To characterize the molecular mechanisms of platelet secretion, we focused on the calcium-induced exocytosis of dense core granules. Platelets contain several known t-SNAREs (soluble N-ethylmaleimide sensitive factor [NSF] attachment protein receptors) such as syntaxins 2, 4, and 7 and SNAP-23 (synaptosomal associated protein 23). By using an in vitro exocytosis assay, we have been able to assign roles for some of these t-SNAREs in dense core granule release. This calcium-induced secretion relies on the SNARE proteins because it is stimulated by the addition of recombinant -SNAP and inhibited by a dominant negative -SNAP–L294A mutant or by anti–-SNAP and anti-NSF antibodies. SNAP-23 antibodies and an inhibitory C-terminal SNAP-23 peptide both blocked dense core granule release, demonstrating a role for SNAP-23. Unlike other cell types, platelets contain a significant pool of soluble SNAP-23, which does not partition into Triton X-114. Of the anti-syntaxin antibodies tested, only anti–syntaxin 2 antibody inhibited dense core granule release. Immunoprecipitation studies showed that the 2 t-SNAREs syntaxin 2 and SNAP-23 do form a complex in vivo. These data clearly show that SNAPs, NSF, and specific t-SNAREs are used for dense core granule release; these data provide a greater understanding of regulated exocytosis in platelets.
Platelet activation and the release reaction can be summarized as a progression through 3 steps: (1) an activating event caused by contact with an agonist such as thromboxane, platelet activating factor, collagen, or thrombin; (2) the generation of intracellular signals through such molecules as G proteins, phospholipase C, phospholipase A, and protein kinase C, and calcium efflux from the dense tubular system or influx from the outside; and (3) a set of cellular responses that include cytoskeletal rearrangement and exocytosis of storage granules (reviewed in1). Platelets contain 3 types of granules: dense core granules, containing such small molecules as ADP, serotonin, calcium, and pyrophosphate; α-granules, containing such proteins as von Willebrand factor, thromboglobulin, and platelet-derived growth factor (PDGF); and lysosomes, containing acid hydrolases (reviewed in 1). After platelet activation, the contents of these granules are released into the extraplatelet space. The exocytotic pathway in platelets is unique. As the platelet changes shape upon stimulation, the secretory granules become increasingly centralized within a constricting microtubular coil. These granules then either fuse directly with the invaginations of the plasma membrane, called the open canalicular system, or fuse with one another (compound fusion) and then with the open canalicular system.2-4 Although the molecular mechanism of signal transduction has been studied extensively, the mechanism of granule to plasma membrane fusion is still unclear.
The molecular mechanisms of membrane fusion events in other systems have been studied intensively over the past few years.5-7 A growing body of data supports the concept that membrane proteins from both the transport vesicle and target membrane are, at least in part, responsible for the specific fusion of the 2 lipid bilayers. As originally stated,8 the SNARE (soluble N-ethylmaleimide sensitive factor [NSF] attachment protein receptor) hypothesis proposed that a vesicle membrane protein from the synaptobrevin/VAMP (vesicle associated membrane protein) family (v-SNARE) binds specifically to a heterodimeric complex in the target membrane (t-SNARE) made up of 1 member of the syntaxin family and 1 from the synaptosomal associated protein (SNAP)-23/25 family. The resulting heterotrimeric, intermembrane complex is the core complex that is minimally required for membrane fusion.9 It is also clear from this body of work that there are numerous accessory proteins, such as SNAPs and NSF, that “activate” the SNARE proteins so that they attain fusion-competent configurations.10,11 Although the specific details of the SNARE hypothesis continue to evolve,12-14 it has served as a useful guide to dissecting the mechanisms of exocytosis events.
In our initial studies of the molecular machinery of platelet exocytosis, we reported that platelets contain the general accessory proteins α-SNAP, γ-SNAP, and NSF, as well as 2 specific plasma membrane t-SNAREs, syntaxin 2 and syntaxin 4.15 Subsequent work by others using antibody inhibition experiments has shown that syntaxin 4 mediates α-granule release.16 As to the heterodimeric partner t-SNARE, SNAP-25 was undetectable in platelets, so SNAP-23 became a good candidate. Several studies have indicated that SNAP-23 can heterodimerize with each of the plasma membrane syntaxins (i.e., 1, 2, 3, and 4).17-20 SNAP-23 is expressed in numerous tissues, where it has been localized to the plasma membrane,21-24 mast cell granules,25 and endosome.26 It has been detected in platelets and was suggested to play a role in α-granule release.16 As for v-SNAREs, only 1 has been described in platelets, VAMP-3/cellubrevin.27 Although it is clear from botulinum toxin–based studies that at least 1 v-SNARE is required for α-granule exocytosis,16 it is unclear whether it is VAMP-3/cellubrevin. Taken together, these data show that many new insights into the mechanisms of platelet exocytosis are being made, especially regarding α-granule release, yet it is clear that there are other, yet to be identified, components that play a role in dense core granule and lysosome exocytosis.
In this report, we demonstrate that dense core granule secretion is mediated by the general membrane-fusion components such as α-SNAP and NSF. Using an in vitro exocytosis assay, we also provide evidence that the t-SNAREs SNAP-23 and syntaxin 2, but not syntaxin 4 or 7, are involved in dense core granule exocytosis from platelets.
Materials and methods
Antibodies and reagents
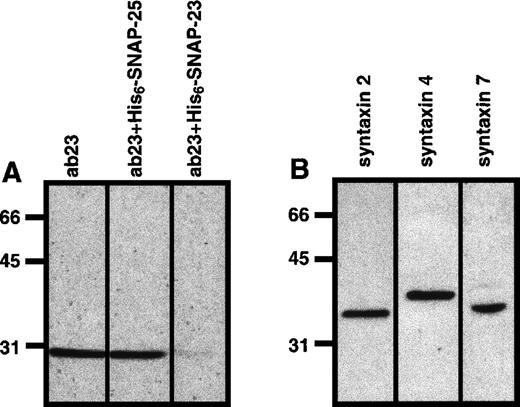
Polyclonal anti–α-SNAP, anti–SNAP-23, and anti–syntaxin 2 and 4 antibodies were generated by immunizing rabbits with appropriate recombinant proteins.26,28 The lack of cross-reactivity with SNAP-25 by our anti–SNAP-23 antibody is demonstrated in Figure1A. Anti–syntaxin 7 antibodies were also produced in our laboratory using a recombinant protein generated from a human expressed sequence tag (Accession N31 042; Genome Systems Inc, St Louis, MO). All antibodies were affinity purified with the appropriate recombinant protein or protein G Sepharose (Pharmacia, Piscataway, NJ). Fab fragments of ab23 (Fab23) were prepared using the ImmunoPure Fab preparation kit (Pierce, Rockford, IL). It should be noted that the platelet forms of the 3 syntaxins differ in apparent molecular weight (Figure 1B). These data demonstrate the specificity of the antibody reagents used in this study because none of our syntaxin-specific antibodies cross-reacted with an inappropriate syntaxin. The anti-NSF monoclonal antibody 2E529 30 was prepared from ascites and purified on protein G Sepharose (Pharmacia).
t-SNAREs in the platelet.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting analyses were used to demonstrate the presence of SNAP-23 (A) and syntaxins 2, 4, and 7 (B) in platelets. One hundred micrograms of whole platelet lysate was separated by SDS-PAGE and then transferred to nitrocellulose. The resulting blots were probed with anti–syntaxin 2 (syntaxin 2), anti–syntaxin 4 (syntaxin 4), and anti–syntaxin 7 (syntaxin 7), and the immunodecorated proteins were detected by ECL. For A, the anti–SNAP-23 antibody was preincubated with 500 μg of either recombinant SNAP-23 or SNAP-25, as indicated.
t-SNAREs in the platelet.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting analyses were used to demonstrate the presence of SNAP-23 (A) and syntaxins 2, 4, and 7 (B) in platelets. One hundred micrograms of whole platelet lysate was separated by SDS-PAGE and then transferred to nitrocellulose. The resulting blots were probed with anti–syntaxin 2 (syntaxin 2), anti–syntaxin 4 (syntaxin 4), and anti–syntaxin 7 (syntaxin 7), and the immunodecorated proteins were detected by ECL. For A, the anti–SNAP-23 antibody was preincubated with 500 μg of either recombinant SNAP-23 or SNAP-25, as indicated.
Reduced streptolysin O (SLO) was purchased from Murex (Dartford, UK). The SNAP-23 C-terminal peptide (ANARAKKLIDS) was a generous gift from Dr. David Castle (University of Virginia, Charlottesville, VA). Thrombin, apyrase VII, heparin, prostaglandin I2, sodium pyruvate, P-nitrophenyl-N-acetyl-β-D-glucosaminide, and NADH were purchased from Sigma (St. Louis, MO). [1,2-3H(N)]-hydroxytryptamine ([3H]5-HT) was purchased from NEN (Boston, MA). All other chemicals were of reagent grade.
Complementary DNAs encoding α-SNAP and α-SNAP mutant (L294A) were inserted into the vector pQE-9 (Qiagen, Chatsworth, CA) using theHindIII and BamHI restriction sites. Constructs were transfected into Escherichia coli M15 pREP4 cells. The transfected cells were selected with ampicillin (100 μg/mL) and kanamycin (50 μg/mL). The constructs were confirmed by dideoxyribose nucleic acid sequencing. Production of recombinant proteins was performed as described previously.31
Freshly banked platelets were procured as units from the Central Kentucky Blood Center (Lexington, KY).
Immunoprecipitation and western blotting
For co-immunoprecipitation experiments, ab23 was covalently coupled to protein G Superose using dimethyl pimelimidate.8 Fifty micrograms of detergent-solubilized platelet extract was incubated with the antibody beads at 4°C for 2 hours. Supernatants were collected, the beads were washed 5 times with phosphate-buffered saline (PBS) and 1% Triton X-100, and the bound material was eluted with 100 mmol/L glycine, pH 2.5, 1% Triton X-100. The supernatants and bound material were subjected to Western blotting analysis with the indicated antibodies. For all Western blotting experiments, the Enhanced Chemiluminescence (ECL) detection system (Pierce, Rockford, IL) was used with secondary antibodies covalently coupled to horseradish peroxidase to visualize the immunodecorated proteins. Protein concentrations were determined using the bicinchoninic acid assay (Pierce).
Subcellular fractionation of platelets
One unit of platelets was sedimented (700g) and then resuspended in 50 mL of platelet wash buffer (137 mmol/L NaCl, 2.7 mmol/L KCl, 3 mmol/L NaH2PO4 [pH 7.4], 5.5 mmol/L D-glucose, 1 mmol/L MgCl2, and 0.35% bovine serum albumin [BSA]). The platelets were resedimented and resuspended in 5 mL of homogenizing buffer, 25 mmol/L HEPES (pH 7.0, 38 mmol/L KCl, 108 mmol/L NaCl, 1 mmol/L dithiothreitol [DTT], and 1 × magic mix containing 1 mmol/L o-phenanthroline, 10 mmol/L EGTA, 1 mmol/L leupeptin, 40 μg/mL antipain, 0.12 U/mL aprotinin, 1 μmol/L pepstatin, 1 mmol/L benzamidine, and 40 μg/mL chymostatin). The platelets were disrupted by 5 freeze-thaw cycles and then clarified by centrifugation at 100 000g for 1 hour. The supernatant (cytosol) was collected. The pellet was washed with homogenizing buffer plus 1 mol/L KCl and 1 mmol/L DTT and recentrifuged. The resulting pellet was resuspended in either 100 mmol/L Na2CO3, pH 11.5, or in 1% Triton PBS and incubated on ice for 30 minutes, then subjected to centrifugation at 100 000g for 1 hour. The supernatants (soluble fractions) and pellets (insoluble fractions) were analyzed by western blotting.
Triton X-114 partitioning was performed as described by Bordier.32 The cytosol and membrane fractions (the fractions after 5 freeze-thaw cycles) were diluted (1:10 v:v) into homogenization buffer containing 1 × magic mix and 1% Triton X-114, and incubated on ice for 1 hour. The Triton X-114–insoluble material was sedimented. The supernatants were warmed at 37°C for 5 minutes and then subjected to centrifugation for 3 minutes. The aqueous phase and detergent phase were collected, precipitated with 12% trichloroacetic acid, and subjected to Western blotting using ab23 and anti–syntaxin 4 antibody.
Preparation of [3H]5-HT–labeled platelets
One unit of freshly banked platelets was incubated at room temperature for 5 minutes in the presence of 10 ng/mL prostaglandin I2 and then sedimented at 700g for 15 minutes at room temperature. The platelets were resuspended in 2 to 5 mL of the platelet-poor plasma, and the concentration of platelets was measured. The concentration of platelets was adjusted to 109to 5 × 1010 platelets/mL by the addition of extra platelet-poor plasma. For [3H]5-HT labeling, platelets were incubated at 37°C for 40 minutes in the presence of 0.2 μCi/mL [3H]5-HT. The labeled platelets were washed twice in Ca++-free Tyrode's solution (154 mmol/L NaCl, 2.7 mmol/L KCl, 1 mmol/L MgCl2, 5.6 mmol/L D-glucose, 7 mmol/L NaHCO3, 0.6 mmol/L NaH2PO4, 5 mmol/L sodium PIPES [pH 6.5], 0.35% BSA, 5 mmol/L EGTA [pH 6.5 adjusted with KOH], 0.03 mg/mL apyrase, and 50 U/mL heparin). The platelets were washed 1 more time with the same medium without heparin (but with apyrase). Finally, the platelets were resuspended in assay buffer (120 mmol/L sodium glutamate, 5 mmol/L potassium glutamate, 20 mmol/L HEPES/NaOH, pH 7.4, 2.5 mmol/L EDTA, 2.5 mmol/L EGTA, 3.15 mmol/L MgCl2, and 1 mmol/L DTT), and the concentration of the platelets was adjusted to 109/mL. Note that the inclusion of DTT in this buffer did not affect the efficiency of the subsequent release reactions and could be eliminated if the streptolysin O used was fully reduced (data not shown).
Permeabilization of platelets with SLO and assay of 5-HT, hexosaminidase, lactate dehydrogenase (LDH), and PDGF release
Fifty microliters of platelets (107-108platelets) in assay buffer was mixed with 50 μL of assay buffer containing 8 mmol/L ATP, 1.6 U/mL SLO, and antibodies or recombinant proteins for 10 minutes at room temperature. The reactions were further incubated on ice for 30 minutes. After the samples had been warmed to 25°C for 5 minutes, CaCl2 was added to give the desired final concentration,33 and the reactions were incubated at 25°C for another 5 minutes. The reactions were stopped by placing the samples on ice for 4 minutes, followed by centrifugation at 13 000g for 1 minute. The supernatants were collected and assayed as described later (see Figure 4B).
[3H]5-HT release was measured by scintillation counter. Hexosaminidase was measured as described by Holmsen and Dangelmaier.34 Five milliliters of citrate-phosphate buffer, pH 4.5, and 2.5 mL of 10 mmol/L substrate (P-nitrophenyl-N-acetyl-β-D-glucosaminide) were mixed and aliquoted (100 μL) into 96-well plates, and 5 μL of the reaction supernatant was added. After incubation at 37°C for 18 hours, 60 μL of 0.08N NaOH was added to stop the reaction. The absorbance was read in a Titertek Multiscan Plus ELISA plate reader (Labsystems, Stockholm, Sweden) with a 405-nm filter. In these assays, the no-enzyme background was subtracted (OD405 = 0.040). For the LDH assay, 700 μL of 0.2 mol/L Tris-HCl, pH 7.4, was mixed with 100 μL of 3 mmol/L NADH and 100 μL of 10 mmol/L pyruvate. Forty-five microliters of the supernatant from the SLO experiment was added into the prewarmed reaction buffer, and the decrease in absorbance at 340 nm versus time was recorded and converted to enzyme activity.
Release of α-granules was measured by quantitative enzyme-linked immunosorbent assay (ELISA) for the α-granule protein PDGF. Supernatants from the SLO-permeabilized platelets were added in triplicate to wells of a microtiter high-protein-binding ELISA plate (Costar, Cambridge, MA) containing 200 μL of 15 mmol/L Na2CO3 and 35 mmol/L NaHCO3, pH 9.6. The samples were dried onto the plate by overnight incubation at 37°C. The wells were washed twice with ELISA wash buffer (3.5 mmol/L NaH2PO4, 31 mmol/L Na2HPO4, 15.4 mmol/L NaCl, and 0.5% Tween-20). Blocking was done for 1 hour at room temperature with 3% BSA (Sigma) in 1 × PBS. Anti–PDGF-BB (R&D Systems, Minneapolis, MN) primary antibody was diluted to 1 μg/mL in blocking solution, added to the wells, and incubated for 2 hours at room temperature. The wells were washed 4 times, and anti-goat secondary antibody (coupled to horseradish peroxidase), diluted in blocking solution, was added and allowed to incubate for 1 hour at room temperature. The wells were again washed 4 times, and 200 μL of 1 mmol/L ABTS (2,2′-azino-di-[3-ethylbenzthiazoline sulfonate) was added. Samples were quantified spectrophotometrically at 405 nm.
Results
Biochemical characterization of platelet SNAP-23
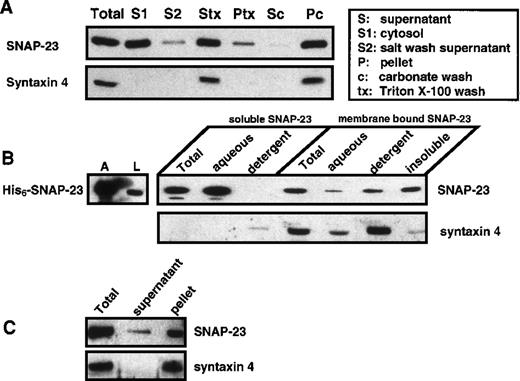
SNAP-25 and SNAP-23 are homologous proteins sharing 59% amino acid identity.17 Both appear to be anchored to the cytosolic side of the plasma membrane by thioester-linked acyl groups.35-39 SNAP-23 was shown in numerous cases to be membrane associated.21-26 To test whether SNAP-23 behaves as a peripheral or integral membrane protein in platelets, we performed a subcellular fractionation experiment (Figure2A). Platelets were disrupted and the membranes were pelleted by centrifugation at 100 000g for 30 minutes. The supernatant (S fraction) was collected, and the membrane pellet was washed with 1 mol/L KCl. The supernatant was collected (S2 fraction), and the salt-washed pellet was resuspended in either 1% Triton X-100 in PBS or in 0.1 mol/L sodium carbonate, pH 11.5, and subjected to further centrifugation. The supernatants (Stx, supernatant of Triton X-100 solubilization; Sc, supernatant of carbonate wash) were collected, and the pellets (Ptx, Triton-insoluble pellet; Pc, carbonate wash–insoluble pellet) were resuspended in 0.2% sodium dodecyl sulfate (SDS) in PBS. SNAP-23 is present in both cytosolic and membrane fractions. The membrane-bound SNAP-23 is 1 mol/L KCl and sodium carbonate–resistant but can be partially solubilized by Triton X-100. Syntaxin 4 served as an integral membrane protein control for this fractionation experiment; it was enriched in the membrane fraction, was not released by sodium carbonate, and was soluble in Triton X-100. The presence of soluble SNAP-23 was also demonstrated using SLO-treated platelets (Figure 2C). A portion of the SNAP-23, but none of the syntaxin 4, diffused out of the permeabilized cells and was detected in the supernatant.
Distribution of SNAP-23 in platelets.
(A) Platelets were disrupted by freeze-thaw cycles and fractionated into a soluble fraction (S1) and pellet by ultracentrifugation. The pellet was incubated with 1 mol/L KCl on ice for 30 minutes. The supernatant (S2) was collected, and the resulting pellet was either solubilized with 1% Triton X-100 in PBS or washed again with 200 mmol/L Na2CO3 for 30 minutes. The supernatants (Triton-soluble, Stx; and carbonate-released, Sc) were collected and the pellets (Ptx and Pc) were resuspended in SDS-loading buffer. All of the above fractions were subjected to western blotting using ab23 and anti–syntaxin 4 antibody. Based on comparison with the starting material, approximately 38.2%, 16.5%, 33.5%, 31.1%, 14.2%, and 8.5% of the total platelet protein was in S1, S2, Sc, Stx, Ptx, and Pc, respectively. (B) The initial soluble and membrane fractions from A were incubated with 1% Triton X-114 for 30 minutes on ice. The aqueous and detergent phases were separated by warming the samples to 37°C, followed by centrifugation. The aqueous and detergent phases of both the soluble fraction and membrane pellet as well as the Triton X-114–insoluble pellet were analyzed by western blotting using ab23 and anti–syntaxin 4 antibody. His6–SNAP-23 was also subjected to Triton X-114 partitioning, and the aqueous and detergent phases were analyzed by western blotting using ab23. (C) Soluble SNAP-23 is released from platelets after the treatment with 0.8 U/mL SLO. Platelets (108) were treated with 0.8 U/mL SLO for 10 minutes and then pelleted by centrifugation. The supernatant (S) and the pellet (P) were subjected to western blotting with ab23 and anti–syntaxin 4.
Distribution of SNAP-23 in platelets.
(A) Platelets were disrupted by freeze-thaw cycles and fractionated into a soluble fraction (S1) and pellet by ultracentrifugation. The pellet was incubated with 1 mol/L KCl on ice for 30 minutes. The supernatant (S2) was collected, and the resulting pellet was either solubilized with 1% Triton X-100 in PBS or washed again with 200 mmol/L Na2CO3 for 30 minutes. The supernatants (Triton-soluble, Stx; and carbonate-released, Sc) were collected and the pellets (Ptx and Pc) were resuspended in SDS-loading buffer. All of the above fractions were subjected to western blotting using ab23 and anti–syntaxin 4 antibody. Based on comparison with the starting material, approximately 38.2%, 16.5%, 33.5%, 31.1%, 14.2%, and 8.5% of the total platelet protein was in S1, S2, Sc, Stx, Ptx, and Pc, respectively. (B) The initial soluble and membrane fractions from A were incubated with 1% Triton X-114 for 30 minutes on ice. The aqueous and detergent phases were separated by warming the samples to 37°C, followed by centrifugation. The aqueous and detergent phases of both the soluble fraction and membrane pellet as well as the Triton X-114–insoluble pellet were analyzed by western blotting using ab23 and anti–syntaxin 4 antibody. His6–SNAP-23 was also subjected to Triton X-114 partitioning, and the aqueous and detergent phases were analyzed by western blotting using ab23. (C) Soluble SNAP-23 is released from platelets after the treatment with 0.8 U/mL SLO. Platelets (108) were treated with 0.8 U/mL SLO for 10 minutes and then pelleted by centrifugation. The supernatant (S) and the pellet (P) were subjected to western blotting with ab23 and anti–syntaxin 4.
Because this was the first evidence of cytosolic SNAP-23, we sought to determine whether this was due to a difference in the biochemical properties of platelet SNAP-23. Platelets were fractionated into soluble and membrane fractions, and each fraction was subjected to Triton X-114 partitioning to determine whether SNAP-23 retained the hydrophobicity of a membrane protein (Figure 2B). The aqueous phase and the detergent phase were collected and subjected to Western blotting using ab23 and anti–syntaxin 4 antibody. As a control, 90% of the recombinant, unacylated, His6–SNAP-23 partitioned into the aqueous phase, as expected for a hydrophilic protein. Soluble platelet SNAP-23 was enriched in the aqueous phase, whereas most of the membrane-bound SNAP-23 was in the detergent phase or in the detergent-insoluble pellet. The soluble SNAP-23 and membrane-bound SNAP-23 appeared to have a difference in hydrophobicity. As an internal control, the true integral membrane protein, syntaxin 4, was enriched in the detergent phase of the membrane fraction.
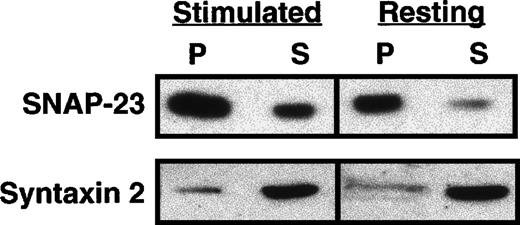
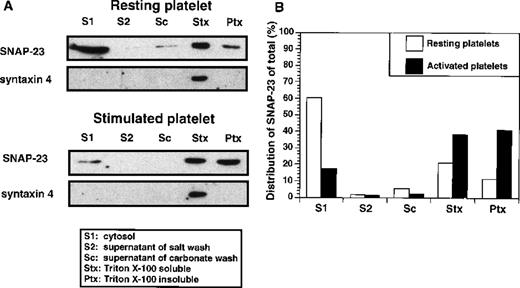
To determine whether the soluble pool of SNAP-23 changes during platelet activation, we performed a similar sequential fractionation of thrombin-treated (1 U/mL) and resting platelets (Figure3). The membrane pellets were washed sequentially with high salt and sodium carbonate, and each time the supernatants were collected. Finally, the pellets were resuspended in Triton X-100 and divided into Triton-soluble and -insoluble fractions. All of the fractions were subjected to Western blotting using ab23 and anti–syntaxin 4 antibody. After stimulation of the platelets, the soluble fraction of SNAP-23 decreased (from 60% to 17% of total), with a subsequent increase (from 11% to 41% of total) in the Triton X-100–insoluble fraction (Figure 3B). Although this could be due to simple trapping, it was not seen for syntaxin 4 from the same samples.
Distribution of SNAP-23 in resting and activated platelets.
(A) Platelets were resuspended in Ca++-free Tyrode's buffer or Tyrode's (1 mmol/L Ca++) buffer containing 1 U/mL thrombin for 5 minutes. The resting and activated platelets were disrupted by freeze-thaw and fractionated by centrifugation. The supernatant (S1) was collected, and the pellets were washed sequentially with 1 mol/L KCl, 200 mmol/L Na2CO3, and 1% Triton X-100, as in Figure 2(S2, Sc, and Stx). The supernatants and Triton X-100–insoluble pellet (Ptx) were analyzed by Western blotting using ab23 and anti–syntaxin 4 antibody. (B) The Western blotting image was scanned and digitized by using NIH 1.6 program (available at rsb.info.nih.gov/nih.image). The pixel number of each band was normalized as a percentage of total pixel number in all lanes of the treatment group.
Distribution of SNAP-23 in resting and activated platelets.
(A) Platelets were resuspended in Ca++-free Tyrode's buffer or Tyrode's (1 mmol/L Ca++) buffer containing 1 U/mL thrombin for 5 minutes. The resting and activated platelets were disrupted by freeze-thaw and fractionated by centrifugation. The supernatant (S1) was collected, and the pellets were washed sequentially with 1 mol/L KCl, 200 mmol/L Na2CO3, and 1% Triton X-100, as in Figure 2(S2, Sc, and Stx). The supernatants and Triton X-100–insoluble pellet (Ptx) were analyzed by Western blotting using ab23 and anti–syntaxin 4 antibody. (B) The Western blotting image was scanned and digitized by using NIH 1.6 program (available at rsb.info.nih.gov/nih.image). The pixel number of each band was normalized as a percentage of total pixel number in all lanes of the treatment group.
Description of a permeabilized platelet exocytosis assay
To study the roles of the fusion machinery proteins in platelet exocytosis, it was first necessary to develop an in vitro exocytosis assay using permeabilized platelets. The initial step was to determine the conditions and concentration of SLO needed to permeabilize platelet plasma membrane without affecting granule integrity. SLO was incubated at various concentrations with platelets for 10 minutes at 25°C or 37°C, followed by chilling on ice for 30 minutes and then reincubation at 25°C or 37°C for another 10 minutes. Permeabilization of the plasma membrane was measured by the appearance of LDH in the media. The integrity of the dense core granules, lysosomes, and α-granules was followed by measuring the appearance of [3H]5-HT, hexosaminidase, and PDGF, respectively (Figure4). LDH appeared in the supernatant at SLO concentrations of 0.6 to 0.8 U/mL at both 25°C and 37°C. However, at 37°C, [3H]5-HT was released when the concentration of SLO exceeded 0.4 U/mL. This leakage was delayed at 25°C, so the optimal permeabilization condition appeared to be 0.8 U/mL of SLO at 25°C. The integrity of lysosome and α-granule membrane was also maintained under these conditions (Figure 4).
Streptolysin O permeabilization of platelets.
(A) Increasing amounts of SLO (0-1.2 U/mL final concentration) were added to the assay buffer containing 108 platelets. Platelets were incubated at 25°C or 37°C for 10 minutes, chilled on ice for 30 minutes, and further warmed to 25°C or 37°C for 10 minutes. The platelets were sedimented and the supernatants were collected. The activity of lactate dehydrogenase (LDH) and hexosaminidase and the amounts of [3H]5-HT and PDGF in the supernatant were measured (see “Materials and Methods”) and compared with the total activity of Triton X-100–solubilized platelets (n = 4). (B) The time line represents the standard reaction scheme used in the permeabilized platelet exocytosis assay.
Streptolysin O permeabilization of platelets.
(A) Increasing amounts of SLO (0-1.2 U/mL final concentration) were added to the assay buffer containing 108 platelets. Platelets were incubated at 25°C or 37°C for 10 minutes, chilled on ice for 30 minutes, and further warmed to 25°C or 37°C for 10 minutes. The platelets were sedimented and the supernatants were collected. The activity of lactate dehydrogenase (LDH) and hexosaminidase and the amounts of [3H]5-HT and PDGF in the supernatant were measured (see “Materials and Methods”) and compared with the total activity of Triton X-100–solubilized platelets (n = 4). (B) The time line represents the standard reaction scheme used in the permeabilized platelet exocytosis assay.
The next step was to optimize the Ca++ concentration needed to stimulate exocytosis (Figure 5A). [3H]5-HT–labeled platelets were first treated with 0.8 U/mL SLO and then chilled on ice for 30 minutes. This 4°C step is required to allow equilibration of potential activators and inhibitors into the platelets without further permeabilization of granule membranes. SLO is inactive at 4°C.40 When this step was performed at room temperature, there was an increase in the Ca++-independent release of granule stores (data not shown), suggesting that the granules were permeabilized during the incubation. After rewarming to 25°C for 5 minutes, the platelets were treated with increasing concentrations of Ca++ for 5 minutes. Exocytosis was stopped by chilling the samples on ice for 4 minutes, and then the platelets were removed by centrifugation at 13 000g for 1 minute. The supernatants were analyzed for [3H]5-HT release. As shown in Figure 5A, dense core granule secretion did not occur until the calcium concentration reached 10 μmol/L. This is similar to the intracellular calcium concentration reached upon stimulation of intact platelets with thrombin.41 This calcium concentration curve was also similar to that reported from other permeabilized platelet exocytosis assays.42,43 As the calcium concentration increased, exocytosis reached a plateau but the standard error between samples increased. This suggests that at the high calcium concentrations, artifactual membrane fusion events were randomly occurring. For this reason and for agreement with previous studies,41-43 we chose to use 10 μmol/L as the calcium concentration to stimulate exocytosis. The calcium-stimulated exocytosis requires energy. When apyrase was added without the addition of ATP (30 μg/mL of apyrase, 0.5 U per assay point, enough to degrade 0.5 μmol of ATP in 15 minutes), the release of [3H]5-HT was eliminated (Figure5A).
The effects of Ca++, -SNAP, and NSF on [3H]5-HT release.
(A) Increasing Ca++(10−8-10−3 mol/L final) was used to induce exocytosis in the presence of 50 μg/mL of wild-type α-SNAP or mutant α-SNAP–L294A. The released [3H]5-HT was measured as in Figure 4. In an additional titration, 30 μg/mL of apyrase was added to deplete ATP from the reaction (n = 6). (B) In a separate, summary experiment, the effects of 50 μg/mL bovine wild-type α-SNAP, 60 μg/mL anti–α-SNAP antibody, and 60 μg/mL rabbit IgG on the 10 μmol/L Ca++-triggered [3H]5-HT secretion were compared with the control (100%) (n = 6). (C) [3H]5-HT–labeled and SLO-permeabilized platelets were incubated with increasing amounts of the 2E5 monoclonal antibody (anti-NSF; 0-0.32 mg/mL). The release of [3H]5-HT was measured as before and normalized to the control (no addition) (n = 5). (D) The 2E5 inhibitory effect can be reversed by the addition of recombinant NSF. Radiolabeled and SLO-permeabilized platelets were incubated on ice for 30 minutes with buffer (control), 80 μg/mL 2E5, 80 μg/mL plus 0.75 mg/mL recombinant NSF, or 0.75 mg/mL recombinant NSF alone. Platelets were activated by 10 μmol/L Ca++, and the release of [3H]5-HT was measured and normalized to the control group (n = 5).
The effects of Ca++, -SNAP, and NSF on [3H]5-HT release.
(A) Increasing Ca++(10−8-10−3 mol/L final) was used to induce exocytosis in the presence of 50 μg/mL of wild-type α-SNAP or mutant α-SNAP–L294A. The released [3H]5-HT was measured as in Figure 4. In an additional titration, 30 μg/mL of apyrase was added to deplete ATP from the reaction (n = 6). (B) In a separate, summary experiment, the effects of 50 μg/mL bovine wild-type α-SNAP, 60 μg/mL anti–α-SNAP antibody, and 60 μg/mL rabbit IgG on the 10 μmol/L Ca++-triggered [3H]5-HT secretion were compared with the control (100%) (n = 6). (C) [3H]5-HT–labeled and SLO-permeabilized platelets were incubated with increasing amounts of the 2E5 monoclonal antibody (anti-NSF; 0-0.32 mg/mL). The release of [3H]5-HT was measured as before and normalized to the control (no addition) (n = 5). (D) The 2E5 inhibitory effect can be reversed by the addition of recombinant NSF. Radiolabeled and SLO-permeabilized platelets were incubated on ice for 30 minutes with buffer (control), 80 μg/mL 2E5, 80 μg/mL plus 0.75 mg/mL recombinant NSF, or 0.75 mg/mL recombinant NSF alone. Platelets were activated by 10 μmol/L Ca++, and the release of [3H]5-HT was measured and normalized to the control group (n = 5).
To optimize the release time, we analyzed the time course of the dense core granule secretion using 10 μmol/L Ca++ to activate secretion. The release of [3H]5-HT started immediately after the addition of Ca++ and was complete by 3 minutes (data not shown). All of our later secretion assays were stopped after 5 minutes of Ca++ treatment, when the release of [3H]5-HT should be complete. For all subsequent assays, 0.8 U/mL of SLO was used to permeabilize the platelets following the scheme outlined in Figure 4B, and incubation with 10 μmol/L Ca++ for 5 minutes was used to stimulate exocytosis. These same assay conditions were used to test the level of exocytosis in fresh platelets as compared with the freshly banked platelets reported here. In 3 separate preparations, fresh platelets released 49% ± 2.3% (n = 9 data points) of the total cellular [3H]5-HT, and freshly banked platelets released 45% ± 0.9% of the total [3H]5-HT (n = 20 data points). Because there was no apparent difference between fresh and freshly banked platelets, the remaining experiments were performed with freshly banked cells. Also under these assay conditions, it is possible to measure Ca++ and GTP-γ–S-stimulated release from all 3 platelet granule stores, as well as a Ca++-induced increase in fibrinogen receptor affinity and centralization of platelet granules caused by cytoskeletal rearrangements (Chen, Lemmons, and Whiteheart, unpublished data).
Platelet dense core granule secretion is facilitated by SNAPs and NSF
Platelets contain α-SNAP, γ-SNAP, and NSF.15 To test whether SNAPs are involved in platelet secretion, we added 50 μg/mL of wild-type α-SNAP and dominant negative α-SNAP–L294A mutant44 to the Ca++ titration assay during the SLO incubation step (Figure 5A). After 30 minutes of incubation on ice, increasing amounts of Ca++ were added to the reaction, and the release of [3H]5-HT was measured (Figure 5A). Wild-type α-SNAP appeared to increase the extent of [3H]5-HT secretion from 43% to 68% of the total. The mutant α-SNAP decreased [3H]5-HT secretion to 27% of the total. The effects of wild-type and mutant α-SNAP were most apparent at 10 μmol/L calcium and less so at higher calcium concentrations. Research has shown that γ-SNAP also stimulates membrane trafficking events, although not as effectively as α-SNAP.45 46 Addition of γ-SNAP (50 μg/mL) did increase [3H]5-HT release, but only by 20% when compared with control (data not shown). Anti–α-SNAP antibodies were also tested for their effect on dense core granule release. In Figure 5B, antibody to α-SNAP almost completely inhibited [3H]5-HT release, whereas the nonspecific antibody control had no effect.
Because α-SNAP has been shown to serve as an adapter for NSF binding,45 we next examined the role of NSF in [3H]5-HT release. A monoclonal anti-NSF antibody (2E5) inhibited dense core granule release (Figure 5C), and this inhibition was reversed by preincubation of the 2E5 antibody with recombinant NSF (Figure 5D). Not unexpectedly, the recombinant NSF alone showed little enhancement of secretion because its size (approximately 480 kDa) makes it unlikely to enter the SLO-induced pores in the platelet membrane. To further confirm this result, we tested the effect of 2E5 on fresh platelets that had been permeabilized with SLO. The anti-NSF antibody reduced calcium-stimulated release from 49% ± 2.3% of total [3H]5-HT to 9% of total in these cells.
SNAP-23 mediates dense core granule secretion
From the above data, it appears that platelet exocytosis uses SNAPs and NSF; therefore, it is likely that SNARE proteins are involved. To address this, we first focused on the t-SNARE SNAP-23. Fab fragments made from ab23 (Fab23) appeared to significantly inhibit Ca++-induced release of [3H]5-HT from dense core granules (Figure 6). Fab23 almost completely inhibited [3H]5-HT release at a concentration of 0.16 mg/mL (Figure 6A). This inhibition of [3H]5-HT release by Fab23 was partially reversed by the addition of recombinant His6–SNAP-23, confirming the specificity of the Fab fragment (Figure 6B). Recombinant His6–SNAP-23 had no effect on [3H]5-HT secretion (Figure 6B), nor did a preimmune IgG fraction. The C-terminal peptides of SNAP-23 and -25 have proved to be good inhibitors of GLUT4 translocation and neurotransmitter release.47 48 The C-terminal peptide of human SNAP-23 inhibits [3H]5-HT secretion by 43% compared with control. On the basis of our data, SNAP-23 is involved in dense core granule secretion.
Anti–SNAP-23 antibody inhibits [3H]5-HT release.
(A) The permeabilized platelet exocytosis was performed in the presence of increasing amounts of Fab23 (0-0.16 mg/mL). After 5 minutes of Ca++ stimulation (10 μmol/L), [3H]5-HT release was measured (n = 3). (B) Secretion triggered by 10 μmol/L Ca++ was analyzed in the presence of Fab23 (0.12 mg/mL), Fab23 (0.12 mg/mL) preincubated with His6–SNAP-23 (0.45 mg/mL), His6–SNAP-23 (0.45 mg/mL), human SNAP-23 C-terminal peptide (0.15 mg/mL), and rabbit IgG purified from preimmune sera (0.15 mg/mL) (n = 6).
Anti–SNAP-23 antibody inhibits [3H]5-HT release.
(A) The permeabilized platelet exocytosis was performed in the presence of increasing amounts of Fab23 (0-0.16 mg/mL). After 5 minutes of Ca++ stimulation (10 μmol/L), [3H]5-HT release was measured (n = 3). (B) Secretion triggered by 10 μmol/L Ca++ was analyzed in the presence of Fab23 (0.12 mg/mL), Fab23 (0.12 mg/mL) preincubated with His6–SNAP-23 (0.45 mg/mL), His6–SNAP-23 (0.45 mg/mL), human SNAP-23 C-terminal peptide (0.15 mg/mL), and rabbit IgG purified from preimmune sera (0.15 mg/mL) (n = 6).
Syntaxin 2 mediates dense core granule secretion
We next focused on the syntaxin family of t-SNARE. Syntaxins 2, 4, and 7 are present in platelets (Figure 1).15 To determine which syntaxin is involved in dense core granule secretion, we tested the antibodies against syntaxins 2, 4, and 7 in the in vitro secretion assay (Figure 7A). Syntaxin 2 antibody dramatically inhibited dense core granule release, reaching 90% inhibition by 0.18 mg/mL. This inhibition was reversed by His6–syntaxin 2 recombinant protein but not by His6–syntaxin 4 (Figure 7B). His6–syntaxin 2, a cytoplasmic domain of syntaxin 2, by itself had no effect on secretion. Neither anti–syntaxin 4 nor anti–syntaxin 7 antibodies affected dense core granule secretion when comparable concentrations were used, nor did a preimmune IgG fraction. An additional monoclonal anti–syntaxin 4 antibody, shown to inhibit α-granule release,16 also had no effect on [3H]5-HT release (data not shown). These data indicate that syntaxin 2 is involved in dense core granule release in platelets.
Anti–syntaxin 2 antibody inhibits [3H]5-HT release.
(A) The permeabilized platelet exocytosis assay was performed in the presence of increasing amounts of anti–syntaxin 2 (▪), 4 (•), and 7 (▴) antibodies and rabbit IgG (⋄ (0-0.18 mg/mL). After stimulation with Ca++ (10 μmol/L), [3H]5-HT release was measured and normalized as a percentage of control release. (B) [3H]5-HT secretion stimulated by 10 μmol/L Ca++ was analyzed in the presence of anti–syntaxin 2 antibody (20 μg/mL), anti–syntaxin 2 antibody (20 μg/mL) preincubated with His6–syntaxin 2 (0.24 mg/mL), His6–syntaxin 2 alone (0.24 mg/mL), anti–syntaxin 2 antibody (0.02 mg/mL) preincubated with His6–syntaxin 4 (0.24 mg/mL), and IgG purified from preimmune sera (20 μg/mL). The [3H]5-HT release was measured and normalized as a percentage of control release (n = 9).
Anti–syntaxin 2 antibody inhibits [3H]5-HT release.
(A) The permeabilized platelet exocytosis assay was performed in the presence of increasing amounts of anti–syntaxin 2 (▪), 4 (•), and 7 (▴) antibodies and rabbit IgG (⋄ (0-0.18 mg/mL). After stimulation with Ca++ (10 μmol/L), [3H]5-HT release was measured and normalized as a percentage of control release. (B) [3H]5-HT secretion stimulated by 10 μmol/L Ca++ was analyzed in the presence of anti–syntaxin 2 antibody (20 μg/mL), anti–syntaxin 2 antibody (20 μg/mL) preincubated with His6–syntaxin 2 (0.24 mg/mL), His6–syntaxin 2 alone (0.24 mg/mL), anti–syntaxin 2 antibody (0.02 mg/mL) preincubated with His6–syntaxin 4 (0.24 mg/mL), and IgG purified from preimmune sera (20 μg/mL). The [3H]5-HT release was measured and normalized as a percentage of control release (n = 9).
SNAP-23 and syntaxin 2 interact in platelets
Because both SNAP-23 and syntaxin 2 are involved in dense core granule secretion, according to the SNARE hypothesis, they should form a complex in the platelet. To test this, we performed an immunoprecipitation experiment to trap this complex. SNAP-23 has been shown to bind to each of the plasma membrane syntaxins in vitro17; however, co-immunoprecipitation from dilute detergent-solubilized extracts seems to be the only method available to demonstrate that a SNAP-23–containing complex exists in vivo. Thrombin-activated and resting platelets were solubilized with 1% Triton X-100 PBS and subjected to immunoprecipitation by ab23 covalently coupled to protein G beads. The bound and free materials were recovered and subjected to Western blot with ab23 and anti–syntaxin 2 antibody. In both activated and resting platelets, weak but detectable complexes were formed; however, there was no obvious change in the amount of complex recovered either before or after thrombin stimulation (Figure 8).
SNAP-23 and syntaxin 2 can form a complex.
Fifty micrograms of Triton X-100–solubilized platelet extracts from resting and thrombin-activated (1 U/mL for 5 minutes) platelets was subjected to immunoprecipitation (IP) with ab23 coupled to protein G beads. The precipitated material (P) and the unbound supernatant material (S) were analyzed by Western blotting with ab23 and anti–syntaxin 2 antibody. The immunodecorated proteins were detected by ECL as described earlier. Hexosaminidase release was measured to confirm the activation of platelets.
SNAP-23 and syntaxin 2 can form a complex.
Fifty micrograms of Triton X-100–solubilized platelet extracts from resting and thrombin-activated (1 U/mL for 5 minutes) platelets was subjected to immunoprecipitation (IP) with ab23 coupled to protein G beads. The precipitated material (P) and the unbound supernatant material (S) were analyzed by Western blotting with ab23 and anti–syntaxin 2 antibody. The immunodecorated proteins were detected by ECL as described earlier. Hexosaminidase release was measured to confirm the activation of platelets.
Discussion
In our previous studies of platelet exocytosis, we identified many of the putative components of the platelet secretory machine, such as α-SNAP, γ-SNAP, NSF, p115/TAP, VAMP-3/cellubrevin, and 2 of the known syntaxins, syntaxins 2 and 4.15 27 In this report, we increase this list by demonstrating the presence of 2 additional t-SNAREs, SNAP-23 and syntaxin 7. As our focus turns from identification to functional studies, we have adapted a permeabilized platelet exocytosis assay to dissect the roles of these secretory machinery proteins. Using this assay, we have shown that α-SNAP and NSF play a role in the Ca++-induced secretion of dense core granules. Using t-SNARE–specific antibodies, we have further demonstrated that syntaxin 2 and SNAP-23 are involved in dense core granule release. These data confirm the fact that platelet exocytosis is mediated by SNARE proteins and offer new insight into the proteins that facilitate the exocytosis events of the platelet.
SNAP-23, like its homologue SNAP-25, behaves as an integral membrane protein in all of the cell types examined.21,22,24,25,47,49This is presumably because the protein is bound to the membrane through a series of thioester-linked palmitoyl groups, attached to a conserved cluster of cysteine residues in the center of both SNAP-23 and SNAP-25.35-39 The distribution of SNAP-23 in platelets is unique in that not all of it is associated with membranes. Subcellular fractionation and SLO permeabilization experiments show that a portion of platelet SNAP-23 is soluble. This soluble SNAP-23 no longer partitions into the detergent phase of a Triton X-114 extraction, suggesting that it has lost its hydrophobicity. This loss of hydrophobicity could result from a difference in the acyl groups attached to platelet SNAP-23, or it could result from a loss of or incomplete addition of palmitoyl groups. Given the inherent instability of thioester bonds, it seems possible that SNAP-23 irreversibly deacylates as platelets age. It is interesting that the pool of soluble SNAP-23 decreases upon platelet activation. This change in distribution could indicate some functional relocation of the molecule, as has been proposed for SNAP-23 in 3T3-L1 and mast cells,25,50 or simple trapping in the aggregated cytoskeleton.51 At this stage, the functional relevance of the formation and redistribution of soluble SNAP-23 is unknown; however, soluble SNAP-23 is not found in the megakaryoblastic leukemia cell line, Meg-O1s (Chen and Whiteheart, unpublished data). Further detailed experimentation will be required to determine the significance of this observation.
To gain insight into the machinery of platelet secretion, we adapted a previous technology42,43 in which the pore-forming bacterial toxin SLO was used to permeabilize platelets. Based on the titration presented in Figure 4, it is clear that permeabilization at 25°C with 0.8 U/mL SLO allows access to the cytosol without significantly damaging the granule membranes. Under these conditions, cells can be permeabilized and then incubated on ice with activators or inhibitors to equilibrate these reagents into the cells. This cold step decreases the activity of SLO and therefore lessens the permeabilization of granule membranes.40 When the equilibration step is performed at room temperature, granule contents leak, thereby decreasing the calcium-dependent secretion signal. Chilling, however, can stimulate changes in platelet shape and α-granule secretion,52,53 presumably through a cold-induced leakage of calcium from intraplatelet stores.54-56 The inclusion of EGTA in the permeabilization buffer appears to eliminate this effect because no release of [3H]5-HT is seen in the absence of added calcium (See Figures 4A and 5A), nor is there any apparent cytoskeletal rearrangement as detected by electron microscopy (Lemons and Whiteheart, unpublished data). Once the permeabilized platelets are rewarmed to 25°C, they can be quickly stimulated with Ca++ and secretion can be measured. Calcium (10−8-10−3 mol/L) was titrated into the permeabilized platelet assay and, as shown in Figure 5A, secretion occurred only when the Ca++ concentration reached 10 μmol/L. As Ca++ was increased, release of granule contents did not increase but became more erratic, as suggested by the increase in the standard errors. For this reason as well as the fact that stimulation of platelets in vivo usually results in an increase in intracellular Ca++ to greater than 1 μmol/L,57 we chose to use 10 μmol/L Ca++ to stimulate exocytosis from the permeabilized platelets. In all cases, content release was dependent on ATP (Figure 5A) and temperature (data not shown). In the presence of apyrase (0.5 U), no Ca++-induced release of granule contents occurred. In addition, granule release occurs rapidly after the addition of Ca++ and appears to plateau by 3 minutes (data not shown). This is very similar to the kinetics of thrombin-induced secretion in vivo.58 For these experiments, we chose to use freshly banked platelets because there was no apparent difference in the degree of calcium-stimulated release between these cells and freshly prepared platelets (see “Results”).
The major difference between the properties of the permeabilized cell system and intact platelets is the extent of content release. The extent of release in the permeabilized cells is approximately half of that seen for thrombin-induced intact platelets or for cells that have been permeabilized for only a short time (approximately 2 minutes43). However, it is similar to the efficiency of the calcium-dependent release (approximately 52%) when the cells were permeabilized for 30 minutes.43 The difference in efficiency between intact and permeabilized cells is probably due to the dilution of important cytosolic components as they diffuse from the platelets. Padfield et al43 indicated that this was a likely explanation for their apparent inability to recapitulate GTP-γ–S-stimulated release in their permeabilized platelet system. From the data presented in this manuscript, it is clear that increasing the total concentration of at least 1 cytosolic component, α-SNAP, by adding recombinant protein to the assay can increase the efficiency of [3H]5-HT release (Figure 5A). This is consistent with the explanation that proteins (at least of a certain molecular weight) can freely diffuse both in and out of the permeabilized cells and affect secretion efficiency. Additionally, it should be noted that in our system, exocytosis was measured at room temperature rather than 37°C, which also might account for some decrease in release efficiency.
Using the above assay, our first task was to demonstrate a role for SNAPs in platelet exocytosis. In other systems, such as intercisternal Golgi transport45 and neurotransmission in squid,46 increasing α-SNAP over endogenous levels leads to an increase in membrane fusion events. In platelets, the addition of α-SNAP caused an almost 2-fold increase in the extent of [3H]5-HT release. The importance of α-SNAP was further demonstrated by the fact that, at the concentration used, the dominant-negative α-SNAP mutant (α-SNAP–L294A44) and the anti–α-SNAP antibody both inhibited dense core release. In it interesting that in the Ca++ titration of Figure 5A, the effects of the wild-type and mutant SNAPs are lost as the Ca++ level is increased above 10 μmol/L, suggesting that other factors (such as calpain activation) affect granule release. NSF was also shown to play a role in [3H]5-HT release because anti-NSF antibodies almost completely blocked secretion, and inhibition was reversed by the addition of recombinant NSF (Figures 5C and 5D). These data indicate that these 2 general membrane fusion machinery components, SNAP and NSF, are involved in exocytosis, thus confirming the similarity between the platelet release reaction and other regulated secretion events.
Given these data, we next attempted to determine which t-SNAREs play a role in dense core granule release. Figures 6A and 6B show that SNAP-23 antibodies inhibit dense core granule release. The inhibition was reversed by the addition of recombinant SNAP-23, supporting the specificity of the effect. To determine which syntaxin is involved in dense core granule release, we used syntaxin-specific antibodies as inhibitors. Only anti–syntaxin 2 antibodies inhibited dense core granule release (Figure 7A). This inhibition was reversed by competition with syntaxin 2 protein but not with syntaxin 4 protein (Figure 7B), supporting the antibody specificity. Syntaxin 4 has been shown to be involved in α-granule exocytosis16 (and demonstrated in our laboratory; Chen et al, in preparation), but neither the syntaxin 4 antibody used by Flaumenhaft et al16nor the one produced by our group had any inhibitory effect on dense core granule release. It is interesting that dense core granule and α-granule as well as lysosome release use SNAP-23 (Chen, Lemons, and Whiteheart, unpublished data), and this suggests that the specificity of the 3 platelet exocytosis events is controlled by the syntaxin component of the t-SNARE heterodimer. This also suggests that SNAP-23 is, in fact, a general t-SNARE that pairs with either syntaxin 2 or 4. As for the v-SNAREs in platelets, it is clear that some synaptobrevin/VAMP is required for α-granule release,16but it remains to be determined which v-SNARE is involved. At present, only 2 potential v-SNAREs have been identified in platelets27 (also Rutledge et al, in preparation).
Further experimentation will be required to clarify which SNAREs are involved in which platelet exocytosis events. However, from the data presented here and in other reports,16 it appears that syntaxin 2 controls dense core release, syntaxin 4 contributes to α-granule release, and SNAP-23 plays a role in both secretion events (Lemons and Whiteheart, unpublished data). As we get closer to a complete mapping of the secretory machinery of platelets, the next series of questions will focus on how these proteins and their interactions are regulated. In this respect, SNARE binding proteins such as Munc18's19 and pantophysin59,60and additional regulatory proteins such as the rabs61 and p115/TAP62 will become important subjects for study. The ultimate goal is to determine how extracellular signals impinge on the secretory machinery to mediate granule release.
Note: During the revision of this manuscript, a report by Poiger and Reed63 appeared that suggested a role for NSF in platelet exocytosis.
Acknowledgments
The authors would like to thank Dr David Castle for his generous gift of the SNAP-23 peptide. They also thank the staff of Central Kentucky Blood Center for their assistance and the members of the Whiteheart lab for their helpful discussions. They would especially like to thank Dr Susan A. Buhrow for her expert editing of this manuscript.
Supported by National Institutes of Health Grant HL56652 to S.W.W.
Reprints:Sidney W. Whiteheart, Department of Biochemistry, University of Kentucky College of Medicine, 800 Rose Street, Lexington, KY 40536; e-mail: whitehe@pop.uky.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 4. Streptolysin O permeabilization of platelets. / (A) Increasing amounts of SLO (0-1.2 U/mL final concentration) were added to the assay buffer containing 108 platelets. Platelets were incubated at 25°C or 37°C for 10 minutes, chilled on ice for 30 minutes, and further warmed to 25°C or 37°C for 10 minutes. The platelets were sedimented and the supernatants were collected. The activity of lactate dehydrogenase (LDH) and hexosaminidase and the amounts of [3H]5-HT and PDGF in the supernatant were measured (see “Materials and Methods”) and compared with the total activity of Triton X-100–solubilized platelets (n = 4). (B) The time line represents the standard reaction scheme used in the permeabilized platelet exocytosis assay.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/3/10.1182_blood.v95.3.921.003k17_921_929/6/m_bloo00317004x.jpeg?Expires=1767701418&Signature=hhcVLvf6JUBMYWpOCxh1CkLeB1CdNoiDUyvFEcvsBDrj9QKLW-cgE7ADvum2P4xIEJDYUz4eK~fzoRp7BSpzp67yIwTQ1VsGcFwAHSd~sTMjoal2XJJoYc8IGC24u6WRvLWKncCr7qn-dLDQZ1Ng6LClTbMe~PEiSU1msWLnjWh2Z25DSJZJzba6inOM-9GOhYe3k68fN00pj9JrUWRFhSas3hFztvKprvYhw4WA46qxVHj2nJU097fWuRJb2rVieR8nB4LLy0ZS4mGctrAyIAAfvkZ5XDwZWtOI2H3~V6hPi4XVt0pjuGX7BNflJl5MptTdM6nkuH9H3KnE2VUubg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. The effects of Ca++, -SNAP, and NSF on [3H]5-HT release. / (A) Increasing Ca++(10−8-10−3 mol/L final) was used to induce exocytosis in the presence of 50 μg/mL of wild-type α-SNAP or mutant α-SNAP–L294A. The released [3H]5-HT was measured as in Figure 4. In an additional titration, 30 μg/mL of apyrase was added to deplete ATP from the reaction (n = 6). (B) In a separate, summary experiment, the effects of 50 μg/mL bovine wild-type α-SNAP, 60 μg/mL anti–α-SNAP antibody, and 60 μg/mL rabbit IgG on the 10 μmol/L Ca++-triggered [3H]5-HT secretion were compared with the control (100%) (n = 6). (C) [3H]5-HT–labeled and SLO-permeabilized platelets were incubated with increasing amounts of the 2E5 monoclonal antibody (anti-NSF; 0-0.32 mg/mL). The release of [3H]5-HT was measured as before and normalized to the control (no addition) (n = 5). (D) The 2E5 inhibitory effect can be reversed by the addition of recombinant NSF. Radiolabeled and SLO-permeabilized platelets were incubated on ice for 30 minutes with buffer (control), 80 μg/mL 2E5, 80 μg/mL plus 0.75 mg/mL recombinant NSF, or 0.75 mg/mL recombinant NSF alone. Platelets were activated by 10 μmol/L Ca++, and the release of [3H]5-HT was measured and normalized to the control group (n = 5).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/3/10.1182_blood.v95.3.921.003k17_921_929/6/m_bloo00317005x.jpeg?Expires=1767701418&Signature=tyEcXUXCRqX~hQTeM-VfYRoUGn1BmUwOyAG49Bnuub~xcqVwBk-jvpFwtvk-CiZTYH8WupYLIF1c4oOM9~tf1gXfLtBefr~wkogiSFB3nlbZKsRv-VKF7TphICqpnrMkU1YxC10J6xm0AFGrlHyasSriU8pMrZtCu86RhQSFJ46MQfmJk1jsIFVgNYKVhQEUxQTTlc5~qPANY7lToXmj7d0b5QTRxIEZyTxXb5ybaKMIpb1KcUrldfGgoszS4IW0uOcWVB-lq9SFo5Y4JC1EnPrBKQOcftk9VT8YJM70xbDrxRfRo1AnCplm2S1zuizCaWibTxHYwCdwJV5lR8Snlg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Anti–SNAP-23 antibody inhibits [3H]5-HT release. / (A) The permeabilized platelet exocytosis was performed in the presence of increasing amounts of Fab23 (0-0.16 mg/mL). After 5 minutes of Ca++ stimulation (10 μmol/L), [3H]5-HT release was measured (n = 3). (B) Secretion triggered by 10 μmol/L Ca++ was analyzed in the presence of Fab23 (0.12 mg/mL), Fab23 (0.12 mg/mL) preincubated with His6–SNAP-23 (0.45 mg/mL), His6–SNAP-23 (0.45 mg/mL), human SNAP-23 C-terminal peptide (0.15 mg/mL), and rabbit IgG purified from preimmune sera (0.15 mg/mL) (n = 6).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/3/10.1182_blood.v95.3.921.003k17_921_929/6/m_bloo00317006x.jpeg?Expires=1767701418&Signature=IbmOXx6mayLO2bqly8EA0LVCEJy-OGLyp856IgJIS~6E9nsw-rhTBeRCK-Ir4-FFICnmEO1MFp7rxGAL-ynnSh1HmiCOYxMUVL8JMBp7U0wL2usiPY3u8DYIfQEK9HT~qDNUjj3HCPlSykxmqE7~n8FGrkKTe1mf4pNXbKZRQI1VjUB-HRYg7l0rL7rbhB2Al0n1DiRAWkDFLLg~jalJymyp8o5nz95hLY529Olg1nSGXORpHBJDVWUkEowROOs3epVS3dgoUY7Fi414eC29ZCmNz8FKXqpHwLmZC6hwsIBpzQL7ZZ3dtOQa4JQ7ExJjcj7VhCPlDUcUBUGEhRhA-A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Anti–syntaxin 2 antibody inhibits [3H]5-HT release. / (A) The permeabilized platelet exocytosis assay was performed in the presence of increasing amounts of anti–syntaxin 2 (▪), 4 (•), and 7 (▴) antibodies and rabbit IgG (⋄ (0-0.18 mg/mL). After stimulation with Ca++ (10 μmol/L), [3H]5-HT release was measured and normalized as a percentage of control release. (B) [3H]5-HT secretion stimulated by 10 μmol/L Ca++ was analyzed in the presence of anti–syntaxin 2 antibody (20 μg/mL), anti–syntaxin 2 antibody (20 μg/mL) preincubated with His6–syntaxin 2 (0.24 mg/mL), His6–syntaxin 2 alone (0.24 mg/mL), anti–syntaxin 2 antibody (0.02 mg/mL) preincubated with His6–syntaxin 4 (0.24 mg/mL), and IgG purified from preimmune sera (20 μg/mL). The [3H]5-HT release was measured and normalized as a percentage of control release (n = 9).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/3/10.1182_blood.v95.3.921.003k17_921_929/6/m_bloo00317007x.jpeg?Expires=1767701418&Signature=Kk4ctX8psMUtZ8g~YrIiEbHATDB6PcwXBvrAumAf~V5WOXT1VaV2WioZU8ER8~rMuyzPlDmuFEJECjTGAlnM6wxoZwpeL2pywbfNcGAXGJSBUkTUFJ7GJ2oDOnEHQcUx3aPqMd~PkllJRnXjnxEw1bNvK57Z9woeRCP~k6axhe3UDK0d0U99OlLYBk1CmP0ucXmpBkjSiiS3TT2b4qAojTgD7RPmXpoS15F9I7uJwajgPQSXLnrneP0zh0w1HT-fuM-JtaJxpGJ-z1Z9p7CVKWMuTMPd0uKPM417jvbrcoXbfGvqt-bCE3suVYzc0OHUxts8llBTYduOlUffGro8Bw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)