High-frequency microsatellite instability (MSI), defined as more than 20% unstable loci, is an inconsistent finding in hematologic malignancies; consequently, the significance of deficient DNA mismatch repair (MMR) to their pathogenesis has been questioned. To further investigate the relationship between MMR deficiency and genomic instability in hematologic malignancies, this study evaluatedMSH2−/− murine lymphomas for insertion/deletion (ID) mutations within the transforming growth factor (TGF)-beta receptor type II (TβR-II) gene and MSI at 10 neutral microsatellites. The lymphomas displayed ID mutations within short mononucleotide runs of TβR-II at a high frequency, whereas nonmalignant tissue from corresponding animals lacked mutations. Loss ofTβR-II transcripts and protein was seen in 6 of 7 murine lymphomas harboring acquired TβR-II mutations. In the analysis of paired nonmalignant and tumor DNA samples, low-frequency but not high-frequency MSI was found. Low-frequency MSI occurred in 8 of 20 lymphomas and 12 displayed microsatellite stability. MSI was even less frequent in nonmalignant tissue as only 3 of 20 samples displayed low-frequency MSI and 17 displayed stability. Evaluation of 20 single cell clones from the MSH2−/− lymphoma cell lines R25 and L15 identified high-frequency MSI in 4 and 2 clones, respectively. The remaining clones showed low-frequency MSI or stability. These findings suggest that acquired TβR-IImutations represent important inactivating events in tumor pathogenesis following MSH2 deficiency. Furthermore, for some hematolymphoid malignancies, the evaluation of cancer-associated genes for ID mutations may represent a more sensitive marker of MMR deficiency than evaluation of neutral microsatellites for high-frequency MSI.
Cancers displaying high-frequency microsatellite instability (MSI), defined as more than 20% unstable loci, are considered to have a defective DNA mismatch repair (MMR) system.1,2 This relationship has been conclusively demonstrated in hereditary non-polyposis colorectal carcinoma (HNPCC) and in some sporadic endometrial, colon, gastric, and pancreatic cancers not associated with HNPCC.2-5 In contrast, tumors displaying low-frequency MSI often appear to have intact MMR gene systems.1-3 High-frequency MSI has not been a consistent finding in hematolymphoid malignancies and thus the significance of MMR deficiency to the molecular pathogenesis of these tumors has been questioned.6-9
Despite the inconsistent finding of high-frequency MSI in hematolymphoid malignancies, strong evidence implicates defects in MMR genes with their development. We reported that MSH2 knockout mice (MSH2−/−) uniformly develop aggressive lymphomas of thymic origin at an early age that recapitulate the human entity precursor T-cell lymphoblastic lymphoma (LBL).10Others have shown that PMS2 and MLH1 knockout mice are prone to develop lymphomas.11,12 In humans, coding region mutations in hMSH2 and hMLH1 have been identified in primary tumor samples from patients with LBL and in acute lymphoblastic leukemia (ALL) cell lines, respectively.10,13Zhu et al found 14 of 43 (32.6%) primary acute myelogenous leukemia (AML) samples lack hMSH2 protein expression.14 Additionally, ID mutations within the BAX(G)8 mononucleotide run were reported in 4 of 29 (14%) human hematolymphoid tumor cell lines not selected for MMR deficiency.15
Nonmalignant tissue harvested from mice lacking MSH2 exhibit hypermutability. MSH2−/− mice carrying thelacI reporter transgene show an increase in spontaneous transition, transversion, and insertion/deletion (ID) mutations and hypermutability on exposure to DNA methylating agents when compared with MSH2+/+ or MSH2+/− mice.16,17Furthermore, the finding of identical ID mutations within short mononucleotide runs (3-5 base pairs) in multiple tissues and in different animals extends the spectrum of sequences that are altered as a consequence of MMR deficiency beyond microsatellites.16
In humans, the TβR-II gene contains a mononucleotide (A)10 repeat within the 5′ end of the coding region that appears to be a consistent target for inactivating ID mutations in gastrointestinal cancers deficient in MMR.18,19 A (GT)3 repeat within the 3′ end of the coding region of TβR-II is also a reported target of mutational inactivation in a minority of MMR-deficient human colon cancers.18 To further investigate the relationship between MMR deficiency and genomic instability we evaluated the thymic lymphomas arising in MSH2−/−mice for ID mutations within the TβR-II gene and the frequencies of MSI at 10 neutral microsatellites. Compared with nonmalignant tissues, the murine lymphomas displayed a high frequency of ID mutations within short mononucleotide runs in theTβR-II gene. A high level of concordance was found betweenTβR-II ID mutations and loss of TβR-II mRNA transcripts and protein. In addition, the murine lymphomas and nonmalignant tissues showed low-frequency but not high-frequency MSI at neutral microsatellites. These findings indicate that disruption of theTβR-II gene is likely an important inactivating molecular event in lymphomagenesis following MMR deficiency. Furthermore, for some hematolymphoid malignancies, the evaluation of genes implicated in tumor pathogenesis for ID mutations may represent a more sensitive marker of MMR deficiency than evaluation of neutral microsatellites for high-frequency MSI.
Materials and methods
Specimens
Details for the generation of MSH2−/− mice and characterization of the lymphomas they develop have been previously reported.10 20 Briefly, the median time to tumor development was 3.8 months and all tumors represented a single histopathologic entity closely resembling human precursor T-cell LBL. By histology, the tumors were comprised of more than 95% malignant cells. Tumor specimens and histologically nonmalignant tail (germline) tissue from each animal were snap frozen in liquid nitrogen and stored at −70°C before DNA and RNA extraction. A portion of each tumor specimen and nonmalignant small intestine tissue was fixed in formalin and embedded in paraffin blocks. The cell lines L15 and R25 were established from thymic LBLs from 2 differentMSH2−/− mice. Twenty single cell clones from each cell line were generated by limiting dilution and expanded into 5 × 105 cells (approximately 19 cell divisions) in tissue culture wells. Cells were harvested by centrifugation and stored as a cell pellet at −70°C before DNA extraction.
Detection of ID mutations in the TβR-II gene by denaturing polyacrylamide gel analysis
High-fidelity polymerase chain reaction (PCR) amplification was performed with PfU polymerase (Invitrogen, San Diego, CA) using genomic DNA isolated from tumor samples and corresponding nonmalignant tissue. The primers JR-1 5′-GAAGATGCCGCTTCTCCCAA-3′, nucleotides 377 to 396, and JR-2 5′-GCTGGTGGTGTATTCTTCCG-3′, nucleotides 486 to 505, were used to amplify a region of the murine TβR-II gene (GenBank Accession # S69 114) containing the mononucleotide (A) repeat corresponding to the (A)10 repeat of the human TβR-II gene. The primers JR-3 5′-GAGACTTTGACCGAGTGCTG-3′, nucleotides 1580 to1599, and JR-4 5′-CCATCTTCTGGAATCTTCTC-3′, nucleotides 1700 to1719, amplified the area of the gene containing the 3′ (GT)3 repeat tract. The blunt-ended PCR products generated byPfU polymerase were resolved on 6% denaturing polyacrylamide, 8.3 mol/L urea gels, transferred to nylon membranes and UV cross-linked. The PCR products were hybridized to internal oligonucleotide probes, JR-14 5′-GGCGAGACTTTCTTCATGTG-3′, nucleotides 425 to 444, or JR-15 5′-GCAGAGCGCTTCAGTGAGCT-3′, nucleotides 1640 to 1659, end-labeled using T4 polynucleotide kinase and γ32P-dATP.
Sequence analysis
The PCR products were cloned using the Zero-Blunt cloning system (Invitrogen). Ten clones from 2 separate PCR experiments were sequenced using the ABI Prism Dye Terminator Cycle Sequencing kit with AmpliTaq FS (Perkin Elmer ABI, Foster City, CA) and analyzed on a 310 Genetic Analyzer (Perkin Elmer ABI).
Reverse transcriptase PCR (RT-PCR)
Total cellular RNA was isolated from tumor samples by the RNAzol B method according to the manufacturer's instructions (Cinna/Biotecx, Friendswood, TX). Single-stranded complementary DNA (cDNA) was prepared from total cellular RNA by reverse transcription according to the manufacturer's recommendations (Perkin-Elmer, Branchburg, NJ). To evaluate expression of TβR-II messenger RNA (mRNA), cDNA was amplified using the primers JR-5 5′-CTCACCTACCACGGCTTCAC-3′, nucleotides 353 to 372, and JR-6.5′-CTCAGCTTCTGCTGCCGGTG-3′, nucleotides 611 to 630. The cycling conditions were 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute for 30 cycles. The primers JR-7 5′-GCTGCTTTCAGGTTTATGAG-3′, nucleotides 701 to 720, and JR-8 5′-GATGCAAGCTAAAAGACATA-3′, nucleotides 899 to 918, were used to evaluate the expression of TβR-I mRNA as a control (GenBank Accession # L15 436). Cycling conditions for these sets of experiments were 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute for 30 cycles. PCR products were visualized by electrophoresis in a 1.8% agarose gel containing ethidium bromide. All experiments were performed in duplicate.
Immunohistochemistry
Immunohistochemical detection of TβR-II was performed using an affinity purified rabbit polyclonal antibody to human TβR-II prepared against a synthetic peptide corresponding to amino acid residues 246 to 266 (L-21; Santa Cruz Biotechnology, Santa Cruz, CA) that cross-reacts to mouse TβR-II. Staining was done on formalin-fixed paraffin embedded tissue sections using the ImmunoCruz Staining System (Santa Cruz Biotechnology). In brief, sections were de-paraffinized and heated at 95°C in 10 mM sodium citrate for 5 minutes. Quenching of endogenous peroxidase activity and blocking of nonspecific protein binding was accomplished using reagents supplied by the manufacturer. Sections were sequentially incubated with prediluted L-21 primary antibody for 2 hours, a biotinylated secondary antibody for 30 minutes, and horseradish peroxidase-strepavidin complex for 30 minutes with phosphate-buffered saline (PBS) washing steps between incubations. After a final wash step, the immunostaining reaction was revealed by incubation with a substrate prepared from substrate buffer, DAB chromogen and peroxidase substrate supplied by the manufacturer. Slides were then counterstained in Gill's hematoxylin.
Western blot analysis
Protein samples were separated by electrophoresis on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel, transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA), and blocked in 5% nonfat milk powder and 0.1% Tween 20 in buffered saline (PBS-T). The membranes were then incubated with a diluted (1:500 in 1% milk powder, PBS-T) rabbit polyclonal antibody, corresponding to amino acids 1 to 567 of human TβR-II that cross-reacts with mouse TβR-II (H-567; Santa Cruz Biotechnology) overnight at 4°C. Following incubation with the primary antibody, the membranes were incubated with a horseradish peroxidase conjugated antirabbit antibody (1:2000 in 1% milk powder PBS-T) for 2 hours at room temperature. The membranes were then developed using a chemiluminescence detection system (NEN Life Science Products, Boston, MA) and exposure to Hyperfilm MP (Amersham Pharmacia Biotech, Piscataway, NJ). The membranes were then incubated in stripping buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 100 mM 2-mercaptoethanol) for 30 minutes at 50°C, washed several times in PBS-T and re-probed with a rabbit polyclonal antibody corresponding to an internal domain of TβR-I of human origin that cross-reacts with mouse TβR-I (V-22; Santa Cruz Biotechnology) with detection as for TβR-II.
Microsatellite instability analysis
DNA was extracted from tumor and corresponding nonmalignant tissue from 20 different MSH2−/− mice and from 20 single cell clones from each cell line using DNAzol (Gibco-BRL, Bethesda, MD) according to the manufacturer's instructions. To assay for MSI, DNA was genotyped at 10 loci: D1Mit4, D2Mit16, D3Mit11, D4Mit11, D5Mit10, D6Mit8, D7Mit12, D8Mit4, D9Mit17, and D10Mit2. PCR was performed using approximately 50 ng genomic DNA, with primers and conditions as recommended by the protocol supplied by Research Genetics for MouseMapPairs (Huntsville, AL). Equal volumes of PCR reaction products and STOP solution (Sequenase kit Version 2.0, USB, Cleveland, OH) were mixed, boiled, and placed on ice. Then 2 μL of the mixture was loaded onto 6% polyacrylamide gels containing 8.3 mol/L urea and electrophoresed at 70 W. Products were transferred to prewetted nylon membranes and the DNA was fixed by UV cross-linking. Microsatellite alleles were detected by hybridization to end-labeled sense primers using T4 polynucleotide kinase and γ32P-dATP and exposure to x-ray film. High-frequency MSI was defined by the presence of unambiguous band shifts in more than 20% of loci evaluated. Low-frequency MSI was defined as 20% or less unstable loci and microsatellite stability as 0% unstable loci.2
Results
ID mutations in TβR-II MSH2−/− murine lymphomas
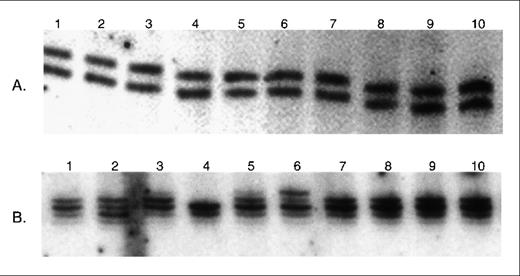
Frameshift ID mutations within the (A)10 repeat of theTβR-II gene occur frequently in MMR-deficient human colon and gastric cancers.18 19 The sequence of the (A) repeat of the murine TβR-II gene 5′-AAAAGAAAAG-3′, nucleotides 411 to 420, differs from the human (A)10 repeat because it contains guanine nucleotides at positions corresponding to the 5th and 10th adenine nucleotides of the human repeat. We analyzed 10 of theMSH2−/− murine lymphomas for ID mutations within a 129 bp DNA fragment containing the (A) repeat by denaturing polyacrylamide gel electrophoresis. Comparison of tumor DNA with corresponding nonmalignant DNA showed mobility shifts consistent with acquired insertion mutations in 6 of 10 (60%) murine lymphomas (Figure1). Sequence analysis confirmed the presence of acquired mutations within the (A) repeat in all 6 lymphomas. In 4 of the 6 lymphomas, the second half of the repeat (nucleotides 416-419) was expanded by 1 adenine nucleotide. The wild-type TβR-II protein consists of 567 amino acids and these frameshift mutations result in the generation of a premature STOP codon predicted to encode a truncated protein of 137 amino acids. In the other 2 lymphomas, acquired insertion mutations occurred in both the first and second halves of the repeat (Figure2). Sequence analysis of the 129 bp DNA fragment from the 4 lymphomas lacking mobility shifts and from nonmalignant tissue from the 6 mice with ID mutations in the tumor DNA showed the wild-type sequence in all cases.
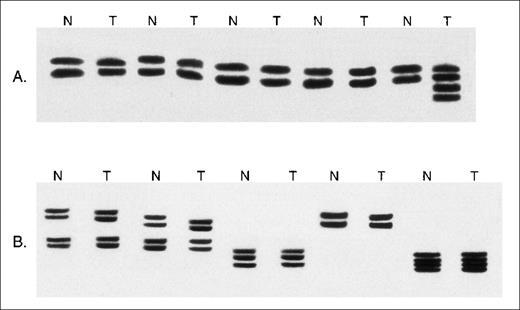
Denaturing polyacrylamide gel analysis of theTβR-II gene in MSH2−/− murine lymphomas.
Examples of the analysis of DNA containing the mononucleotide (A) repeat from paired nonmalignant (N) and whole lymphoma tumor specimens (T). Slow migrating bands consistent with acquired frameshift insertion mutations are seen in the tumors in 3 of the 4 paired specimens.
Denaturing polyacrylamide gel analysis of theTβR-II gene in MSH2−/− murine lymphomas.
Examples of the analysis of DNA containing the mononucleotide (A) repeat from paired nonmalignant (N) and whole lymphoma tumor specimens (T). Slow migrating bands consistent with acquired frameshift insertion mutations are seen in the tumors in 3 of the 4 paired specimens.
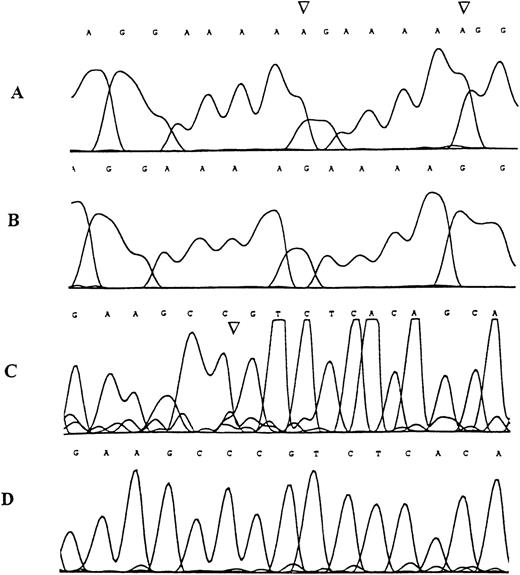
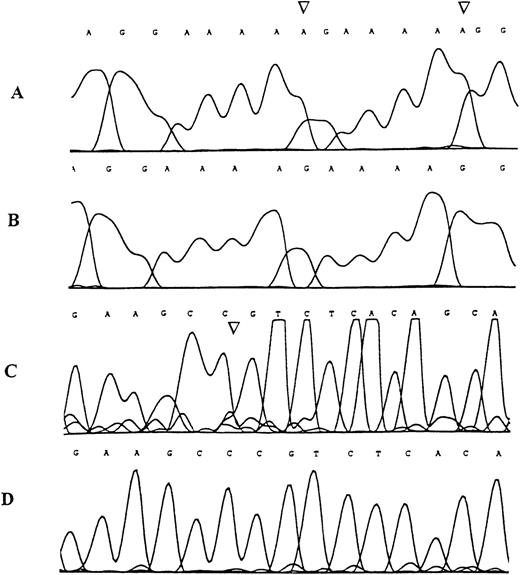
Sequence analysis of the TβR-II gene inMSH2−/− murine lymphomas.
Examples of the genomic sequence profile of clones obtained from tumor DNA specimens demonstrating denaturing polyacrylamide gel band shifts in the region containing the mononucleotide (A) repeat (A and B) and in the region containing the (GT)3 repeat (C and D). (A) Mutant sequence with adenine insertion mutations (arrows), (B) corresponding wild type sequence, (C) mutant sequence with cytosine deletion mutation (arrow), (D) corresponding wild type sequence.
Sequence analysis of the TβR-II gene inMSH2−/− murine lymphomas.
Examples of the genomic sequence profile of clones obtained from tumor DNA specimens demonstrating denaturing polyacrylamide gel band shifts in the region containing the mononucleotide (A) repeat (A and B) and in the region containing the (GT)3 repeat (C and D). (A) Mutant sequence with adenine insertion mutations (arrows), (B) corresponding wild type sequence, (C) mutant sequence with cytosine deletion mutation (arrow), (D) corresponding wild type sequence.
The (GT)3 repeat within the 3′ end of TβR-II coding region is also a reported target of mutational inactivation in a minority of MMR-deficient human colon cancers.18 We analyzed the same 10 MSH2−/− murine lymphomas for acquired ID mutations within a 140 bp DNA fragment of theTβR-II gene containing the (GT)3 repeat. One lymphoma showed a mobility shift by denaturing polyacrylamide gel electrophoresis consistent with an acquired deletion mutation. Sequence analysis confirmed an acquired deletion mutation as 1 of 3 cytosines in nucleotides 1617 to 1619 located just upstream of the (GT)3 repeat was deleted (see Figure 2). This deletion results in a premature STOP codon predicted to code for a truncated protein of 528 amino acids. Sequencing of the 9 lymphomas whose PCR products lacked mobility shifts and nonmalignant tissue from 6 mice showed the wild-type sequence in all cases.
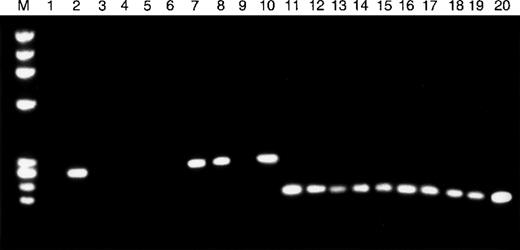
The ID mutations located in the (A)10 repeat of the humanTβR-II gene are associated with reduced levels ofTβR-II mRNA.18 Therefore, we evaluated all 10 murine lymphomas for TβR-II transcripts by RT-PCR. All 6 of the lymphomas harboring ID mutations within the (A) repeat showed an absence of TβR-II mRNA (Figure3). In contrast, TβR-II mRNA was detectable in the lymphoma containing the deletion mutation within the region of the (GT)3 repeat and in lymphomas lacking TβR-IIgene mutations. Immunohistochemical and Western blot analysis confirmed the RT-PCR results. Murine lymphomas without detectable RNA transcripts by RT-PCR lacked detectable protein by either of these methods, whereas those with positive RT-PCR results showed detectable protein. In contrast, all of the murine lymphomas had TβR-I protein by Western blot analysis (Figure 4).
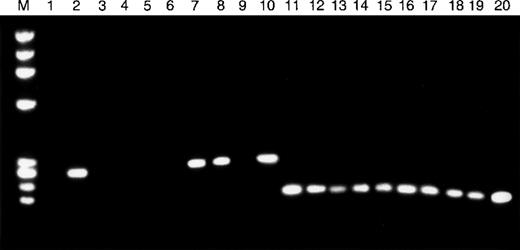
TβR-II mRNA expression in MSH2−/−murine lymphomas.
RT-PCR was used to assess the expression of TβR-II (lanes 1-10) and TβR-I (lanes 11-20) mRNA in 10 whole lymphoma tumor specimens. Tumor specimens with frameshift mutations in the mononucleotide (A) repeat of TβR-II (lanes 1, 3-6, 9) show absence of TβR-II mRNA transcripts, whereas all other tumor specimens (lanes 2, 7, 8, 10) express TβR-II. All tumor specimens, including those with TβR-II frameshift mutations, show TβR-I mRNA transcripts.
TβR-II mRNA expression in MSH2−/−murine lymphomas.
RT-PCR was used to assess the expression of TβR-II (lanes 1-10) and TβR-I (lanes 11-20) mRNA in 10 whole lymphoma tumor specimens. Tumor specimens with frameshift mutations in the mononucleotide (A) repeat of TβR-II (lanes 1, 3-6, 9) show absence of TβR-II mRNA transcripts, whereas all other tumor specimens (lanes 2, 7, 8, 10) express TβR-II. All tumor specimens, including those with TβR-II frameshift mutations, show TβR-I mRNA transcripts.
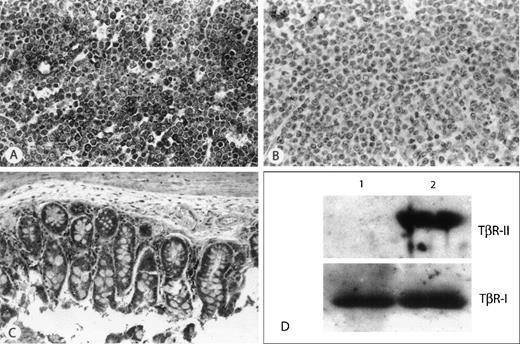
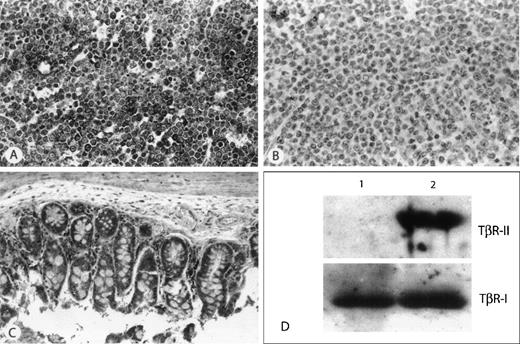
Immunohistochemical and Western Blot detection of TβR-II protein.
(A) Example of a thymic lymphoma with wild-type TβR-II (A) repeat and detectable mRNA by RT-PCR. All tumor cells show strong immunohistochemical staining for TβR-II. (B) Example of a thymic lymphoma harboring an (A) repeat ID mutation, and showing no detectable mRNA by RT-PCR, demonstrates the absence of immunohistochemical staining for protein. (C) Nonmalignant tissue from the small intestine of the animal from panel B shows strong immunohistochemical staining for TβR-II. (D) Representative Western analysis of 2 murine lymphomas. Lane 1, lymphoma harboring TβR-II (A) repeat ID mutation; lane 2, lymphoma showing wild-type TβR-II (A) repeat. Top panel demonstrates absence of TβR-II protein in lane 1. Bottom panel is the same blot re-probed with an antibody to TβR-I as control.
Immunohistochemical and Western Blot detection of TβR-II protein.
(A) Example of a thymic lymphoma with wild-type TβR-II (A) repeat and detectable mRNA by RT-PCR. All tumor cells show strong immunohistochemical staining for TβR-II. (B) Example of a thymic lymphoma harboring an (A) repeat ID mutation, and showing no detectable mRNA by RT-PCR, demonstrates the absence of immunohistochemical staining for protein. (C) Nonmalignant tissue from the small intestine of the animal from panel B shows strong immunohistochemical staining for TβR-II. (D) Representative Western analysis of 2 murine lymphomas. Lane 1, lymphoma harboring TβR-II (A) repeat ID mutation; lane 2, lymphoma showing wild-type TβR-II (A) repeat. Top panel demonstrates absence of TβR-II protein in lane 1. Bottom panel is the same blot re-probed with an antibody to TβR-I as control.
Microsatellite instability in MSH2−/− murine lymphomas
A common strategy to detect underlying MMR deficiency is the comparison of germline DNA with tumor DNA for MSI. We evaluated the thymic lymphomas arising in 20 MSH2−/− mice for instability at 10 neutral microsatellites. In the analysis of 200 paired nonmalignant and tumor DNA samples high-frequency MSI was not observed. Low-frequency MSI occurred in 8 of 20 lymphomas with none showing alterations at more than one microsatellite. Twelve of 20 lymphomas displayed microsatellite stability (Figure5). MSI was even less frequent in nonmalignant tissue because none of the 20 tail tissue samples displayed high-frequency MSI, 3 of 20 displayed low-frequency MSI, and 17 of 20 displayed microsatellite stability.
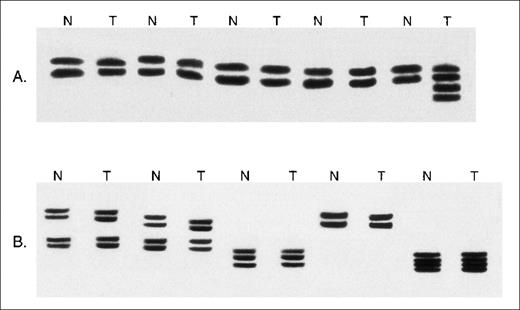
Microsatellite analysis of MSH2−/− murine lymphomas.
(A) Examples of the analysis of DNA from paired nonmalignant (N) and whole lymphoma tumor specimens (T) at the D5Mit10 marker. Novel microsatellite alleles are present in 1 tumor specimen. (B) Examples of the analysis of DNA from paired normal tissue (N) and whole tumor tissue (T) at the D7Mit12 marker. No microsatellite instability is seen.
Microsatellite analysis of MSH2−/− murine lymphomas.
(A) Examples of the analysis of DNA from paired nonmalignant (N) and whole lymphoma tumor specimens (T) at the D5Mit10 marker. Novel microsatellite alleles are present in 1 tumor specimen. (B) Examples of the analysis of DNA from paired normal tissue (N) and whole tumor tissue (T) at the D7Mit12 marker. No microsatellite instability is seen.
A more sensitive strategy to detect MSI uses DNA from single cell clones.21 We evaluated DNA from 20 single cell clones from the MSH2−/− murine thymic lymphoma cell lines R25 and L15 for instability at the same 10 neutral microsatellites used to detect MSI in whole tumor DNA (Figure 6). For the R25 cell line, high-frequency MSI was found in 4 of 20 clones, low-frequency MSI in 14 of 20 clones, and stability in 2 of the 20 clones. In the 4 high-frequency MSI clones, instability was seen in 6 of the microsatellite loci in 1 clone, 5 of the loci in 2 clones, and 4 of the loci in 1 clone. For the L15 cell line, high-frequency MSI was less common than in the R25 cell line and was observed in 2 of 20 clones. Low-frequency MSI was seen in 16 of 20 clones and stability in 2 clones.
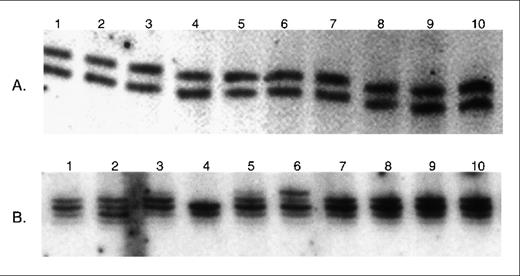
Microsatellite analysis of MSH2−/− murine lymphoma single cell clones.
(A) Examples of analysis of DNA from single cell clones derived from the R25 cell line at D6Mit8 marker. No microsatellite instability is seen. (B) Examples of analysis of DNA from single cell clones derived from the L15 cell line at D8Mit14 marker. Novel microsatellite alleles are present in 2 of the clones (lanes 2 and 6).
Microsatellite analysis of MSH2−/− murine lymphoma single cell clones.
(A) Examples of analysis of DNA from single cell clones derived from the R25 cell line at D6Mit8 marker. No microsatellite instability is seen. (B) Examples of analysis of DNA from single cell clones derived from the L15 cell line at D8Mit14 marker. Novel microsatellite alleles are present in 2 of the clones (lanes 2 and 6).
Discussion
We previously reported that MSH2−/− mice uniformly develop aggressive thymic lymphomas of a single histopathologic subtype at an early age and demonstrate aberrant activation of T-cell associated oncogenes, RBTN-2 andTAL-1.10 In this study, we used theMSH2−/− murine lymphomas as a model to investigate the spontaneous mutations arising in these tumors in vivo as a consequence of MMR deficiency. A high frequency of ID mutations within the TβR-II gene was observed in the tumors of different animals, whereas the corresponding nonmalignant tissue lacked mutations. These findings suggest that in addition to the molecular activation of RBTN-2 and TAL-1, acquiredTβR-II mutations represent important molecular inactivating events in tumor pathogenesis following MSH2 deficiency. In contrast, when tumor and nonmalignant MSH2−/−tissues were compared for the frequency of MSI at neutral microsatellites no appreciable difference was observed because both showed low-frequency but not high-frequency MSI. This lack of difference implies that analysis of cancer-associated genes for ID mutations may be a more sensitive indicator of underlying MMR deficiency than analysis of neutral microsatellites for MSI.
It has been previously established that MSH2 and PMS2knockout mice exhibit hypermutability consistent with deficient MMR.16,17 22 Nonmalignant tissues harvested fromMSH2−/− and PMS2−/− mice bearing the neutral reporter transgenes lacI, supF, orsupFG1 demonstrate a high frequency of base transitions, transversions, and ID mutations in short mononucleotide runs. In the present study, we evaluated a known tumor suppressor gene and found that the TβR-II ID mutations occurred in short mononucleotide runs. Our findings corroborate that short mononucleotide runs may represent potential mutational hotspots in the setting of MMR deficiency. The high frequency of ID mutations within the (A)10 repeat of TβR-II in human MMR-deficient cancers further supports this contention.
Escape from TGF-β-mediated growth inhibition, due to mutations inTβR-II or loss of TβR-II surface protein expression (or both), is a defined pathogenetic mechanism in many cancers, including some hematolymphoid malignancies.23,24 In MMR-deficient human cancers, ID mutations within the TβR-II (A)10 repeat are predominantly deletions and are associated with reduced levels ofTβR-II mRNA transcripts that are thought to be secondary to decreased mRNA stability.18 In contrast to ID mutations in the TβR-II (A)10 repeat in human cancers, all of the ID mutations within the murine TβR-II mononucleotide A repeat were insertions. The reason for this difference is not entirely clear but may reflect sequence differences because the murine repeat contains guanine nucleotides at positions corresponding to the 5th and 10th adenine nucleotides of the human repeat. A high frequency of insertion mutations has been reported in other mononucleotide runs in the setting of MMR deficiency. For example, in human MMR-deficient gastrointestinal cancers, the BAX (G)8 and hMSH6 (C)8 mononucleotide runs show a high frequency of insertion mutations.19 25Similar to studies of MMR-deficient human cancers, theMSH2−/− murine lymphomas showed concordance between the presence of ID mutations within the mononucleotide A repeat ofTβR-II and loss of TβR-II mRNA transcripts and protein as detected by RT-PCR, immunohistochemistry, and Western blot analysis. These ID mutations were detected on 1 allele. Thus, other genetic or epigenetic changes may be present to account for the loss of transcripts and protein. For example, our study analyzed 269 of the 1704 bp in the TβR-II coding region and it is possible other mutations may be present. Furthermore, epigenetic changes such as gene inactivation through promoter hypermethylation remain to be investigated. Our findings strengthen the association between acquired abnormalities in TβR-II following MSH2 deficiency and tumorigenesis and suggest that some hematolymphoid malignancies may share similar pathogenetic mechanisms as some solid cancers.
The lack of TβR-II ID mutations in the nonmalignant tail tissue of MSH2−/− mice is not entirely clear but may in part reflect tissue-specific differences in mutational frequencies. In studies of MSH2−/− mice bearing thelacI transgene, ID mutations were greatest in tissues characterized by having a comparatively high cell turnover rate such as the small intestine and thymus.16 17
In contrast to the high frequency of acquired TβR-II ID mutations in MSH2−/− murine lymphomas, only low-frequency MSI was detected in the analysis of neutral microsatellites. Others have also found a lack of high-frequency MSI in some tumors arising in the setting of bona fide MMR deficiency.1,26-28 Tumor growth kinetics and tumor age are believed to be important determinants for frequency of MSI. Shibata et al used xenografts of single cell clones of the human mutator phenotype colorectal cell line HCT 116 to simulate mutator phenotype tumors with rapid growth rates and found a low frequency of altered microsatellite alleles despite a high intrinsic mutation rate.26,27 In comparison, slow-growing adenomas displayed greater microsatellite allele diversity.26,27 In MSH2−/−,APC +/− mice, intestinal adenomas develop at a young age that lack MSI.28 The short mitotic history of these adenomas has been suggested as a possible explanation for the lack of MSI.1,28 Thus, in rapidly growing tumors, absence of high-frequency MSI may not preclude underlying MMR deficiency. Taken together, we reason that the lack of high-frequency MSI in theMSH2−/− murine lymphomas may have its basis in the rapid growth characteristics of these aggressive high-grade lymphomas and their short mitotic history, because the median time to tumor development is 3.8 months.10
Analysis of single cell clones is a more sensitive method to detect MSI compared with analysis of whole tumor samples.21 In keeping with the increased sensitivity, we observed high-frequency MSI in a minority of single cell clones obtained from the R25 and L15 cell lines, 20% and 10%, respectively. Although analysis of single cell clones increases the ability to detect MSI, it remains an impractical method to screen human cancers for underlying MMR deficiency.
Our finding that links MMR deficiency with frameshift mutations within the TβR-II gene may not be generalized to all subtypes of hematolymphoid malignancies. Childhood ALL represents a subtype of hematolymphoid malignancy with a distinct biologic behavior associated with a generally favorable outcome. Molenaar et al found mutations within the repeat tracts of the BAX and TβR-II genes in 3 of 6 adult ALL cell lines but not in 55 cases of childhood ALL.29 Thus, in humans, MMR deficiency and TβR-IIframeshift mutations may represent pathogenetic events more important to the development of adult leukemia/lymphoma.
High-frequency MSI has been an inconsistent feature in human hematolymphoid malignancies and consequently MMR gene defects have been thought not to significantly contribute to their molecular pathogenesis. Acquired alterations within neutral microsatellites likely do not confer a selective growth advantage and may be difficult to detect amid the large background of normal alleles, even when using a more sensitive method such as the analysis of single cell clones.1,21,26-28 The lack of high-frequency MSI inMSH2−/− lymphomas implies that MSI at neutral microsatellites may not always reflect MMR status. Rather, we observed a high frequency of ID mutations within a cancer-associated gene,TβR-II, in the MSH2−/− lymphomas. Whether these ID mutations represent commonly occurring early events in tumor pathogenesis or events that contribute to clonal expansion and dominance remains to be determined.1 Irrespective of the potential role of these mutations in cancer development or progression, we submit that for some rapidly growing hematolymphoid malignancies, the evaluation of cancer-associated genes for ID mutations may represent a more sensitive marker of MMR deficiency than evaluation of neutral microsatellites for high-frequency MSI.
Acknowledgments
The authors would like to thank Mr Bob van den Beuken and Mr Todd Reichert for assistance in preparing the figures, and Dr T Al-Tweigeri for his support.
Supported by grants from the Medical Research Council of Canada, the Health Service Utilization and Research Commission of Saskatchewan, Saskatchewan Cancer Agency, and Saskatchewan Health. R.L., A.M., and J.F.D. are recipients of Medical Research Council of Canada Awards.
Reprints:John F. DeCoteau, Department of Pathology, Royal University Hospital, University of Saskatchewan, 103 Hospital Drive, Saskatoon, Saskatchewan, Canada, S7N 0W8; e-mail: decoteauj@sdh.sk.ca.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.