Abstract
The non-Hodgkin lymphoma (NHL) subtype anaplastic large-cell lymphoma (ALCL) is frequently associated with a t(2;5)(p23;q35) that results in the fusion of the ubiquitously expressed nucleophosmin (NPM) gene at 5q35 to the anaplastic lymphoma kinase (ALK) gene at 2p23, which is not normally expressed in hematopoietic tissues. Approximately 20% of ALCLs that expressALK do not contain the t(2;5), suggesting that other genetic abnormalities can result in aberrant ALK expression. Here we report the molecular characterization of an alternative genetic means of ALK activation, the inv(2)(p23q35). This recurrent abnormality produces a fusion of the amino-terminus of 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), a bifunctional homodimeric enzyme that catalyzes the penultimate and final steps of de novo purine nucleotide biosynthesis, with the intracellular portion of the ALK receptor tyrosine kinase. RT-PCR analysis of 5 ALCL tumors that contained the inv(2) revealed identical ATIC-ALK fusion cDNA junctions in all of the cases. Transient expression studies show that theATIC-ALK fusion transcript directs the synthesis of an approximately 87-kd chimeric protein that is localized to the cytoplasm, in contrast to NPM-ALK, which typically exhibits a cytoplasmic and nuclear subcellular distribution. ATIC-ALK was constitutively tyrosine phosphorylated and could convert the IL-3–dependent murine hematopoietic cell line BaF3 to cytokine-independent growth. Our studies demonstrate an alternative mechanism for ALK involvement in the genesis of NHL and suggest that ATIC-ALK activation results from ATIC-mediated homodimerization. In addition, expected decreases in ATIC enzymatic function in ATIC-ALK–containing lymphomas may render these tumors more sensitive to antifolate drugs such as methotrexate.
Roughly 40% to 60% of anaplastic large cell lymphomas (ALCLs) aberrantly express truncated and constitutively active anaplastic lymphoma kinase (ALK) fusion proteins, most frequently as the t(2;5)(p23;q35)-associated nucleophosmin (NPM)-ALK chimera.1-6 Because ALK expression is normally restricted to neural tissues,7,8 immunostaining of lymphomas with ALK-specific antibodies is a convenient and reliable means to identify ALK-positive tumors.1,2,9 ALK immunostaining of NPM-ALK–positive ALCL cases shows a characteristic cytoplasmic and nuclear distribution of the chimeric ALK protein that is due to hetero-oligomerization of NPM-ALK and normal NPM, a shuttle protein that normally moves ribonucleoproteins between the cytoplasm and nucleus but that can aberrantly transport NPM-ALK to the nucleus.10 Large clinico-pathologic studies of ALCL have shown about 80% of cases to exhibit this cytoplasmic and nuclear staining pattern indicative of the presence of NPM-ALK, whereas the remaining 20% express ALK proteins only in the cytoplasm of tumor cells,1 2 suggesting that a fusion partner other than NPM might be found in these cases.
Coincident with the identification of “cytoplasmic only” ALK-positive ALCLs, a novel chromosomal abnormality, inv(2)(p23q35), was recently discovered in cases possessing this staining pattern.11 The inv(2)(p23q35) had previously been unrecognized as a recurrent aberration, most probably because identification of the inversion by classical cytogenetics is difficult, the involved terminal bands on the arms of chromosome 2 having similar sizes and banding patterns. Fluorescence in situ hybridization (FISH) analysis of inv(2) cases has confirmed the involvement of theALK locus at 2p23, and the clinical and pathologic characteristics of inv(2)-positive ALCLs appear to be very similar to typical t(2;5)/NPM-ALK–containing cases.11 Here we report the molecular characterization of the inv(2)(p23q35), identifying the novel fusion ATIC-ALK that shares functional qualities with NPM-ALK, which correlate with lymphomagenic capabilities.
Materials and methods
Clinical cases
Clinical, pathologic, and cytogenetic data for ALCL patients 1 through 3, all of whom had tumors shown to contain the inv(2), have been reported previously.11 Patients 4 and 5 were both female, aged 13 and 15 years, respectively, and had ALCL that was ALK-positive by immunostaining, with expression limited to the cytoplasm of tumor cells. Interphase FISH of these 2 cases indicatedALK alteration without NPM rearrangement, and suggested the presence of the inv(2) (R.S., S.G. and B.S., unpublished data).
Long-distance inverse–polymerase chain reaction (LDI-PCR)
High-molecular-weight DNA from the tumor of patient 2 was digested to completion with EcoRI or XbaI and ligated at low concentration (0.8 ng/μL) to promote the formation of monomeric circles. The DNA was then purified using a Wizard column (Promega) and long-distance inverse–polymerase chain reaction (LDI-PCR) was performed in a nested reaction using primers designed to anneal to theALK exon encoding the juxtamembrane portion of the protein: Internal IN5I, 5′-AGGTGCGGAGCTTGCTCAGCTTGTAC-3′; Internal IN3I, 5′-CTGCTTTGCTGGCAAGA CCTCCTCCA-3′; External IN5E, 5′-TCTGGGGTCGACATGGCTTGCAGCTCCTGGTGCTT CCGGCGG-3′; External IN3E, 5′-GAGGTGCCGCGGAAAAACATCACCCTCATTCG-3′. Aliquots of the reactions were separated in a 0.8% agarose gel, excised, and purified using the Qiaquick Gel Extraction kit (Qiagen), then ligated into the TA cloning vector pCR2.1 (Invitrogen) for further analysis.
PAC clone isolation and fluorescence in situ hybridization analysis
PAC clones from the 2q35 ATIC gene locus were identified using a primer pair (2q35A,5′-AGGCATGGAGGCCTTGGAATC-3′ and 2q35B,5′-CAGATGCAGCTTCTGCTCTC-3′) derived from intronic sequences located within 1.8 kilobase (kb) adjacent to the 2q35 breakpoint in the tumor from patient 2. These primers, which amplify a 1.35-kb product, were used to screen a human PAC library (Genome Systems) and 2 clones (clone addresses 213D24 and 218E3) were identified. Although sequence analysis of the portion of these clones immediately surrounding the location of the 2q35 breakpoint failed to identify ATIC exons (see text), hybridizations performed subsequently using an ATIC oligonucleotide from the 5′ coding region of the gene (5′-ACCTGACCGCTCTTGGTTTG-3′) confirmed the presence of the locus. FISH with PAC213D24 on metaphases and interphases from inv(2)-positive tumors was performed as previously described.11
5′ RACE and ATIC-ALK RT-PCR
Identification of the ALK partner gene of the inv(2)(p23q35) was performed with total RNA from the tumor of patient 1 using RACE. The RNA (1 μg) was reverse transcribed using MMLV reverse transcriptase (Gibco BRL) and ALK oligonucleotide primer 5′-TGATGATCAGGGCTTCCATGAGG-3′ and the purified first strand cDNA was then tailed, using terminal deoxynucleotidyl transferase and dATP. Second strand synthesis was performed using Klenow polymerase and oligonucleotide 5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTT-3′, and anchored PCR was performed for 35 cycles with primers 5′-CCAGTGAGCAGAGTGACG-3′ and 5′-AGCACACTTCAGGCAG CGTCTTC-3′. The resulting PCR products were cloned into pCR2.1 and sequenced. RT-PCR to detect ATIC-ALK was performed using an RT step as described previously; a seminested PCR was performed with primers flanking the fusion cDNA junction (initiallyATIC primer 5′-CGCTCGCCCTGAACCCAGTG-3′ andALK primer 5′-CGAGGTGCGG AGCTTGCTCAGC-3′, then the latter oligonucleotide and ATIC primer 5′-GTGTCCACGGAGATGCAGAG-3′) that generate a 315- base pair (bp) final product.
Construction of ATIC-ALK fusion cDNA for expression studies
The 5′ portion of ATIC-ALK was amplified using cDNA prepared from the inv(2)-positive ALCL tumor of patient 1 with primers 5′-TCCTACCTGCGCACGTGGT-3′ and 5′-AGCACACTTCAGGCAGCGTCTTC-3′, and the 1059-bp product was cloned into TA vector pCR2.1. This cDNA insert, which contained sequences encoding the entire portion of ATIC present in the fusion and part of ALK, was digested with EcoRI and NarI to release a 929-bp ATIC-ALK fragment, which was then ligated with a 1943-bp NarI/NotI fragment from the normalALK cDNA8 that encodes the remaining carboxy-terminal ALK residues. The entire ATIC-ALK cDNA was cloned into mammalian expression plasmid pcDNA3 (Invitrogen) as anEcoRI(5′)/NotI(3′) insert, then sequenced in its entirety. In vitro transcription/translation (TNT kit, Promega) of this construct generated the expected 87-kd ATIC-ALK protein.
Immunoprecipitation and immunoblotting
COS7 cells were electroporated with pcDNA3-ATIC-ALK, pcDNA3-NPM-ALK, or empty vector (80 μg of each construct DNA, 8 × 106 cells, 975 μF, 270V), then cultured for 48 hours. The cells were lysed in 1% Triton X-100 lysis buffer (30 mmol/L HEPES [pH 7.4], 150 mmol/L NaCl, and 1% Triton X-100 with 44 μg of aprotinin per milliliter, 1.4 mmol/L phenylmethylsulfonyl fluoride, 2 mmol/L EDTA, 1 mmol/L sodium orthovanadate) and the lysates incubated with 10 μL of rabbit polyclonal antiserum anti-ALK #11(ref.8) or normal rabbit serum (NRS). Immunoprecipitated proteins were resolved by reducing SDS-polyacrylamide gel electrophoresis (PAGE), then transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon P, Millipore). Immunoblotting was performed with antiphosphotyrosine antibody 4G10 (1:3500 dilution) (Upstate Biotechnology), and proteins were visualized by chemiluminescence (ECL, Amersham Life Science).
Subcellular protein localization (immunofluorescence microscopy)
COS7 cells were electroporated with pcDNA3-ATIC-ALK, pcDNA3-NPM-ALK or empty vector, grown on coverslips for 72 hours, then fixed with 3.7% paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes and permeabilized with cold acetone at −20°C for 7 minutes. The cells were washed 3 times for 5 minutes each in PBS, incubated with monoclonal antibody ALK1 (undiluted)9 for 45 minutes at 37°C, washed again with PBS, then incubated for 45 minutes with fluorescein isothiocyanate-conjugated donkey antimouse IgG (Santa Cruz Biotechnology). The cells were again washed, and the nuclei counterstained with propidium iodide (Sigma). The immunofluorescent staining was examined with a Leica TCS NT confocal microscope. The optical sections were viewed in green and red channels.
BaF3 transfections and cell culture
BaF3 (8 × 106) were electroporated with pcDNA3-ATIC-ALK, pcDNA3-NPM-ALK, or empty vector (80 μg DNA, 975 μF, 270V), then selected in IL-3-containing media with 1 mg/mL G418. G418-resistant pools were tested for ATIC-ALK or NPM-ALK expression, then seeded at 2 × 105 cells/mL in growth media with or without IL-3. Viable cells were counted daily for 7 days.
Results
Genomic cloning and fluorescence in situ hybridization of the inv(2)
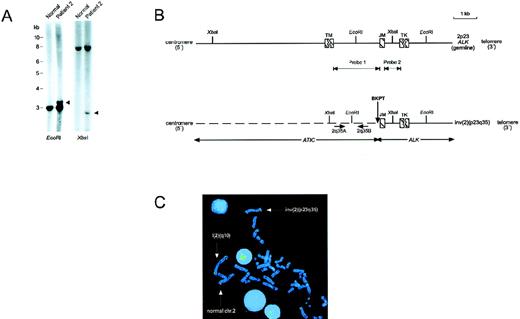
We cloned the inv(2)(p23q35) genomic breakpoints by using a LDI-PCR strategy with primers homologous to the ALK exon that encodes the juxtamembrane exon of the receptor (see “Materials and Methods”). Southern hybridization analysis of DNA from an ALCL tumor specimen that contained the inv(2) using ALK intron probes indicated that the 2p23 genomic breakpoint occurred in the approximately 2-kb intron, separating the exons encoding the transmembrane and juxtamembrane portions of ALK, the same location of the ALK breakpoints as in the t(2;5)12 (Figure1A and B). Sequence analysis of chimeric PCR products amplified from XbaI- and EcoRI-digested tumor DNA templates revealed the ALK genomic breakpoint in this case to be located 148 bp 5′ of the beginning of the juxtamembrane exon (Figure 1B). Examination of the sequence of the heterologous DNA segment juxtaposed to ALK that we amplified by LDI-PCR (an approximately 2.0-kb region) failed to identify exons of the putative 2q35 ALK partner gene. To isolate a larger portion of the 2q35 locus, we performed PCR-based screening of a human PAC clone library, isolating 2 independent clones, PAC213D24 and PAC218E3. Fluorescence in situ hybridization (FISH) of metaphases and interphases prepared from the inv(2)(p23q35)-containing ALCL tumors of 2 patients using PAC213D24 confirmed its location to 2q35 and demonstrated that it spans the 2q35 breakpoints in these cases (Figure1C and data not shown). Sequence analysis of an approximately 8-kbKpnI restriction fragment subcloned from PAC213D24 that spanned the breakpoint identified in patient 2 also failed to reveal potential exons of the involved 2q35 gene.
Genomic cloning of inv(2)(p23q35).
(A) Southern blot hybridization of ALCL tumor DNA from patient 2 usingALK probe 2 (see panel B), demonstrating rearranged restriction fragments (arrowheads). (B) Schematic of a portion of the germlineALK locus, and of the chimeric genomic fragment generated by the inv(2)(p23q35). The exact sizes of the exons encoding the transmembrane (TM) and tyrosine kinase (TK) domains of ALK have not been determined, and are therefore shown as interrupted cross-hatched boxes. The position of the inv(2) genomic breakpoint in the tumor of patient 2 is illustrated. The genomic location of ATIC exons was not determined; however, no exons were identified by sequencing of the approximately 2.0-kb segment of the ATIC locus extending from the XbaI restriction site to the breakpoint location. The location of the primer pair (2q35A, 2q35B) from the 2q35 gene locus used to identify clones PAC213D24 and PAC218E3 by PCR-based library screening is shown. JM, ALK juxtamembrane-encoding exon. (C) FISH analysis of inv(2). FISH of a metaphase chromosome spread from the inv(2)-positive ALCL tumor of patient 1 with PAC213D24, demonstrating its localization to 2q35 on the normal chromosome 2 and i(2)(q10) (which is a secondary chromosomal abnormality seen in inv(2)-positive cases that is formed by the joining of 2 long arms produced by the inversion11), and splitting of the clone by the inv(2)(p23q35).
Genomic cloning of inv(2)(p23q35).
(A) Southern blot hybridization of ALCL tumor DNA from patient 2 usingALK probe 2 (see panel B), demonstrating rearranged restriction fragments (arrowheads). (B) Schematic of a portion of the germlineALK locus, and of the chimeric genomic fragment generated by the inv(2)(p23q35). The exact sizes of the exons encoding the transmembrane (TM) and tyrosine kinase (TK) domains of ALK have not been determined, and are therefore shown as interrupted cross-hatched boxes. The position of the inv(2) genomic breakpoint in the tumor of patient 2 is illustrated. The genomic location of ATIC exons was not determined; however, no exons were identified by sequencing of the approximately 2.0-kb segment of the ATIC locus extending from the XbaI restriction site to the breakpoint location. The location of the primer pair (2q35A, 2q35B) from the 2q35 gene locus used to identify clones PAC213D24 and PAC218E3 by PCR-based library screening is shown. JM, ALK juxtamembrane-encoding exon. (C) FISH analysis of inv(2). FISH of a metaphase chromosome spread from the inv(2)-positive ALCL tumor of patient 1 with PAC213D24, demonstrating its localization to 2q35 on the normal chromosome 2 and i(2)(q10) (which is a secondary chromosomal abnormality seen in inv(2)-positive cases that is formed by the joining of 2 long arms produced by the inversion11), and splitting of the clone by the inv(2)(p23q35).
Identification of ATIC as the 2q35 partner gene ofALK
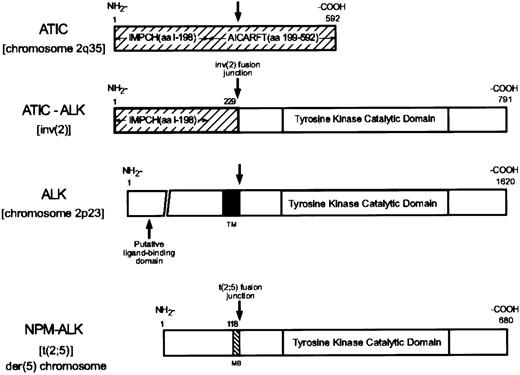
Because our analysis of the 2q35 genomic breakpoint region of the inv(2) did not identify the involved gene, we performed 5′ RACE using ALK-specific primers with total RNA prepared from the inv(2)-containing ALCL tumor specimen of patient 1. Sequence of the 5′ RACE products obtained in these studies identified an in-frame fusion of a 687-bp open reading frame to the nucleotides encoding the carboxy-terminal 562 amino acids (aa) of ALK, which constitute the entire intracellular portion of the receptor and are the residues present in the NPM-ALK chimeric protein6 (Figure2). Database searches showed the 5′ sequences fused with the ALK intracellular region to encode the amino-terminal 229 aa of 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (AICAR transformylase [AICARFT]/IMPCHase, or ATIC), formerly known as pur H.13-15 The 592 aa ATIC protein contains 2 non-overlapping and structurally independent regions extending from amino acids 1-198 and 199-592 that possess full IMPCH or AICARFT enzymatic activity, respectively (Figure 2). IMPCH catalyzes the final, and AICARFT the penultimate, steps in de novo purine nucleotide biosynthesis that lead to the production of inosinate (IMP), the precursor for adenylate (AMP) and guanylate (GMP) and ultimately, the adenine and guanine nucleotides of RNA and DNA, and ATP and GTP.16 The sequence of the portion of ATIC present in ATIC-ALK was identical to that of the 592 aa protein reported by Sugita et al,14 which begins with the residues MAPGQLALF-, rather than the 591 aa protein isoform described by Rayl et al13 that contains the amino-terminal residues MSSLSALF-. The juxtaposition of 5′ ATICsequences with the 3′ portion of ALK by the inv(2) indicates that, like the ALK locus,5 theATIC gene is transcribed in a centromeric to telomeric orientation on normal chromosome 2.
Diagrammatic representations of ATIC, ATIC-ALK, ALK, and NPM-ALK.
IMPCH, inosine monophosphate cyclohydrolase; AICARFT, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase; MB, NPM metal binding domain; TM, ALK transmembrane domain.
Diagrammatic representations of ATIC, ATIC-ALK, ALK, and NPM-ALK.
IMPCH, inosine monophosphate cyclohydrolase; AICARFT, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase; MB, NPM metal binding domain; TM, ALK transmembrane domain.
ATIC-ALK RT-PCR analysis
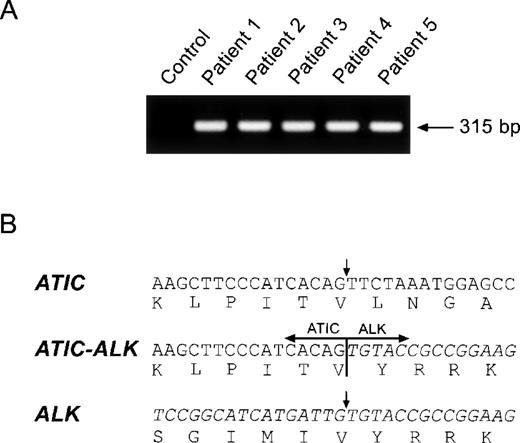
To establish that ATIC-ALK fusion by the inv(2) is a recurrent abnormality in ALCL, we performed RT-PCR analysis of total RNA prepared from 5 ALCL tumor specimens in which evidence suggestive for the presence of the inv(2) had been obtained by conventional karyotypic and/or FISH examination. All of these cases exhibited anti-ALK immunostaining restricted to the cytoplasm of the tumor cells rather than the cytoplasmic and nuclear ALK staining characteristic of NPM-ALK–positive ALCL.1 2 As shown in Figure 3, each of these tumors was demonstrated to express identical ATIC-ALK fusion transcripts. By contrast, expression of the reciprocal (ALK-ATIC) fusion cDNA was not detectable by RT-PCR (data not shown).
(A) ATIC-ALK RT-PCR analysis and (B) cDNA fusion junction.
(A) Total RNA prepared from the tumor specimens of 5 ALCL patients was examined by ATIC-ALK RT-PCR; all cases expressed the expected 315-bp ATIC-ALK product. Total RNA prepared from theNPM-ALK-positive ALCL cell line SUP-M2 was used as a negative control. (B) ATIC-ALK fusion cDNA sequences were identical in each of the 5 lymphomas tested.
(A) ATIC-ALK RT-PCR analysis and (B) cDNA fusion junction.
(A) Total RNA prepared from the tumor specimens of 5 ALCL patients was examined by ATIC-ALK RT-PCR; all cases expressed the expected 315-bp ATIC-ALK product. Total RNA prepared from theNPM-ALK-positive ALCL cell line SUP-M2 was used as a negative control. (B) ATIC-ALK fusion cDNA sequences were identical in each of the 5 lymphomas tested.
Biochemical and functional characterization of ATIC-ALK
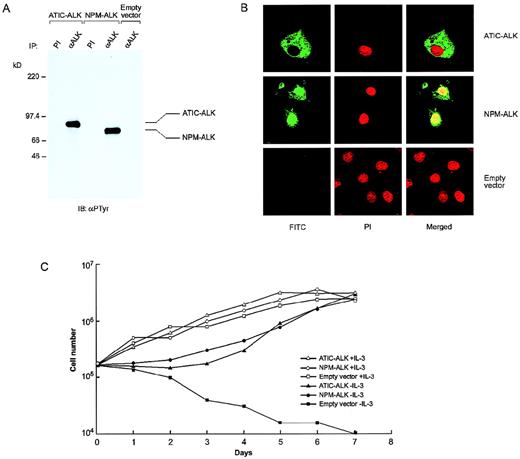
The ATIC-ALK fusion cDNA encodes a 791aa chimeric protein with a predicted mass of 87.2 kd. Consistent with this prediction, expression in COS7 cells of the full-length ATIC-ALK coding sequence produced a protein of approximately this size that was constitutively tyrosine phosphorylated (Figure4A), suggesting that the ATIC portion can induce dimerization of the chimera and resultant ALK activation similar to NPM in NPM-ALK.10 As noted above, ALCL tumor cells that contain the inv(2) exhibit anti-ALK staining that is restricted to the cytoplasm, in contrast to the cytoplasmic and nuclear staining observed in NPM-ALK-positive ALCL1,2,11; subcellular localization of ATIC-ALK in COS7 showed a cytoplasm–only expression pattern, in keeping with this clinical-histologic observation (Figure 4B). To assay the biologic activity of ATIC-ALK, we generated stable transfectants of the IL-3-dependent murine pro-B cell line BaF3, which is known to be rendered factor-independent by NPM-ALK.17 As shown in Figure 4C, BaF3 cells expressing ATIC-ALK proliferated in the absence of IL-3 with similar kinetics to cells containing NPM-ALK, indicating that ATIC-ALK should possess similar transforming capabilities.11 17-19
Functional characterization of ATIC-ALK.
(A) An antiphosphotyrosine antibody (αPTyr) immunoblot of transfected COS7 cell lysates immunoprecipitated using either preimmune (PI) serum or anti-ALK #11 (αALK) rabbit polyclonal antiserum demonstrates constitutive tyrosine phosphorylation of the approximately 87-kd ATIC-ALK and 80-kd NPM-ALK chimeric proteins. (B) Immunofluorescent microscopy of COS7 cells transiently transfected with pcDNA3-ATIC-ALK, pcDNA3-NPM-ALK, or empty vector and stained with the ALK1 monoclonal antibody. Subcellular localization of the expressed proteins, as indicated by green fluorescent signal from the FITC (fluorescein isothiocyanate)-labeled secondary antibody, shows ATIC-ALK to be present in the cytoplasm only, whereas NPM-ALK is found in both the cytoplasm and nucleus. Propidium iodide (PI) staining of DNA was performed simultaneously to identify nuclei. (C) ATIC-ALK confers factor-independent growth to BaF3 cells. Stably transfected BaF3 pools expressing ATIC-ALK or NPM-ALK were assessed for growth in the presence or absence of IL-3, together with empty vector-containing cell pools. Viable cell counts were performed in triplicate using trypan blue at 24-hour intervals, with each point being the average of the triplicate determinations.
Functional characterization of ATIC-ALK.
(A) An antiphosphotyrosine antibody (αPTyr) immunoblot of transfected COS7 cell lysates immunoprecipitated using either preimmune (PI) serum or anti-ALK #11 (αALK) rabbit polyclonal antiserum demonstrates constitutive tyrosine phosphorylation of the approximately 87-kd ATIC-ALK and 80-kd NPM-ALK chimeric proteins. (B) Immunofluorescent microscopy of COS7 cells transiently transfected with pcDNA3-ATIC-ALK, pcDNA3-NPM-ALK, or empty vector and stained with the ALK1 monoclonal antibody. Subcellular localization of the expressed proteins, as indicated by green fluorescent signal from the FITC (fluorescein isothiocyanate)-labeled secondary antibody, shows ATIC-ALK to be present in the cytoplasm only, whereas NPM-ALK is found in both the cytoplasm and nucleus. Propidium iodide (PI) staining of DNA was performed simultaneously to identify nuclei. (C) ATIC-ALK confers factor-independent growth to BaF3 cells. Stably transfected BaF3 pools expressing ATIC-ALK or NPM-ALK were assessed for growth in the presence or absence of IL-3, together with empty vector-containing cell pools. Viable cell counts were performed in triplicate using trypan blue at 24-hour intervals, with each point being the average of the triplicate determinations.
Discussion
“Variant” rearrangements at the 2p23 ALK locus other than the inv(2) have been identified in ALCL.20-22 To date, 2 of these, the t(1;2)(q25;p23) and t(2;3)(p23;q21), have been molecularly investigated, having been recently shown to generate the fusion TPM3-ALK that alters sequences of the nonmuscle tropomyosin gene TPM320 and TFG-ALK that involves TRK-fused gene (TFG),31respectively, both previously reported as fusion partners with theNTRK1 receptor tyrosine kinase gene in human papillary thyroid carcinomas.23 32 Further study will be required to determine the incidence of ATIC-ALK, TPM3-ALK, TFG-ALK, and possible other ALK fusions in the 20% of ALK-positive/NPM-ALK–negative ALCLs that occur, although our finding of ATIC-ALK in 5 such cases suggests that the fusion is likely to be frequent.
A large number of clinical studies have shown that aberrant expression of ALK is present in about 40% to 60% of ALCLs, the remaining cases being of unknown pathogenesis.1-4 Recognition of ALK-positive ALCL appears to be of considerable clinical importance given that those patients with ALCL containing aberrant ALK expression have a dramatically better prognosis when administered appropriate therapy, compared with those with ALK-negative disease (for example, with 5-year disease-free survival rates of more than 80% versus less than 40% reported).2 24 It remains to be determined unequivocally whether ALK expression in ALCL due to fusions such as ATIC-ALK, TPM3-ALK, and TFG-ALK similarly connotes a superior outcome because the majority of ALK-positive lymphomas studied thus far in this regard were NPM-ALK–positive.
A shared feature of all identified fusion partners of oncogenic truncated tyrosine kinases, including the NPM portion of NPM-ALK, is their ability to mediate oligomerization of the chimeric proteins, mimicking receptor aggregation and kinase activation normally controlled by ligand binding.10,25-27 Equilibrium sedimentation studies, as well as crystal structure, have shown ATIC to exist as a homodimer.15 Preliminary results have shown that an amino-terminal 230 aa fragment of ATIC also forms dimers (G.P. Beardsley and J. Vergis, Yale University School of Medicine, personal communication); our studies suggest that the NH2-terminal segment of the protein found in ATIC-ALK likewise serves this function.
A role for altered ATIC enzymatic activity in lymphomagenesis due to ATIC-ALK fusion is unclear but unlikely. Removal of the ATICgene promoter on the allele altered by the inv(2) from the downstream portions of the gene would be predicted to decrease AICARFT enzymatic activity in lymphoma cells because the reciprocal ALK-ATICfusion is not expressed, whereas IMPCH enzymatic function would likely be either decreased to a similar degree if absent in ATIC-ALK, or unchanged if retained. None of these possible changes would be predicted to enhance lymphoma cell growth. Indeed, given that neither the IMPCH nor AICARFT enzymatic steps are rate-limiting in de novo purine biosynthesis—5-phosphoribosyl-1-pyrophosphate (PRPP) aminotransferase, the first enzyme in the pathway holding this distinction28—the level of purine synthesis in cells containing ATIC-ALK is likely to be altered minimally. It is intriguing to speculate, however, whether ATIC-ALK–positive lymphoma cells might be sensitized to purineless death induced by antifolate therapeutic agents such as methotrexate, 5,10-dideazatetrahydrofolate, or 5-deazacyclotetrahydrofolate, each of which can inhibit AICARFT,29 30 given the expected decrease in this activity due to the inv(2).
Acknowledgments
We thank V. Audino and P. Vandiveer for manuscript preparation; K. G. Murti for help with subcellular protein localization (immunofluorescence microscopy); C. Becher and D. Schuster for excellent technical assistance; M. Tiemann and H. Kreipe for providing histopathologic diagnoses and cryopreserved tumor specimens; and G. P. Beardsley for stimulating discussion and comments regarding the manuscript.
I.W. and S.W.M. share senior authorship.
Supported in part by National Cancer Institute (NCI) Grant No. CA69129 (S.W.M.), NCI Cancer Center CORE Grant No. CA21765, the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children's Research Hospital, the Belgian Program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming (I.W.), the Deutsche Krebshilfe Grant 10-0992-Sch13, Wilhelm Sandler Stiftung Grant 95.003.2, and the Interdisciplinary Center for Clinical Cancer Research, University of Kiel. J.C. is an “Aspirant” of the“FWO-Vlaanderen.” S.G. holds a scholarship of the Hensel-Stiftung. B.S. holds a Hermann and Lilly Schilling professorship.
Reprints:Stephan W. Morris, St. Jude Children's Research Hospital, Department of Pathology, Room 5024, Thomas Tower, 332 N Lauderdale, Memphis, TN 38105-2794; e-mail:steve.morris@stjude.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.