Granulocyte colony-stimulating factor (G-CSF) can effectively mobilize hematopoietic stem and progenitor cells from bone marrow into blood, thereby allowing peripheral blood stem cells (PBSCs) to be used for transplantation. The efficiency of PBSC mobilization response to G-CSF is a multigene trait. DBA/2 (high-responder) and C57BL/6 (low-responder) mice were used for a genetic analysis of G-CSF–induced progenitor release. Significant linkages were found on chromosome 2 by analyzing segregation distortion among the high responders of 500 backcross mice and on chromosome 11 by using the quantitative trait locus analysis of 26 strains of BXD recombinant inbred mice.
Granulocyte colony-stimulating factor (G-CSF) is a major regulator of neutrophilic granulocyte production in the bone marrow and the subsequent functional activation of mature neutrophils.1,2 As such, recombinant human G-CSF (rhG-CSF) has been used extensively to promote granulocyte formation in chronic idiopathic neutropenia, in chemotherapy-induced myelosuppression, and following allogenic or autologous bone marrow transplantation. G-CSF also has the capacity to mobilize stem and progenitor cells to the peripheral blood, and peripheral blood stem cells (PBSCs) have proven to be a superior population for transplantation.3 There is wide individual variation in PBSC mobilization in response to G-CSF,1-3 and similar quantitative variation has been noted in the responses of inbred strains of mice.4 Analysis has shown that multiple mechanisms may operate to determine the release of stem and progenitor cells to the blood in response to agents such as G-CSF. These mechanisms include alteration in adhesion interactions between stromal, endothelial, and hematopoietic cells in the early phases of the response3 5-8 and less well-characterized events to sustain subsequent PBSC elevations.
We analyzed PBSC responses in various mouse strains to identify genetic loci associated with quantitative differences in the response to injection of G-CSF.
Materials and methods
Mice
B6D2F1 (F1) mice were bred by mating female C57BL/6J (B6) and male DBA/2J/WEHI (D2) mice (Walter Eliza Hall Institute of Medical Research, Victoria, Australia). Two independent backcross experiments were performed comprising 260 mice (165 male and 95 female) bred from D2 female and F1 male mice and 245 mice (122 male and 123 female) bred from F1 and D2 mice. We used 26 male mice from the BXD/Ty recombinant inbred (RI) strains (The Jackson Laboratory, Bar Harbor, ME). All mice were maintained and bred under specific pathogen-free conditions and used in all experiments at 7-11 weeks of age.
Phenotyping
We diluted rhG-CSF (filgrastim; Amgen, Thousand Oaks, CA) in sterile normal saline containing 5% bovine newborn calf serum (Hyclone, Logan, UT). Twice daily for 5 days, mice were injected intraperitoneally with 40 μg/kg rhG-CSF. Blood was harvested on the sixth day and cultured in semisolid 1-mL agar cultures as previously described.4 After 7 days each culture was fixed and stained as described.4 Triplicate plates were counted, and the average value was represented as colonies per μL of peripheral blood colony-forming cells (PB-CFCs).
Genotyping
Mice were genotyped at an average 10-centimorgan (cM) resolution with fluorescent markers as described (Burt et al, unpublished data, January 1999). The genotypes of 26 BXD RI strains of mice were downloaded (Mouse Genome Informatics; The Jackson Laboratory,), supplemented with our own data, and analyzed (MapManager QTb19ppc; http://mcbio.med.buffalo.edu/mapmgr.html).
Results
Linkage analysis of backcross mice
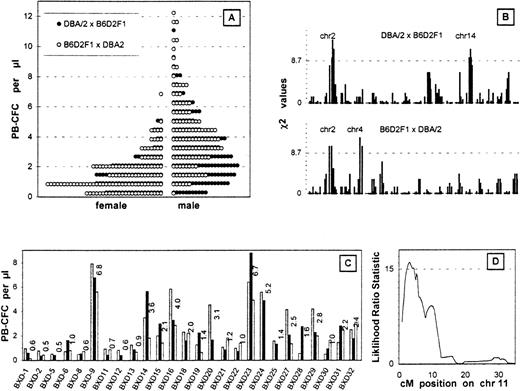
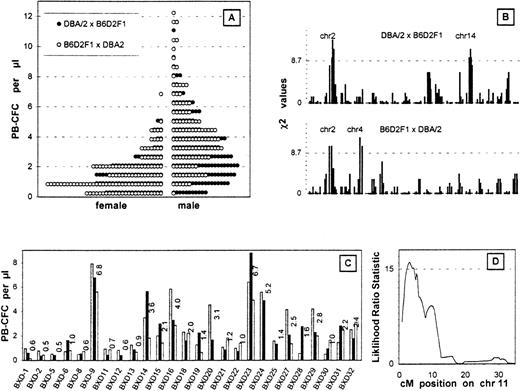
G-CSF mobilized the progenitor cells into the peripheral blood more effectively in D2 mice than in B6 mice. After 5 days of injections, the average PB-CFC counts were 5-6 colonies/μL in male D2 mice and 0-1 colonies/μL in male B6 mice.4 Figure1A shows the PB-CFC counts in the backcross mice. Female PB-CFC counts were almost half the value of the male counts; no differences were seen between the reciprocal female crosses; and males bred with F1 and D2 mice showed slightly higher PB-CFC counts than females bred with D2 and F1 mice.
Analysis of the backcross mice.
(A) PB-CFC values after 5 days of G-CSF injection. The histograms are the data of 165 males and 95 females bred from D2 and F1 mice and 122 males and 123 females bred from F1 and D2 mice binned by an interval of 0.6 colonies/μL. (B) Linkage analysis of PB-CFC values across the entire genome of both backcross cohorts. The χ2 analysis measures the support for Mendelian inheritance ratios of 1:2:1 at each locus. Only high responders were used for the analysis. The dotted line depicts 32 males and 20 females bred from D2 and F1 mice and 24 males and 24 females bred from F1 and D2 mice. A threshold for suggestive linkage is shown by the dotted line. Chromosomes showing significant or suggestive linkage are indicated. (C) Phenotype of BXD RI strains using 2-3 mice from each strain is plotted as individual histograms. (D) Interval mapping on chromosome 11 using the BXD RI lines. This quantitative trait analysis was performed using the ln PB-CFC values (MapManager QTb19ppc). The threshold for significant linkage is shown by the dotted line.
Analysis of the backcross mice.
(A) PB-CFC values after 5 days of G-CSF injection. The histograms are the data of 165 males and 95 females bred from D2 and F1 mice and 122 males and 123 females bred from F1 and D2 mice binned by an interval of 0.6 colonies/μL. (B) Linkage analysis of PB-CFC values across the entire genome of both backcross cohorts. The χ2 analysis measures the support for Mendelian inheritance ratios of 1:2:1 at each locus. Only high responders were used for the analysis. The dotted line depicts 32 males and 20 females bred from D2 and F1 mice and 24 males and 24 females bred from F1 and D2 mice. A threshold for suggestive linkage is shown by the dotted line. Chromosomes showing significant or suggestive linkage are indicated. (C) Phenotype of BXD RI strains using 2-3 mice from each strain is plotted as individual histograms. (D) Interval mapping on chromosome 11 using the BXD RI lines. This quantitative trait analysis was performed using the ln PB-CFC values (MapManager QTb19ppc). The threshold for significant linkage is shown by the dotted line.
The top 20% of responders or the high responders from each group were analyzed for the frequencies of homozygous D2 alleles. Linkage was determined when a deviation in segregation from the normal mendelian ratios in the high responders was observed (Figure 1B). One statistically significant linkage was found on chromosome 2, which was seen in each of the backcrosses (mice bred with F1 and D2 and mice bred with D2 and F1) (Table1). Two suggestive linkages were seen on chromosomes 4 and 14 (Figure 1B).
BXD recombinant inbred mice
We calculated associations between the G-CSF–induced PB-CFC values of the 26 BXD RI strains (Figure 1C) and their genotypes at each locus across the genome (MapManager QTb19ppc, ). RI strains the inbred products of an F1 intercross and hence are homozygous for either parental chromosome in approximately equal doses. Significant linkage was seen on chromosome 11; the highest likelihood ratio statistic of 15.1 was observed at D11Ncvs42, and 16.0 was observed at the peak position (3 cM) after the interval mapping (Figure 1D). Since this linkage was not seen in the backcross experiments, the locus is likely to be D2 dominant.
Discussion
Genetics is a powerful tool in studying phenotypes that only manifest at a whole animal level. Loci which contain genes that control the phenotype can be identified, as demonstrated in this study, with 2 loci being implicated in the control of progenitor release from the marrow. In addition, relationships to other phenotypes can be identified. A strong inverse correlation has been reported between the lifespan of different strains and the number of autonomously cycling progenitors (day-7 Cobblestone Area Forming Units [CAFC]) in the femur.9 A putative locus causing the variability of lifespan (small variance in D2) was mapped near the centromere on chromosome 11,10 a region which is coincident with our result using the same BXD RI mapping strains for mapping PB-CFC mobilization. It is therefore formally possible that these 2 phenotypes are controlled by the same gene.
Possible candidate genes for PB-CFC mobilization include histidine decarboxylase (chromosome 2), a gene known to be induced by G-CSF in bone marrow11 and found to have possibly relevant sequence polymorphisms between D2 and B6 mice (M. Hasegawa, unpublished data, September 1998; Genbank accession number AF109 137).11-13 The integrin subunit α4 (Genbank accession number AF109 136), another possible candidate on chromosome 4, and Transcobalamin II (Genbank accession number AF090 686),14 15 a chromosome 11 candidate (M. Hasegawa, unpublished data, September 1998), were found to have identical coding regions between the D2 and B6 mice.
In summary, 2 loci on chromosomes 2 and 11 control the progenitor mobilization response to G-CSF, with possible contributions from loci on chromosomes 4 and 14. The complexity of this multigene trait will be biologically dissected by the breeding of mice congenic for each locus.
Acknowledgments
We thank Sandra Mifsud and the supporting staff at Animal Services, the DNA Sequencing Lab, and Australian Genome Research Facility, Melbourne, Australia.
Supported by grants from the Chugai Research Institute for Molecular Medicine Inc., Chugai, Japan; The Wellcome Trust (S. Foote), London, United Kingdom; and the National Health and Medical Research Council (NHMRC), Australia.
The sequences reported in this paper have been deposited in the Genbank database under accession numbers AF090 686 (Tcn2), AF109 136 (Itga4), and AF109 137 (B6 Hdc).
Reprints:Simon J. Foote, The Walter and Eliza Hall Institute of Medical Research, Post Office, Royal Melbourne Hospital, 3050 Victoria, Australia; e-mail: foote@wehi.edu.au.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.