK 293T cells were maintained in Dulbecco's minimal essential medium plus 10% fetal calf serum, whereas K562 cells were maintained in RPMI 1640 plus 10% fetal calf serum. HEK 293 and HEK 293T (SV40 T-antigen transformed) are human embryonic kidney cell lines that are readily transfected.
Plasmids and antibodies
The pMT3-HA-SAPK-p46β plasmid was provided by Dr J. Kyriakis (Harvard, Boston, MA). The pcDNA3-Ras-N17 was a kind gift of Dr S. Gutkind (National Institutes of Health, Bethesda, MD). Dr S. Gutkind provided the Crk-I and Crk-II expression vectors after obtaining permission from Dr M. Matsuda (National Institute of Health, Tokyo, Japan). The CrkL expression vector was obtained from Dr J. Groffen, (Children's Hospital, Los Angeles, CA), and the Bcr-Abl expression vector was obtained from Owen Witte (UCLA, Los Angeles, CA). The pcDNA-HA-GCKR, pCRIII-GCKR, pCRIII-GCKRT178A, and pCRIII-GCKR(AS) were described previously.7 The pcDNA-HA-GCKR deletion constructs 1-699, 1-599, 1-496, 1-396, 386-846, and 399-846 were created by inserting the appropriate polymerase chain reaction (PCR) product from pcDNA-HA-GCKR into pcDNA-H(A) The pEBG GST-CrkL1–109 was created by insertion of the appropriate PCR product in frame with the GST coding sequence in pEBG-GST (kindly provided by U. Siebenlist, National Institutes of Health). The orientation and veracity of each of the constructs were verified by nucleotide sequencing. The anti-HA (12CA), anti-pY4G10, and the anti-Crk and CrkL antibodies were purchased from Boehringer Mannheim (Indianapolis, IN), Upstate Biotechnology (Lake Placid, NY), and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. The GCKR antiserum was generated in rabbits by immunizing with a peptide (RKETEARDEMC) coupled to Keyhole limpet hemocyanin as described.7
In vitro kinase assays
Equal numbers of HEK 293 cells were plated on 10-cm plates at an approximate density of 5 × 105 cells/plate the day before transfection. The HEK 293T and HEK 293 cells were transiently transfected using a calcium phosphate method, and the K562 cells were transfected using Superfect (Qiagen, Valencia, CA). Transfected DNA levels were normalized with control plasmids. Forty-eight hours after the transfection, HA-immunoprecipitates were subjected to in vitro kinase assays using myelin basic protein or c-jun1–79 as substrates for the GCKR and SAPK assays, respectively.7 13Before the in vitro kinase assay, the HA-immunoprecipitates were washed 3 times with kinase lysis buffer (20 mmol/L Hepes, pH 7.4, 2 mmol/L EGTA, 50 mmol/L β-glycerophosphate, 1% Triton X-100, 1 mmol/L Na3V04, and 10% glycerol) to which a protease inhibitor cocktail tablet was added; Boehringer Mannheim), 3 times with a LiCl wash buffer (500 mmol/L LiCl, 100 mmol/L Tris, pH 7.4, 0.1% Triton X-100, and 1 mmol/L dithiothreitol), and 3 times with kinase buffer (20 mmol/L MOPS, pH 7.2, 2 mmol/L EGTA, 10 mmol/L MgCl2, and 0.1% Triton X-100). The GCKR (1:300 dilution), HA, and phosphotyrosine immunoblots were performed using standard methodology. The signals were detected by enhanced chemiluminescence (ECL; Amersham, Piscataway, NJ).
Coimmunoprecipitations
The coimmunoprecipitations were performed using lysates (20 mmol/L Tris, pH 8, 137 mmol/L NaCl, 2 mmol/L EDTA, 1% Triton X-100, 1 mmol/L sodium orthovanadate, plus protease inhibitors) prepared from HEK 293T cells coexpressing HA-GCKR or HA-GCKR truncation mutants (2 μg) and Crk-I, Crk-II, or CrkL or prepared from K562 cells. Anti-HA, anti-Crk, anti-CrkL, anti-Erk-1, anti-myc, anti-GCKR, or the 4G10 monoclonal antibody was added, and the immunoprecipitates were collected with the appropriate secondary antibody-coupled magnetic beads (Dynal, Glastrup, Denmark). They were washed 3 times in lysis buffer, twice in lysis buffer with 0.5 mol/L NaCl, fractionated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by immunoblotting with the appropriate antibody. The Bcr-Abl and GCKR coimmunoprecipitations in the presence or absence of CrkL were performed with lysates prepared from HEK 293T cells transfected with the indicated expression vectors.
Results
Crk-I, Crk-II, and CrkL all associate with GCKR
GCKR has 1 proline-rich sequence that could serve as an interaction site with Crk proteins, though a similar site in HPK1, P2, served as an interaction site for the C-terminal SH3 domain of Grb2.15In addition there are several other proline-rich sites in the regulatory domain of GCKR, but none fit the consensus sequence for a Crk SH3-domain-binding site. To test whether Crk or CrkL interacted with GCKR, we first coexpressed Crk-I or Crk-II along with an HA-tagged version of GCKR in HEK 293T cells. We examined Crk immunoprecipitates for the presence of HA-GCKR and HA immunoprecipitates for the presence of Crk proteins (Figure 1). The Crk immunoprecipitates from the Crk-I– and Crk-II–transfected cells contained HA-GCKR. In the converse experiment, the HA immunoprecipitates from the Crk-I– and Crk-II–transfected cells contained Crk-I and Crk-II, respectively. In addition some endogenous Crk-II could be detected coimmunoprecipitating with HA-GCKR in the Crk-I–transfected cells. An anti-Flag monoclonal antibody failed to immunoprecipitate either HA-GCKR or Crk proteins. Similar experiments revealed that CrkL and GCKR also coimmunoprecipitated (shown as a control in Figure 3 below).
Crk-I and Crk-II coimmunoprecipitate with GCKR. HEK 293T cells were transfected with constructs that directed the expression of HA-GCKR and Crk-I or Crk-II.
Lysates prepared from the transfected cells were immunoprecipitated with HA, FLAG (control antibody), or a Crk antibody as indicated. The immunoprecipitates were fractionated on SDS-PAGE, transferred, and blotted with the HA antibody (top) or the Crk antibody (bottom). Locations of HA-GCKR, Crk-II, and Crk-I are indicated with arrows as the immunoglobulin light chains. These experiments were performed 3 times with similar results.
Crk-I and Crk-II coimmunoprecipitate with GCKR. HEK 293T cells were transfected with constructs that directed the expression of HA-GCKR and Crk-I or Crk-II.
Lysates prepared from the transfected cells were immunoprecipitated with HA, FLAG (control antibody), or a Crk antibody as indicated. The immunoprecipitates were fractionated on SDS-PAGE, transferred, and blotted with the HA antibody (top) or the Crk antibody (bottom). Locations of HA-GCKR, Crk-II, and Crk-I are indicated with arrows as the immunoglobulin light chains. These experiments were performed 3 times with similar results.
Mapping the regions in GCKR and CrkL responsible for their interaction and demonstration of an association between the endogenous proteins
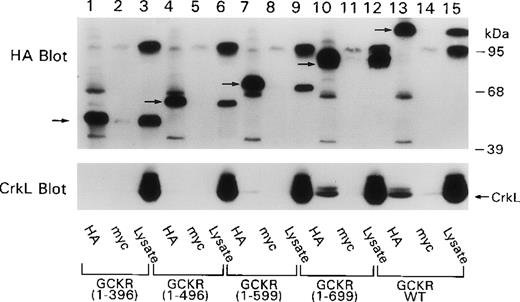
To analyze the site in GCKR that is important for interaction with Crk proteins, we expressed truncated HA-tagged GCKR proteins in HEK 293T cells along with CrkL, and we examined HA immunoprecipitates for the presence of CrkL using a CrkL-specific antibody. We found that CrkL readily coimmunoprecipitated with wild-type GCKR and with GCKR1–696 (Figure 2). However, the next truncation of GCKR, GCKR1–599, resulted in a significant reduction in the interaction. GCKR1–496 very weakly coimmunoprecipitated CrkL, and an interaction between GCKR1–396 and CrkL could only be detected on long exposures. This result was surprising because we had assumed that the P2 proline-rich site in GCKR, which is located at the C-terminus of GCKR1–396 and present in GCKR1–496 and GCKR1–596, would mediate the interaction with GCKR. These results suggested that the P2 site alone was insufficient to mediate a high-affinity interaction with GCKR.
Coimmunoprecipitation of CrkL with GCKR depends on the region in GCKR between amino acids 599 and 699.
HEK 293T cells were transfected with constructs that directed the expression of CrkL and GCKR or truncated versions of GCKR. Cell lysates and myc (control antibody) and HA antibody immunoprecipitates were analyzed by immunoblotting. The blot was sequentially reacted with the HA antibody (top) and the CrkL antibody (bottom). CrkL is indicated by an arrow. The truncated GCKR proteins can be seen in the cell lysate and in the HA immunoprecipitation lanes (arrows). Several nonspecific proteins are also present (top), the most prominent just below 95 kd in the cell lysate lanes.
Coimmunoprecipitation of CrkL with GCKR depends on the region in GCKR between amino acids 599 and 699.
HEK 293T cells were transfected with constructs that directed the expression of CrkL and GCKR or truncated versions of GCKR. Cell lysates and myc (control antibody) and HA antibody immunoprecipitates were analyzed by immunoblotting. The blot was sequentially reacted with the HA antibody (top) and the CrkL antibody (bottom). CrkL is indicated by an arrow. The truncated GCKR proteins can be seen in the cell lysate and in the HA immunoprecipitation lanes (arrows). Several nonspecific proteins are also present (top), the most prominent just below 95 kd in the cell lysate lanes.
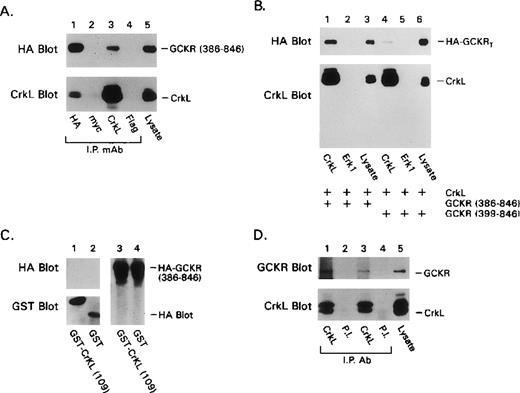
To examine the importance of the N-terminal portion of GCKR in the CrkL interaction, we truncated the first 385 amino acids of GCKR and created GCKR386–846, which lacked the GCKR catalytic domain, but retained the P2 site. The expressed truncated protein readily coimmunoprecipitated with CrkL, indicating that the catalytic domain of GCKR was not required for the interaction with CrkL (Figure3A). Next, by making a short additional truncation, we created a construct GCKR399–846, which expressed a version of GCKR that lacked the P2 site. In contrast to the results with GCKR386–846, GCKR399–846coimmunoprecipitated poorly with CrkL (Figure 3B). Based on this result, we expected that the SH3 domains of CrkL would be indispensable for its interaction with GCKR. When we expressed a truncated version of CrkL that lacked both SH3 domains as a GST-fusion protein in conjunction with either full-length GCKR (data not shown) or with GCKR386–846, we were unable to detect a significant interaction between the transfected proteins (Figure 3C). Although the truncated CrkL protein did not bind GCKR, it did extract several tyrosine-phosphorylated proteins from the cell lysate (data not shown). In sum, these experiments revealed that the amino acids 600-695 and 387-398 in GCKR and the SH3 domains of CrkL are required for an optimal association between the 2 proteins.
Further requirements for the interaction of GCKR and CrkL and demonstration of an association of the endogenous proteins.
(A) The regulatory domain of GCKR interacts with CrkL. Constructs directing the expression of HA-GCKR 386–846 and CrkL were cotransfected into HEK 293T cells. The indicated immunoprecipitates and a cell lysate were examined for either HA or CrkL expression. Anti-myc antibody was used as a control. (B) The P2 motif in GCKR was required to detect a strong GCKR/CrkL interaction. Constructs directing the expression of HA-GCKR386–846 (lanes 1-3); CrkL (lanes 1-6); and HA-GCKR399–846 (lanes 4-6) were transfected into HEK 293T. The indicated immunoprecipitates or cell lysates were analyzed for HA or CrkL expression by immunoblotting. The designation HA-GCKRT indicated both truncated forms of GCKR, whose mobilities on SDS-PAGE were indistinguishable. (C) The CrkL SH2 domain did not contribute to the interaction with the GCKR regulatory domain. Constructs directing the expression of HAGCKR386–846, GST (lanes 2, 4) or GST-CRKL1–109 (lanes 1, 3) were transfected into HEK 293T cells. Cell lysates extracted with glutathione-Sepharose 4B beads (Pharmacia Biotech AB) were analyzed for GST and HA-GCKR expression by immunoblotting. HA-GCKR386–846expression was similar in the GST- and the GST-CRKL1-109-transfected cells (lanes 3, 4). (D) GCKR and CrkL are constitutively associated in K562 cells. CrkL immunoprecipitates of cell lysates prepared from 10 × 106 million K562 cells or cell lysates from 0.5 × 106 cells were analyzed for GCKR expression using a GCKR specific antiserum. A preimmune antiserum (P.I.) was used as a control. Results from 2 different experiments (lanes 1, 2 and lanes 3, 4) are shown. Each of these experiments was preformed at least twice with similar results.
Further requirements for the interaction of GCKR and CrkL and demonstration of an association of the endogenous proteins.
(A) The regulatory domain of GCKR interacts with CrkL. Constructs directing the expression of HA-GCKR 386–846 and CrkL were cotransfected into HEK 293T cells. The indicated immunoprecipitates and a cell lysate were examined for either HA or CrkL expression. Anti-myc antibody was used as a control. (B) The P2 motif in GCKR was required to detect a strong GCKR/CrkL interaction. Constructs directing the expression of HA-GCKR386–846 (lanes 1-3); CrkL (lanes 1-6); and HA-GCKR399–846 (lanes 4-6) were transfected into HEK 293T. The indicated immunoprecipitates or cell lysates were analyzed for HA or CrkL expression by immunoblotting. The designation HA-GCKRT indicated both truncated forms of GCKR, whose mobilities on SDS-PAGE were indistinguishable. (C) The CrkL SH2 domain did not contribute to the interaction with the GCKR regulatory domain. Constructs directing the expression of HAGCKR386–846, GST (lanes 2, 4) or GST-CRKL1–109 (lanes 1, 3) were transfected into HEK 293T cells. Cell lysates extracted with glutathione-Sepharose 4B beads (Pharmacia Biotech AB) were analyzed for GST and HA-GCKR expression by immunoblotting. HA-GCKR386–846expression was similar in the GST- and the GST-CRKL1-109-transfected cells (lanes 3, 4). (D) GCKR and CrkL are constitutively associated in K562 cells. CrkL immunoprecipitates of cell lysates prepared from 10 × 106 million K562 cells or cell lysates from 0.5 × 106 cells were analyzed for GCKR expression using a GCKR specific antiserum. A preimmune antiserum (P.I.) was used as a control. Results from 2 different experiments (lanes 1, 2 and lanes 3, 4) are shown. Each of these experiments was preformed at least twice with similar results.
To examine whether an interaction occurred between endogenous Crk protein and GCKR, we examined whether CrkL and GCKR coimmunoprecipitated using cell lysates prepared from K562 cells. These cells are derived from human erythroleukemia and contain significant levels of CrkL and the Bcr-Abl oncogene. An antipeptide antibody raised against an internal peptide in GCKR was used to immunoblot CrkL immunoprecipitates. We found that they contained significant amounts of GCKR, whereas immunoprecipitates prepared using a control antiserum did not contain GCKR (Figure 3D). Thus, Crk-I, Crk-II, and CrkL can all interact with GCKR; in K562 cells, GCKR and CrkL associate constitutively. However, the constitutive association of GCKR and CrkL in K562 may not indicate that this is the case in other cell types because the presence of Bcr-Abl may alter either GCKR or CrkL.
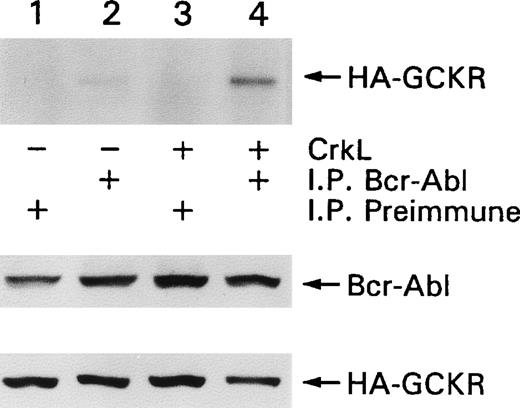
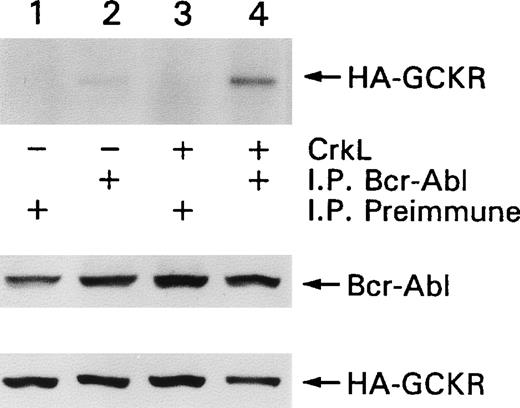
Because we showed an association between Bcr-Abl and GCKR,25 we determined whether the presence of CrkL enhanced the ability to detect an association between those 2 proteins. HEK 293T cells were transfected with constructs expressing Bcr-Abl and GCKR in the presence or absence of CrkL. The coexpression of CrkL enhanced the amount of HA-GCKR immunoprecipitating with Bcr-Abl (Figure4). The levels of Bcr-Abl and HA-GCKR expressed were unaltered by the concomitant expression of CrkL. Thus, CrkL enhanced the association between Bcr-Abl and GCKR.
CrkL enhances the association of Bcr-Abl and GCKR.
HEK 293T cells were transfected with constructs that directed the expression of Bcr-Abl and HA-GCKR in the presence or absence of CrkL (lanes 1, 2 versus lanes 3, 4). Bcr-Abl (lanes 2, 4) or control (lanes 1, 3) immunoprecipitates were analyzed by immunoblotting for HA-GCKR. Cell lysates from the same transfection were analyzed for Bcr-Abl and HA-GCKR by immunoblotting with a Bcr-Abl or an HA antibody. This experiment was preformed twice with similar results.
CrkL enhances the association of Bcr-Abl and GCKR.
HEK 293T cells were transfected with constructs that directed the expression of Bcr-Abl and HA-GCKR in the presence or absence of CrkL (lanes 1, 2 versus lanes 3, 4). Bcr-Abl (lanes 2, 4) or control (lanes 1, 3) immunoprecipitates were analyzed by immunoblotting for HA-GCKR. Cell lysates from the same transfection were analyzed for Bcr-Abl and HA-GCKR by immunoblotting with a Bcr-Abl or an HA antibody. This experiment was preformed twice with similar results.
Overexpression of Crk-I, Crk-II, or CrkL increases GCKR kinase activity
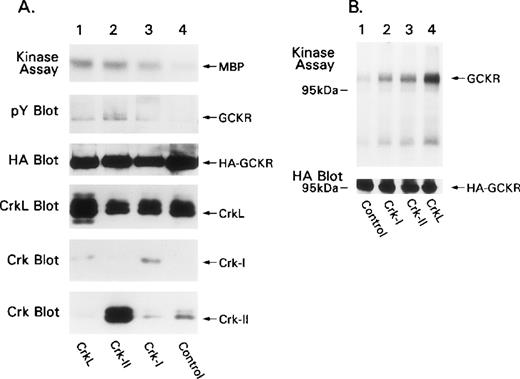
To examine the effects of Crk-I, Crk-II, and CrkL on GCKR kinase activity, we cotransfected HEK 293T with expression constructs for the Crk proteins and HA-GCKR. We subjected HA-GCKR immunoprecipitates to an in vitro kinase assay using the substrate myelin basic protein. We found that Crk-I, Crk-II, and CrkL increased HA-GCKR kinase activity approximately 4.8-, 10.3-, and 11-fold, respectively, compared with HA-GCKR immunoprecipitated from control cells (Figure5A, top). Crk-I appeared not to express as well as Crk-II, though the Crk antibody did not detect Crk-I as well as Crk-II. The coexpression of the Crk proteins with GCKR resulted in minor increases in the amount of GCKR tyrosine phosphorylation detected by phosphotyrosine immunoblotting of HA-GCKR immunoprecipitates (Figure5A, second panel). The expression of Crk proteins enhanced the amount of GCKR phosphorylation in the in vitro kinase assay. Compared with control transfections, Crk-I, Crk-II, and CrkL increased the amount of GCKR phosphorylation 5.1-, 6.2-, and 10.9-fold, respectively (Figure5B, top). The identity of the coimmunoprecipitating phosphorylated band with a more rapid mobility than GCKR is unknown.
Crk proteins activate GCKR kinase activity.
(A) Coexpression of Crk proteins enhances GCKR kinase activity. HEK 293T was transfected with a control or with constructs that direct the expression of Crk-I, Crk-II, or CrkL and HA-GCKR. HA-immunoprecipitates were subjected to an in vitro kinase assay using myelin basic protein (MBP) as a substrate (autoradiograph, labeled as kinase assay). The SDS-PAGE fractionated in vitro kinase assay was analyzed by immunoblotting for phosphotyrosine (pY blot). In addition, cell lysates prepared from the same transfections were analyzed by immunoblotting for HA-GCKR (HA blot), CrkL (CrkL blot), and Crk-I and Crk-II (Crk blot). (B) Coexpression of Crk proteins triggers GCKR phosphorylation. HA-GCKR immunoprecipitates from cells transfected as in A were subjected to an in vitro kinase assay in the absence of an exogenous substrate (autoradiograph, labeled as kinase assay). Cell lysate analyzed for HA-GCKR expression by immunoblotting. Each of these experiments was performed at least twice.
Crk proteins activate GCKR kinase activity.
(A) Coexpression of Crk proteins enhances GCKR kinase activity. HEK 293T was transfected with a control or with constructs that direct the expression of Crk-I, Crk-II, or CrkL and HA-GCKR. HA-immunoprecipitates were subjected to an in vitro kinase assay using myelin basic protein (MBP) as a substrate (autoradiograph, labeled as kinase assay). The SDS-PAGE fractionated in vitro kinase assay was analyzed by immunoblotting for phosphotyrosine (pY blot). In addition, cell lysates prepared from the same transfections were analyzed by immunoblotting for HA-GCKR (HA blot), CrkL (CrkL blot), and Crk-I and Crk-II (Crk blot). (B) Coexpression of Crk proteins triggers GCKR phosphorylation. HA-GCKR immunoprecipitates from cells transfected as in A were subjected to an in vitro kinase assay in the absence of an exogenous substrate (autoradiograph, labeled as kinase assay). Cell lysate analyzed for HA-GCKR expression by immunoblotting. Each of these experiments was performed at least twice.
Crk-I, Crk-II, and CrkL activate GCKR through a Ras-dependent mechanism
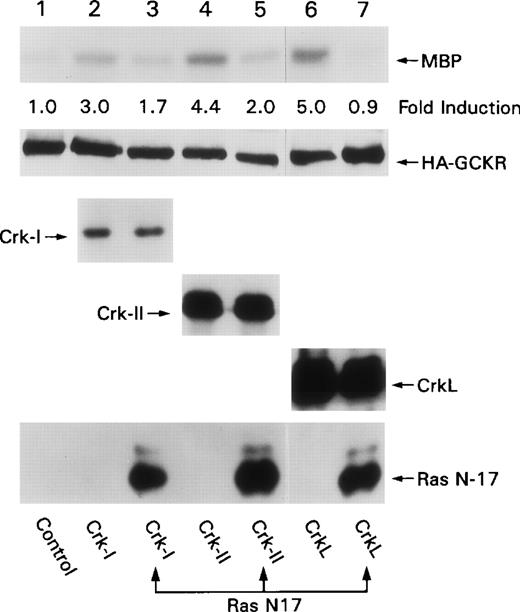
The first SH3 domain of Crk or CrkL binds SOS and C3G.19,20 SOS is a nucleotide exchange factor for Ras, whereas C3G is a nucleotide exchange factor for Rap1. Several pieces of evidence indicate that Ras is important in Crk–induced signaling. A dominant negative form of Ras suppressed v-Crk–induced transformation of NIH 3T3 cells and CrkL-induced transformation of Rat-1 fibroblasts, and it inhibited Crk-II–induced apoptosis of HEK 293 cell.21,26,27 To examine whether Ras was important in Crk-induced GCKR activation, we made use of a dominant negative form of Ras, Ras N-17. We coexpressed expression vectors for Crk-I, Crk-II, or CrkL with HA-GCKR and with either Ras N-17 or a control plasmid. We found that the presence of Ras N-17 resulted in a significant reduction in Crk-induced GCKR activation (Figure 6). The expression of Ras N-17 did not alter the expression of any of the Crk proteins. These data are similar to those we have observed with Bcr-Abl–induced GCKR activation, where Ras N-17 also blocks.25 In contrast, Ras N-17 has no effect on TNF-induced GCKR activation (Shi C-S, unpublished observation).
Ras N-17 blocks Crk-induced GCKR activation. HEK 293T cells were cotransfected constructs directing the expression of HA-GCKR in conjunction with Crk-I (lanes 2, 3), Crk-II (lanes 4, 5), CrkL (lanes 6, 7), or a control plasmid (lane 1) in the presence (lanes 3, 5, 7) or absence of Ras N-17 (lanes 1, 2, 4, 6).
HA-GCKR immunoprecipitates were assayed for activity using an in vitro kinase with MBP as a substrate. The fold induction compared to HA-GCKR immunoprecipitate from the control transfection is indicated. Levels of Crk-I, Crk-II, CrkL, Ras N-17, and HA-GCKR expression as detected by immunoblotting are shown. These experiments were performed twice.
Ras N-17 blocks Crk-induced GCKR activation. HEK 293T cells were cotransfected constructs directing the expression of HA-GCKR in conjunction with Crk-I (lanes 2, 3), Crk-II (lanes 4, 5), CrkL (lanes 6, 7), or a control plasmid (lane 1) in the presence (lanes 3, 5, 7) or absence of Ras N-17 (lanes 1, 2, 4, 6).
HA-GCKR immunoprecipitates were assayed for activity using an in vitro kinase with MBP as a substrate. The fold induction compared to HA-GCKR immunoprecipitate from the control transfection is indicated. Levels of Crk-I, Crk-II, CrkL, Ras N-17, and HA-GCKR expression as detected by immunoblotting are shown. These experiments were performed twice.
The activation of the SAPK pathway by Crk-I, Crk-II, or CrkL is partially blocked by a GCKR antisense construct or a dominant negative form of GCKR
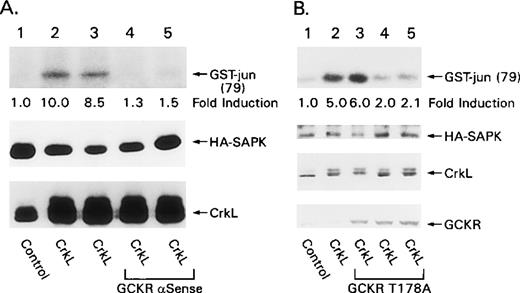
Transient overexpression of v-Crk-, Crk-I-, or Crk-II-activated SAPK in HEK 293 cells, whereas the overexpression of CrkL activated the SAPK pathway in Rat-1 fibroblasts.21-24 Because GCKR is expressed in HEK 293 cells,7 we sought to determine whether GCKR activation contributes to activation of the SAPK pathway after Crk overexpression. For these experiments, we used an antisense GCKR construct known to reduce the endogenous level of GCKR in HEK 293 cells and a dominant negative form of GCKR that inhibits TNF-induced SAPK activation and Bcr-Abl– induced SAPK activation.7 25 We first examined whether the GCKR antisense construct inhibited Crk-I- and Crk-II-induced SAPK activation. HEK 293 cells were cotransfected with either Crk-I or Crk-II in the presence or absence of a construct that expresses antisense GCKR RNA. The coexpression of GCKR antisense partially inhibited both Crk-I– and Crk-II–induced SAPK activation (Figure 7). The antisense construct did not alter the expression of either Crk-I or Crk-II. Similarly, the GCKR antisense construct inhibited SAPK activation by CrkL. In addition, the coexpression of the kinase-deficient form of GCKR partially blocked CrkL-induced SAPK activation (Figure 8). The coexpression of GCKR T178A did not alter the levels of HA-SAPK or CrkL in the transfected cells. In both sets of experiments, we often failed to see a significant inhibition of Crk- or CrkL-induced SAPK activation until we reached a critical level of expression of the inhibitor (antisense- or kinase-dead GCKR). Neither GCKR antisense nor GCKRT178A significantly impaired MEKK1-induced SAPK activation (data not shown).
Evidence for GCKR involvement in Crk-I- and Crk-II-induced SAPK activation.
HEK 293 cells were cotransfected with constructs that direct the expression of HA-SAPK (lanes 1 to 6), Crk-II (lanes 1, 2), Crk-I (lanes 3, 4), or GCKR (lane 5) in the presence (lanes 1, 3) or absence (lanes 2, 4 to 6) of a construct that expressed a GCKR antisense RNA. HA-SAPK immunoprecipitates were subjected to an in vitro kinase assay using GST-jun1–79 as a substrate. The fold induction compared to HA-SAPK alone is indicated. The levels of HA-SAPK, Crk-II, and Crk-I as detected by immunoblotting are shown below. These experiments were performed twice with similar results.
Evidence for GCKR involvement in Crk-I- and Crk-II-induced SAPK activation.
HEK 293 cells were cotransfected with constructs that direct the expression of HA-SAPK (lanes 1 to 6), Crk-II (lanes 1, 2), Crk-I (lanes 3, 4), or GCKR (lane 5) in the presence (lanes 1, 3) or absence (lanes 2, 4 to 6) of a construct that expressed a GCKR antisense RNA. HA-SAPK immunoprecipitates were subjected to an in vitro kinase assay using GST-jun1–79 as a substrate. The fold induction compared to HA-SAPK alone is indicated. The levels of HA-SAPK, Crk-II, and Crk-I as detected by immunoblotting are shown below. These experiments were performed twice with similar results.
Evidence that GCKR is required for CrkL-induced SAPK activation.
(A) A GCKR antisense construct inhibits CrkL-induced SAPK activation. HEK 293 cells were cotransfected with constructs that direct the expression of HA-SAPK (lanes 1 to 5), CrkL (lanes 2 to 5), and a GCKR antisense RNA (lanes 4, 5). HA-SAPK immunoprecipitates were subjected to an in vitro kinase assay using GST-jun1–79 as a substrate. The fold induction compared to the control is shown below the in vitro kinase assay result. Levels of HA-SAPK and CrkL expression are shown as detected by immunoblotting. (B) GCKRT178A inhibits CrkL-induced SAPK activation. HEK 293 cells were cotransfected with constructs that direct the expression of HA-SAPK (lanes 1 to 5), CrkL (lanes 2 to 5), and increasing concentrations of GCKRT178A (1, 2, and 3 μg/mL, lanes 3 to 5, respectively). HA-SAPK immunoprecipitates were subjected to an in vitro kinase assay using GST-jun1–79 as a substrate. The fold induction compared to cells transfected only with the construct directing HA-SAPK expression is indicated below the autoradiograph. Levels of HA-SAPK, CrkL, and GCKRT178A as detected by immunoblotting are shown. Low levels of endogenous GCKR are detected (lanes 1, 2, bottom). These experiments were performed twice with similar results.
Evidence that GCKR is required for CrkL-induced SAPK activation.
(A) A GCKR antisense construct inhibits CrkL-induced SAPK activation. HEK 293 cells were cotransfected with constructs that direct the expression of HA-SAPK (lanes 1 to 5), CrkL (lanes 2 to 5), and a GCKR antisense RNA (lanes 4, 5). HA-SAPK immunoprecipitates were subjected to an in vitro kinase assay using GST-jun1–79 as a substrate. The fold induction compared to the control is shown below the in vitro kinase assay result. Levels of HA-SAPK and CrkL expression are shown as detected by immunoblotting. (B) GCKRT178A inhibits CrkL-induced SAPK activation. HEK 293 cells were cotransfected with constructs that direct the expression of HA-SAPK (lanes 1 to 5), CrkL (lanes 2 to 5), and increasing concentrations of GCKRT178A (1, 2, and 3 μg/mL, lanes 3 to 5, respectively). HA-SAPK immunoprecipitates were subjected to an in vitro kinase assay using GST-jun1–79 as a substrate. The fold induction compared to cells transfected only with the construct directing HA-SAPK expression is indicated below the autoradiograph. Levels of HA-SAPK, CrkL, and GCKRT178A as detected by immunoblotting are shown. Low levels of endogenous GCKR are detected (lanes 1, 2, bottom). These experiments were performed twice with similar results.
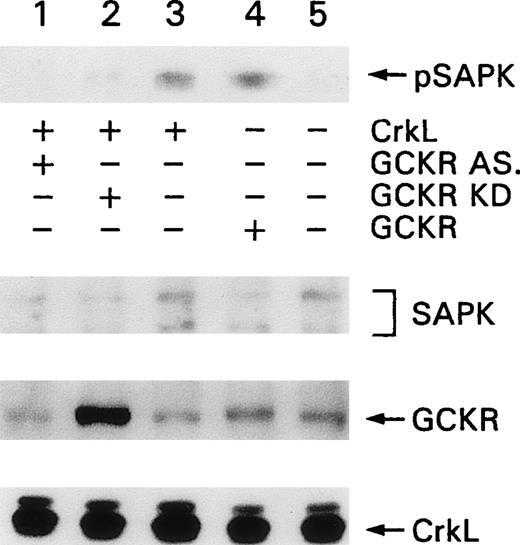
To examine whether CrkL overexpression could enhance SAPK activity and to define the role of endogenous GCKR in SAPK activation by CrkL, we transfected K562 cells with CrkL and measured SAPK activation by immunoblotting for phosphorylated SAPK (pSAPK). We found that the transfection of either GCKR or CrkL enhanced the levels of pSAPK in K562 cells and that the concomitant expression of either the GCKR antisense construct or the kinase inactive form of GCKR inhibited this increase (Figure 9, top). Endogenous SAPK and endogenous and transfected levels of GCKR and CrkL were detected by immunoblotting. The antisense form of GCKR reduced the level of endogenous GCKR by approximately 40%, and the presence of the kinase inactive form of GCKR accounted for the enhanced GCKR band (Figure 9, lanes 1 and 2, fourth panel, respectively).
GCKR involvement in CrkL-induced SAPK activation in K562 cells.
K562 cells were transfected with contructs directing the expression of CrkL (lanes 1 to 3), antisense GCKR (lane 1), kinase inactive GCKR (lane 2), or wild-type GCKR (lane 4). Levels of phosphorylated SAPK were detected with an antibody specific for pSAPK. Levels of SAPK, GCKR, and CrkL in the cell lysates were detected by immunoblotting with SAPK, GCKR, and CrkL-specific antibodies. This experiment was performed twice with similar results.
GCKR involvement in CrkL-induced SAPK activation in K562 cells.
K562 cells were transfected with contructs directing the expression of CrkL (lanes 1 to 3), antisense GCKR (lane 1), kinase inactive GCKR (lane 2), or wild-type GCKR (lane 4). Levels of phosphorylated SAPK were detected with an antibody specific for pSAPK. Levels of SAPK, GCKR, and CrkL in the cell lysates were detected by immunoblotting with SAPK, GCKR, and CrkL-specific antibodies. This experiment was performed twice with similar results.
Discussion
We have provided evidence that the Crk adapter proteins interact with the serine/threonine kinase GCKR and likely regulate its activity. In support of this conclusion, GCKR coimmunoprecipitated with Crk-I, Crk-II, and CrkL after transient transfection in HEK 293T and was found constitutively associated with CrkL in K562 cells. Two regions in GCKR are important for its interaction with CrkL, which were localized to amino acids 386-398 and 599-696. That CrkL-GCKR interaction may modulate GCKR function was supported by experiments showing that overexpression of Crk-I, Crk-II, or CrkL enhanced GCKR kinase activity. Implicating Ras activation in this process, the concomitant expression of a dominant negative form of Ras blocked Crk-triggered GCKR kinase activity. That GCKR activation contributed to Crk-induced SAPK activation was based on data generated with a GCKR antisense and the GCKR T178A constructs.
In the course of these experiments, the GCKR-related kinase HPK1 was shown to bind selectively to the first SH3 domain of c-Crk and CrkL.16 Based on a series of binding experiments, the P2 site in HPK1—referred to as M2—appeared to be important in mediating the binding. GCKR, designated as KHS, was also shown to bind to Crk proteins.16 Based on the shared P2 site with HPK1, that region in GCKR was implicated in mediating the GCKR-Crk interaction. The similarity of the P2 site in GCKR to known Crk SH3–binding sites19 20 suggested to us that this proline-rich site would confer on GCKR the ability to interact with Crk proteins. Indeed, the association of GCKR with Crk-I, Crk-II, and CrkL was readily apparent on cotransfection of the appropriate expression vectors into HEK 293T cells and endogenous CrkL, and GCKR could be coimmunoprecipitated from K562 cells. Consistent with an SH3 domain–mediated interaction between GCKR and the Crk proteins, deletion of the CrkL SH3 domains abolished our ability to detect an interaction between GCKR and CrkL. Although the P2 site in GCKR is likely indispensable for a high-affinity interaction between GCKR and CrkL, the region in GCKR between amino acids 599 and 699 also appears important. This region contains 2 sites that are weakly homologous to the consensus CrkL SH3–binding site, PDRILPRK609 and PLPSPLN673. However, whether these regions contribute to the Crk-GCKR interaction requires further study. An alternative possibility is that the region in GCKR between amino acids 599 and 699 does not directly bind to Crk proteins but that it serves as a homodimerization interface that stabilizes the Crk-GCKR interaction.
CrkL is a prominent substrate of the oncoprotein Bcr-Abl, and the first SH3 domain of CrkL binds Bcr-Abl.20,21 We have previously shown that GCKR is constitutively associated with the Bcr-Abl in K562 cell.25 However, our ability to coimmunoprecipitate the 2 proteins depended on the presence of the CT region, which is distinct from those important for interaction with CrkL. Thus, it may be possible to assemble a trimolecular complex that contains GCKR, Bcr-Abl, and CrkL or other Crk proteins. Consistent with that possibility, CrkL overexpression enhanced the ability to detect an association between Bcr-Abl and GCKR. Furthermore, Crk proteins should link GCKR to other signaling or docking molecules by virtue of their SH2 domains, which can bind tyrosine-phosphorylated proteins such as CBL, HEF1, members of the p130Cas family, or paxillin. These proteins may modulate GCKR activity or provide a mechanism to translocate GCKR to sites of action.
Several studies have reported that the overexpression of Crk proteins leads to the activation of the SAPK pathway but not the MAPK pathway.21-24 In HEK 293T cells, Crk has been reported to activate the SAPK pathway by the guanine–nucleotide exchange protein C3G through a Ras-independent mechanism.21,27Kinase-negative28 forms of the mixed lineage kinase (MLK) and the dual leucine zipper kinase (DLK) inhibited C3G-induced SAPK activation, suggesting their involvement in Crk-induced SAPK activation. In this model Crk recruits C3G, which acts as an exchange factor for a small GTPase whose identity is unknown. The activated GTPase results in MLK3 activation, which leads to MEKK1 and subsequent SAPK activation or to DLK activation and entrance into the SAPK cascade at the level of Sek1/MKK4.29-32 In contrast to these studies in COS-7 cells, Crk-induced SAPK activation depended on the activation of Ras and Rac because dominant negative forms of either small GTPase ablated the Crk-induced SAPK activation.24 In the same study, Crk-induced SAPK was shown to depend on both the SH3 and the SH2 domains of Crk, suggesting that a signaling complex must be assembled and indicating the probable involvement of p130Cas. Consistent with that possibility, overexpression of p130Cas also activated the SAPK pathway, though weakly. In this model Crk recruits SOS, DOCK180, or both, which leads to Ras and Rac activation. Rac activates a PAK kinase or MLK3, which leads to MEKK1 and subsequent SAPK activation.24
In this article, we provide several pieces of evidence for the participation of GCKR in Crk-mediated SAPK activation in HEK 293 cells. First, GCKR is present in COS-7 and HEK 293 cells, and it is a potent SAPK activator in them.7 Second, the transient expression of Crk results in Ras activation, which is either a direct or an indirect GCKR activator,25 thus providing a mechanism by which Crk could activate GCKR. Indeed a dominant negative form of Ras blocked Crk-induced GCKR activation. Third, the interaction of Crk proteins with GCKR links GCKR to other proteins that may regulate its activity. Fourth, a reduction of GCKR protein levels using the GCKR antisense construct impairs Crk-mediated SAPK activation, and a kinase-dead form of GCKR inhibits the ability of Crk proteins to activate SAPK.
How do we reconcile this information with the previously discussed models of Crk-induced SAPK activation? GCKR could act upstream of MLK3, providing a linear pathway initiated by Crk- or CrkL-binding GCKR. The Crk–GCKR complex could recruit through the GCKR C-terminal SH3-binding domain (amino acids 486-490) MLK3/DLK. A similar region in HPK1 has been shown to mediate binding to MLK3.11 In a physiologic setting, tyrosine phosphorylation of a receptor could assemble a complex of Crk, GCKR, and MLK3 (or DLK) at the membrane in which Rac1 or Cdc42 activates the complex by MLK3 dimerization through its leucine zippers. Recently, MLK3 dimerization was shown to activate its kinase activity.33 Alternatively, Crk-mediated Ras activation could lead to GCKR and subsequent SAPK activation pathway through MEKK1. There is evidence that several of the GCK family members signal the SAPK pathway through MEKK1.9,12,34 GCK has been shown to interact directly with MEKK1. This interaction is dependent on the CT region and its third PEST region.34 The CT region is conserved with GCKR, and arguing that GCKR can also activate the SAPK pathway through MEKK1, a construct expressing a MEKK1 dominant negative, impairs GKCR-induced SAPK activation.7
Delineating how GCK family members traffic within the cell and interact with other molecules will be crucial for understanding how MAPK signaling modules couple to their activators. Both direct- and adaptor-mediated interactions between GCK family members and other signaling molecules have been found, and their consequences for GCK family member function is just emerging. Because some MAP3Ks such as c-raf undergo stimulus-induced membrane translocation for their activation, the colocalization of GCK family members with them provides an attractive hypothesis, whereby specific GCK family members activate distinct MAP3Ks. Crk proteins may serve such a role for GCKR.
Acknowledgments
We thank Mary Rust for her fine editorial assistance and Dr Anthony Fauci for his continued support. We thank Dr John Kyriakis for his constructive discussions.
Reprints:John H. Kehrl, Laboratory of Immunoregulation, NIAID, National Institutes of Health, Building 10, Room 11B08, 10 Center Drive, MSC 1876, Bethesda, MD 20892-1876; e-mail:jkehrl@atlas.niaid.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.