The reciprocal translocation between chromosomes 9 and 22 that fuses coding sequences of the Bcr and Abl genes is responsible for a remarkably diverse group of hematologic malignancies. A newly described 230-kd form of Bcr-Abl has been associated with an indolent myeloproliferative syndrome referred to as chronic neutrophilic leukemia. We have cloned the corresponding gene and examined the biologic and biochemical properties of p230 Bcr-Abl after retroviral-mediated gene transfer into hematopoietic cell lines and primary bone marrow cells. p230 Bcr-Abl–expressing 32D myeloid cells were fully growth factor-independent and activated similar signal transduction pathways as the well-characterized p210 and p185 forms of Bcr-Abl. In contrast, primary mouse bone marrow cells expressing p230 required exogenous hematopoietic growth factors for optimal growth, whereas p185- and p210-expressing cells were independent of growth factors. The 3 Bcr-Abl proteins exerted different effects on differentiation of bone marrow cells. p185 induced outgrowth of lymphoid precursors capable of tumor formation in immunodeficient mice. In contrast, p210- and p230-expressing bone marrow cells caused limited outgrowth of lymphoid precursors that failed to form tumors in immunodeficient mice. Removal of cytokines and autologous stroma from Bcr-Abl–expressing bone marrow cultures produced the expansion of distinct lineages by the various Bcr-Abl proteins. p185 drove expansion of cytokine-independent lymphoid progenitors, while p210 and p230 generated cytokine-independent monocyte/myeloid cells. These findings suggest that the different Bcr-Abl fusion proteins drive the expansion of different hematopoietic populations, which may explain the association of the various Bcr-Abl oncoproteins with different spectra of human leukemias.
The Bcr-Abl oncogene is formed by the reciprocal translocation between chromosomes 9 and 22 and is associated with diverse human leukemias.1 The Bcr-Abl oncogene product, Bcr-Abl, is a chimeric protein with deregulated tyrosine kinase activity. Bcr-Abl exerts its leukemogenic effects via both autophosphorylation and phosphorylation of cellular substrates. This ultimately leads to activation of signal transduction pathways involved in the altered biologic behavior of Bcr-Abl–containing cells. These include the Ras, Raf, Erk, Jnk, Myc, Jak/Stat, PI3kinase-Akt, and NF-κB pathways, among others.2-12 More recently, the oncogenic Bcr-Abl proteins have been shown to elicit the ubiquitin-dependent degradation of target proteins.13
The leukemia-associated Bcr-Abl oncogenes vary in the amount of the Bcr gene that is included within the chimera.1The structure of the various Bcr-Abl oncoproteins may influence the disease phenotype associated with their expression. p185 Bcr-Abl (also referred to as p190 Bcr-Abl) contains the dimerization, binding, SH2 and serine/threonine kinase domains of Bcr and is associated with the subset of acute lymphoblastic leukemias (ALL) that are Philadelphia chromosome positive (Ph1+).14,15 ALL is typically an acute, aggressive leukemia that requires prompt treatment with high-dose chemotherapy.16 p210 Bcr-Abl contains additional Bcr sequences coding for Pleckstrin homology (PH) and Dbl-like domains and is associated with nearly all cases of chronic myelogenous leukemia (CML).17 CML begins as a chronic myeloproliferative disorder that is amenable to management with chemotherapeutic or biologic agents, but inevitably progresses to a fatal blast crisis of either myeloid or lymphoid phenotype.
Recently, Pane et al18 have shown that a subset of patients with Ph1+ chronic neutrophilic leukemia (CNL) have a unique breakpoint (designated μ) within the Bcr gene on chromosome 22.19 This breakpoint is 3′ to the more common breakpoints found in patients with CML and the subset of patients with ALL that are Ph1+. Thus, p230 Bcr-Abl contains additionalBcr coding sequences that are not found in the p185 or p210 variants. Specifically, p230 Bcr-Abl contains the calcium-phospholipid binding (CalB) domain and the first third of the domain associated with GTPase activating activity for p21rac (GAPrac). CNL, which is also known in the literature as neutrophilic chronic myelogenous leukemia, is a rare disorder marked by sustained mature neutrophilic expansion with mild hepatosplenomegaly. This disease typically has a more indolent course when compared with classical CML, and progression to blast crisis is uncommon.18 However, a few patients have been reported who exhibit typical CML and carry the μ breakpoint that generates p230 Bcr-Abl.20-22
Thus, the unique protein domains that are contributed by the variousBcr breakpoints, when translocated to the c-Abl gene can lead to diseases with remarkably different phenotypes. To gain insight into the molecular pathology of the various Bcr-Abl–related leukemias, we have cloned the CNL-associated p230 Bcr-Abloncogene and expressed the protein product in hematopoietic cell lines as well as primary mouse bone marrow cells under conditions that favor the outgrowth of either myeloid or lymphoid progenitors. Here, we analyze the biochemical and biologic properties of p230 Bcr-Abl and compare it with the p210 and p185 variants.
Materials and methods
P230 Bcr-Abl cDNA
An overlapping polymerase chain reaction (PCR) approach using a 3′ Bcr primer, incorporating the known p230 μ breakpoint, was used to generate a 1013-base pair (bp) Kpn1/HindIII Bcr-Abl cDNA fragment, spanning the μ breakpoint region.19 The primers (5′-GGAGCAGCAGAAGAGCTGTT-3′) and (5′-GTCAGATGCTACTGGCCGCTGAAGGGCTTTGACGTCGAAGGCT GCC-3′) were used with the EcoR1 fragment of pGEM4[0]cBcr(3′UT) as the substrate23,24 and primers (5′-AAGCCCTTCAGCGGCCAG-3′) and (5′-CTGCTCTCACTCTCACGCAC-3′) were used with the EcoR1 fragment of pGEM4cAbl1b as substrate.23,24 These fragments were annealed and PCR-amplified using (5′-GGAGCAGCAGAAGAGCTGTT-3′) and (5′-CTGCTCTCACTCTCACGCAC-3′). This fragment was sequenced in both directions to ensure that it contained the correct μ Bcr-Abl breakpoint. The amplified fragment was then digested with Kpn I and HindIII and ligated with the EcoR1/Kpn1 cAbl and Kpn1/EcoR1 Bcr fragments into the pSRαMSVtkneo retroviral vector. The generation of pSRαMSVtkneo-p185Bcr-Abl and pSRαMSVtkneo-p210Bcr-Abl has been previously described.24
Cell culture
32D and DAGM cells were grown in Roswell Park Memorial Institute medium (RPMI) containing 10% fetal bovine serum (FBS), 10% WEHI-3B conditioned media (WEHI CM) as the source of IL3, and penicillin/streptomycin (P/S) antibiotics. Mouse bone marrow mononuclear cells (MNCs) were isolated from the femurs of twelve 3- to 4-week-old male Balb/C mice by density gradient through HISTOPAQUE-1077 (Sigma, St Louis, MO). Murine bone marrow MNCs were maintained in Iscove's modified Dulbecco medium (IMDM) with 15% FCS, 0.1% bovine serum albumin (BSA), 25 μmol/L β-mercaptoethanol, and P/S with or without the following cytokines: 100 IU/mL recombinant murine IL3, 100 IU/mL recombinant murine IL6, and 1000 IU/mL (100 ng/mL) recombinant murine stem cell factor (SCF) (all obtained from Intergen, Purchase, NY). Cells were maintained at 37°C, 5% CO2.
Gene transfer
For gene transfer into tissue culture cell lines, retroviruses were generated by transient transfection of 293T cells, as previously described.25 Briefly, the various Bcr-Abl constructs were cotransfected with psvψ2 onto 293T fibroblasts26 by calcium phosphate transfection in the presence of 25 μmol/L chloroquine. Two days after transfection, Bcr-Abl–containing and control retroviral supernatants were collected and incubated with 1 million 32D, FL5-12, or DAGM cells in 10% WEHI-CM and 4 μg/mL polybrene in a volume of 1 mL. Infections were performed at 37°C and 5% CO2 for 3 to 6 hours. Cells were pelleted and resuspended in growth medium. Two days after infection, cells were selected by IL3 deprivation or resistance to G418 (0.3 to 0.7 mg/mL). IL3 deprivation was performed by washing the cells twice with RPMI and resuspending them in RPMI containing 10% FBS and P/S.
For gene transfer into primary mouse bone MNCs, p185 Bcr-Abl, p210 Bcr-Abl, and p230 Bcr-Abl were subcloned into the bicistronic retroviral vector MIGR1.27 Retroviruses generated by transient transfection in 293T cells were used to infect GP + E-86 fibroblasts.26 Three rounds of retroviral infection were performed over 2 days, after which these cells were trypsinized and selected for green fluorescent protein (GFP) expression using fluorescence activating cell sorting (FACS) on a Becton Dickinson (San Jose, CA) FACSTAR-PLUS with excitation at 488 nm. Bcr-Abl and GFP-coexpressing GP+E-86 retroviral producer lines were expanded in DMEM, 10% FBS, and P/S. These retroviral producer lines were resorted every 60 days to maintain a population that contained greater than 90% GFP-positive cells. Murine bone marrow MNCs were isolated by HISTOPAQUE-1077 (Sigma) gradient and between 2 and 5 million cells were plated onto confluent GP+E-86 producer lines. Immediately before gene transfer, GP + E-86 producer lines were gamma-irradiated at 1250 rads to prevent subsequent contamination of bone marrow cultures. Infections were carried out for 48 hours in the presence of 100 IU/mL recombinant murine IL3, 100 IU/mL recombinant murine IL6, and 1000 IU/mL (100 ng/mL) recombinant murine stem cell factor with 4 μg/mL polybrene. The efficiency of gene transfer into primary mouse bone marrow cells between the various Bcr-Abl producer lines varied with the size of the construct, with transfer efficiency for p185 Bcr-Abl being generally 2 to 3 times greater than p230 Bcr-Abl, and p210 Bcr-Abl transfer efficiency generally between that of p185 and p230. Cocultivation with γ-irradiated GP + E-86 producer lines consistently improved gene transfer efficiency into primary murine stem cells by 25- to 50-fold, compared with infection with retroviral supernatants alone. Two days after retroviral infection, designated as “day zero,” the murine bone marrow MNCs were used for experimentation. For in vitro differentiation experiments in the presence of autologous stroma, day zero murine bone marrow MNCs were plated directly after retroviral-mediated gene transfer and grown in the presence of 100 IU/mL recombinant murine IL3, 100 IU/mL recombinant murine IL6, and 1000 IU/mL (100 ng/mL) recombinant murine stem cell factor. We found that cocultivation, followed by plating in the presence of cytokines, reproducibly resulted in the formation of an adherent, confluent stromal layer and a nonadherent hematopoietic layer. Fresh growth media with cytokines was added every 2 days. The nonadherent cells were not passaged into new flasks to maintain contact with the adherent stromal compartment, which we found to be resistant to trypsinization. For in vitro experiments in the absence of autologous stroma, day zero GFP-expressing murine bone marrow MNCs were selected by FACS before plating either in the presence or absence of cytokines. We found that both control and GFP-containing cultures plated after GFP selection developed few, if any, adherent cells.
Western blotting
Primary mouse bone marrow cells were lysed at a concentration of 5 × 107 cells/mL directly in boiling 2 × sample buffer. Lysates were centrifuged at 100 000g for 25 minutes at 4°C and supernatants equivalent to 1 × 107 cells were loaded onto SDS polyacrylamide gels for analysis. After gel electrophoresis, proteins were transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk in Tris-buffered saline (TBS) and blotted with antibodies to Abl (8E9, Pharmingen, San Diego, CA) or phosphotyrosine (4G10, Upstate Biotechnologies, Lake Placid, NY) in 2% nonfat dry milk in TBS, followed by enhanced chemiluminescent detection (Amersham, Piscataway, NJ).
Immunophenotyping
Cultures of Bcr-Abl and GFP-expressing or control GFP-expressing bone marrow cells were resuspended in phosphate-buffered saline (PBS) and 1% FBS. Cells were incubated with anti-CD16/CD32 (“Fc Block,” Pharmingen) at 1.25 μg/mL for 15 minutes on ice, followed by incubation with biotin-conjugated primary antibodies at 3.3 μg/mL for 30 minutes on ice. Cells were washed 3 times with PBS and 1% FBS, followed by incubation with a combination of streptavidin-cy-chrome (Pharmingen) at 1.3 μg/mL and phycoerythrin (PE)-conjugated primary antibodies at 3.3 μg/mL for 15 minutes on ice. Cells were washed twice with PBS and 1% FBS and analyzed on a Becton Dickinson FACStarPLUS, equipped with a 488 nm argon laser and tunable dye laser. For some analyses, dead cells were excluded by staining with 7AAD, in which case streptavidin-allophycocyan (Pharmingen) was used as the secondary reagent. Primary biotin and PE-conjugated antibodies used for this study were purchased from Pharmingen: B220 (clone RA3-6B2), Thy 1.2 (clone 53-2.1), Gr-1 (clone RB6-8C5), Mac-1 (clone M1/70), CD 43 (Ly-48), cKit (clone 2B8), CD 34 (clone RAM 34), CD9 (clone KMC8), and Sca1 (Ly-6A/E). FACS analysis was performed until 10 000 GFP positive events were acquired.
Cell cycle analysis
Cultures of Bcr-Abl and GFP-expressing or control GFP-expressing bone marrow cells were incubated with 10 μmol/L bromodeoxyuridine (Brdu) for 30 minutes in IMDM with 15% FCS, 0.1% BSA, 25 μmol/L β-mercaptoethanol and P/S with or without cytokines at 37°C, and 5% CO2. One million cells were then washed twice with PBS and 1% FBS and fixed with 5 mL 70% ethanol for 30 minutes on ice. This successfully extracted 100% of the GFP from the cells in control experiments. The cells were then stained with FITC-conjugated anti-Brdu antibody and propidium iodide as per the manufacturer's instructions (Becton Dickinson) and analyzed by flow cytometry on a Becton Dickinson FACStarPLUS with laser excitation at 488 nm.
Tumor challenge
Cells were washed twice in RPMI only and resuspended at 1 × 107 cells/mL RPMI containing 0.1% FBS. A total of 1 × 106 cells (100 μL) were injected subcutaneously into the right flanks of 7- to 8-week-old male SCID mice (strain CB17SC-M, Taconic Farms, Germantown, NY).
Results
Cloning and expression of p230 Bcr-Abl.
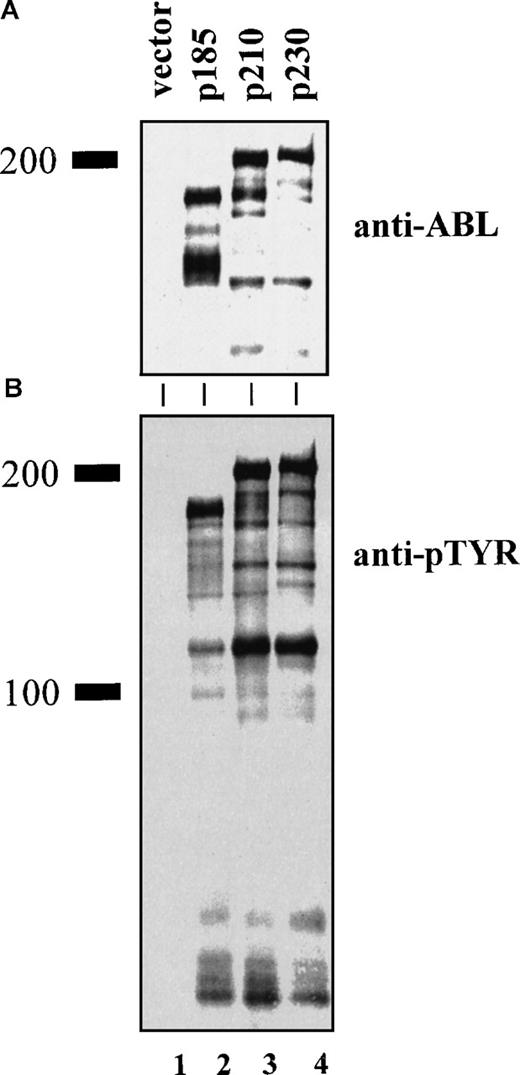
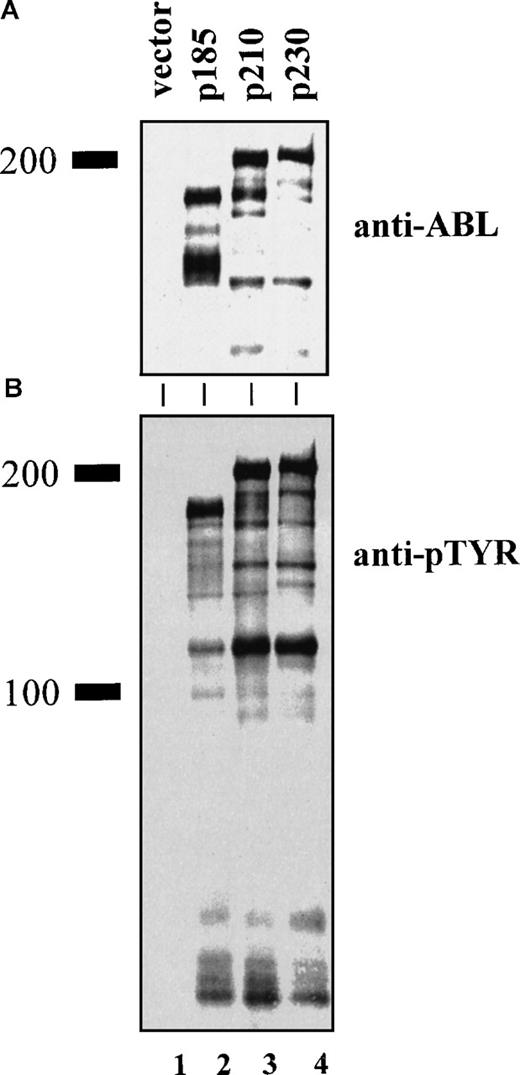
We used an overlapping PCR technique with Bcr and AblcDNA templates to create a Bcr-Abl chimeric cDNA that contained the unique p230 μ breakpoint.1,18,19 After retroviral-mediated gene transfer of the various leukemia-associated Bcr-Abl isoforms into 32D cells, Western blot analysis with an anti-Abl antibody demonstrated expression of proteins of the expected sizes (Figure 1A). Western blotting with an antibody that recognizes the PH domain of Bcr showed that this domain is present in p210 and p230 Bcr-Abl, but not in p185 Bcr-Abl (data not shown). The tyrosine kinase activity of the Bcr-Abl chimeric oncoprotein is critical to its transforming ability.3 2832D cells expressing the various leukemia-associated Bcr-Abl isoforms contained roughly equivalent levels of tyrosine-phosphorylated Bcr-Abl, along with additional phosphoproteins (Figure 1B). This finding indicates that p230 Bcr-Abl is kinase active, and its tyrosine kinase activity is similar to p210 and p185 Bcr-Abl oncoproteins in vivo.
Expression of p230 Bcr-Abl in 32D mouse myeloid cells leads to tyrosine phosphorylation of cellular proteins.
Lysates of 32D cells expressing empty vector (lane 1), p185 (lane 2), p210 (lane 3), and p230 Bcr-Abl (lane 4) were analyzed by Western blotting with antibodies against (A) Abl (8E9, Pharmingen) or (B) phosphotyrosine (4G10, UBI).
Expression of p230 Bcr-Abl in 32D mouse myeloid cells leads to tyrosine phosphorylation of cellular proteins.
Lysates of 32D cells expressing empty vector (lane 1), p185 (lane 2), p210 (lane 3), and p230 Bcr-Abl (lane 4) were analyzed by Western blotting with antibodies against (A) Abl (8E9, Pharmingen) or (B) phosphotyrosine (4G10, UBI).
p230 Bcr-Abl renders 32D cells factor independent and activates similar signaling pathways as p185 and p210 Bcr-Abl in these cells.
Expression of p185 and p210 Bcr-Abl in 32D cells leads to growth factor independence and inhibition of apoptosis.3 The ability of p230 Bcr-Abl to cause growth factor independence of 32D cells was compared with the p210 and p185 Bcr-Abl variants. After IL3 withdrawal, p230 cells grew at a rate equivalent to p210- and p185-expressing cells, whereas control cells underwent apoptosis (data not shown). p230 as well as p210- and p185-expressing 32D cells were resistant to the differentiating effects of GCSF, whereas control cells stopped cycling and differentiated into granulocytes (data not shown). Furthermore, p230 Bcr-Abl–expressing 32D cells were as potent as p185 and p210 Bcr-Abl–expressing 32D cells in their ability to form tumors in SCID mice (Table 1). Thus, the biologic effects of the p230 Bcr-Abl isoform were indistinguishable from those of the more aggressive p210 and p185 isoforms of Bcr-Abl after expression of the proteins in 32D cells. Consistent with these findings, analysis of the signaling properties of p230 Bcr-Abl in 32D cells revealed that p230 activated Ras, Erk, and Jnk to similar levels as those induced by p185 and p210 Bcr-Abl in these cells3,6 29 (data not shown). Similar results were obtained in FL5-12 and DAGM cells expressing the 3 Bcr-Abl isoforms (data not shown). This data indicates that in lineage-restricted hematopoietic cell lines, the p185, p210, and p230 forms of Bcr-Abl activate Ras, Erk, and Jnk to similar extent, and elicit similar survival, proliferative, and tumorigenic effects.
Effect of expression of distinct Bcr-Abl proteins on the differentiation of primary mouse bone marrow cultures expanded in the presence of exogenous cytokines and autologous stroma.
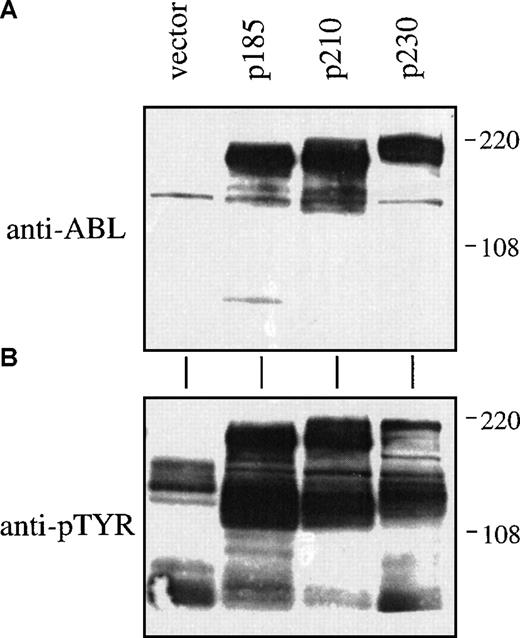
The effects of p230 Bcr-Abl expression were next studied on primary bone marrow cells, the natural target of Bcr-Abl related leukemias. We used a bicistronic retroviral vector to coexpress the leukemia-associated Bcr-Abl cDNAs with the gene for enhanced green fluorescent protein (GFP) as a selectable marker.27 After retroviral-mediated gene transfer in primary bone marrow cells and selection for GFP, we observed equivalent expression of the 3 forms of Bcr-Abl (Figure2A). Furthermore, the levels of in vivo phosphotyrosine were similar for cell lysates expressing the p185, p210, and p230 forms of Bcr-Abl (Figure 2B).
Expression of P230 Bcr-Abl in primary mouse bone marrow cells leads to tyrosine phosphorylation of cellular proteins.
Whole cell lysates were prepared from cytokine and serum-starved primary mouse bone marrow cells by boiling in 2 × sample buffer. Lysates were analyzed by SDS-PAGE and Western blotting with antibodies against (A) Abl or (B) phosphotyrosine.
Expression of P230 Bcr-Abl in primary mouse bone marrow cells leads to tyrosine phosphorylation of cellular proteins.
Whole cell lysates were prepared from cytokine and serum-starved primary mouse bone marrow cells by boiling in 2 × sample buffer. Lysates were analyzed by SDS-PAGE and Western blotting with antibodies against (A) Abl or (B) phosphotyrosine.
We developed hematopoietic culture conditions that would allow us to compare the ability of the 3 Bcr-Abl proteins to drive the differentiation of lymphoid versus myeloid lineages in vitro. Previous work has shown that both p185 and p210 Bcr-Abl drive lymphoid expansion of primary mouse bone marrow cells even under conditions that favor myeloid expansion.30 We assayed the effects of the various leukemia-related Bcr-Abl tyrosine kinases on primary mouse bone marrow cells under 3 different conditions: (1) expansion in the presence of exogenous cytokines and autologous stroma; (2) expansion in the presence of exogenous cytokines alone; and (3) expansion without exogenous cytokines or autologous stroma.
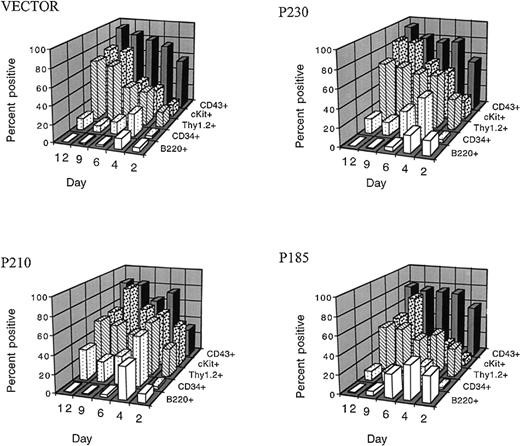
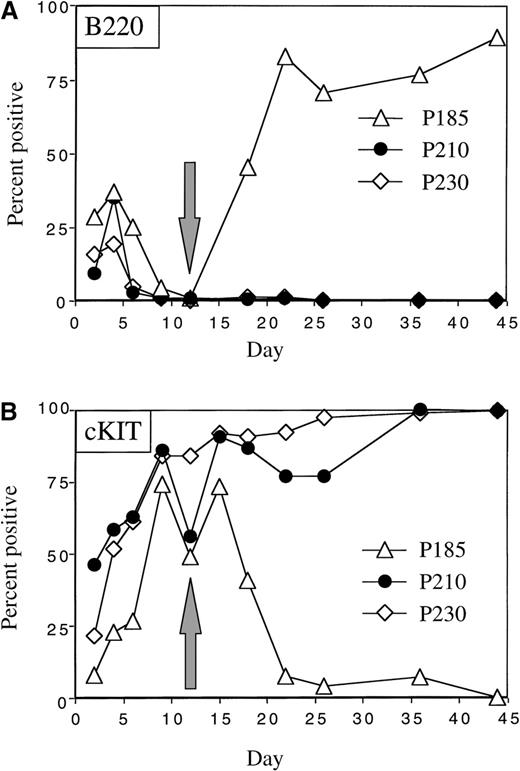
We first examined whether the various Bcr-Abl variants could overcome the differentiation-inhibitory effects of exogenous cytokines in the presence of autologous stroma.31 We found that, when primary mouse bone marrow cells expressing the 3 different Bcr-Abl kinases and control bone marrow cells were expanded in the presence of cytokines and autologous stroma, outgrowth of B220 positive pre-B cells was inhibited up to 12 days in culture (Figure3). This effect was dependent on the presence of autologous stroma and on the duration of cytokine exposure. p185 Bcr-Abl expressing cultures contained slightly more B220 positive cells in the first 6 days of the culture compared with p210, p230, and control bone marrow cells (Figure 3 and Figure4A). However, by day 12, all cultures contained less than 1% B220 positive cells under these conditions. The loss of B220 expression in the bone marrow cultures was accompanied by the expansion of cKit positive cells, such that after 9 days of growth in the presence of cytokines and stroma, more than 70% of control and Bcr-Abl expressing bone marrow cells were cKit positive (Figure 3).
Exogenous hematopoietic growth factors block the differentiation-inducing effects of the p185, p210, and p230 Bcr-Abl proteins in primary bone marrow cells.
After retroviral-mediated gene transfer and expansion in the presence of cytokines and autologous stroma, GFP-positive cells were analyzed for the expression of surface markers associated with B cells (B220), and stem cells (CD34, Thy1.2, cKit, and CD 43) at the indicated times after infection.
Exogenous hematopoietic growth factors block the differentiation-inducing effects of the p185, p210, and p230 Bcr-Abl proteins in primary bone marrow cells.
After retroviral-mediated gene transfer and expansion in the presence of cytokines and autologous stroma, GFP-positive cells were analyzed for the expression of surface markers associated with B cells (B220), and stem cells (CD34, Thy1.2, cKit, and CD 43) at the indicated times after infection.
The presence of cytokines and autologous stroma inhibits the outgrowth of cytokine-independent p210- and p230-expressing pre-B cells.
Two days after retroviral-mediated gene transfer, mouse bone marrow cells were expanded in the presence of autologous stroma and cytokines. After 12 days, nonadherent cells were replated in the absence of cytokines (indicated by the arrows). Cells were serially analyzed for the expression of B220 (A) and cKit (B).
The presence of cytokines and autologous stroma inhibits the outgrowth of cytokine-independent p210- and p230-expressing pre-B cells.
Two days after retroviral-mediated gene transfer, mouse bone marrow cells were expanded in the presence of autologous stroma and cytokines. After 12 days, nonadherent cells were replated in the absence of cytokines (indicated by the arrows). Cells were serially analyzed for the expression of B220 (A) and cKit (B).
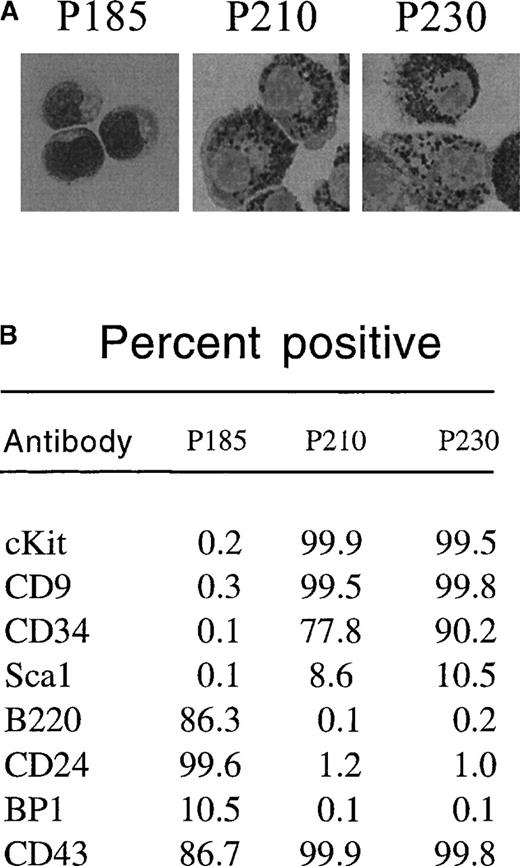
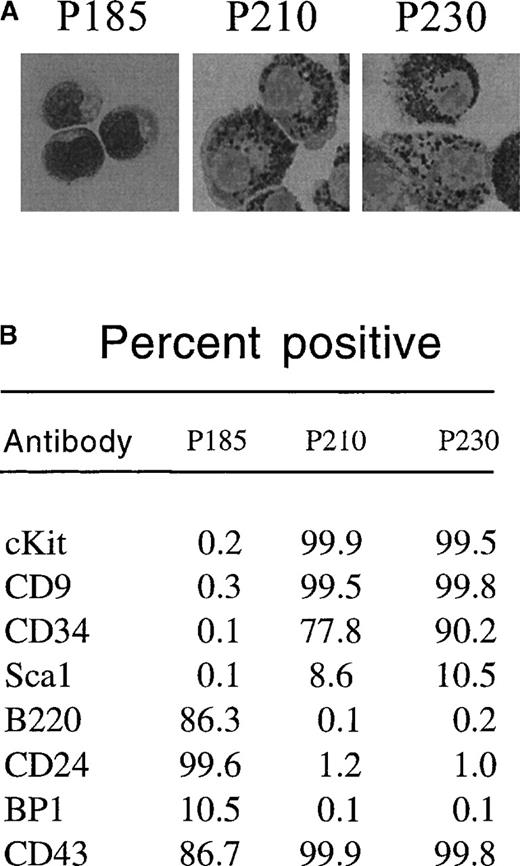
To determine whether p230 Bcr-Abl expression causes cytokine independence in primary cells, we tested the ability of day 12 bone marrow cultures expressing p230 Bcr-Abl to survive in the absence of cytokine and stromal support. Further, we compared the effects of p230 to those of p185 and p210 Bcr-Abl in this assay. We found that p185 Bcr-Abl–expressing primary bone marrow cultures expanded in the presence of cytokines and stroma were able to survive after cytokine and stromal withdrawal, resulting in the expansion of B220-positive pre-B cells (Figure 4A). These cells coexpressed CD24 and CD43 and were BP1 low (Figure 5B). Histologically, these cytokine and stroma-independent p185 Bcr-Abl–expressing bone marrow cells were small and contained few cytoplasmic granules (Figure 5A). Thus, these cells likely represent an immature B-cell progenitor.
p185-expressing primary mouse bone marrow cells expanded with cytokines and stroma differentiate into pre-B cells after cytokine withdrawal, whereas p210-and p230-expressing cells differentiate into myeloid/monocyte cells under the same conditions.
Two days after retroviral-mediated gene transfer, primary mouse bone marrow cells were expanded in the presence of autologous stroma and cytokines. After 12 days, nonadherent cells were replated in the absence of cytokines. Thirty-two days after replating (44 days after retroviral-mediated gene transfer), GFP-positive cells were subjected to cytospin and Wright's stain (A) or flow cytometry (B) to detect expression of the indicated cell surface markers.
p185-expressing primary mouse bone marrow cells expanded with cytokines and stroma differentiate into pre-B cells after cytokine withdrawal, whereas p210-and p230-expressing cells differentiate into myeloid/monocyte cells under the same conditions.
Two days after retroviral-mediated gene transfer, primary mouse bone marrow cells were expanded in the presence of autologous stroma and cytokines. After 12 days, nonadherent cells were replated in the absence of cytokines. Thirty-two days after replating (44 days after retroviral-mediated gene transfer), GFP-positive cells were subjected to cytospin and Wright's stain (A) or flow cytometry (B) to detect expression of the indicated cell surface markers.
The response of p210 and p230 Bcr-Abl–expressing mouse bone marrow cells to cytokine and stromal withdrawal was markedly different from that of p185 Bcr-Abl–expressing bone marrow cells. Whereas cytokine and stromal withdrawal led to the expansion of cells that were cKit negative in p185 Bcr-Abl bone marrow cultures, p210 and p230 Bcr-Abl–expressing bone marrow cells retained cKit expression even after cytokine and stromal withdrawal (Figure 4B and Figure 5B). These cytokine- and stromal-independent p210 and p230 Bcr-Abl–expressing cells coexpressed CD9 and CD34, were positive for cKit (CD 117) (Figure5B), were larger than the pre-B cells generated from p185-expressing cultures, and contained abundant azurophilic and basophilic granules (Figure 5A). All of these properties are consistent with differentiation into cells of the myeloid or monocyte lineages.32-38 Thus, under the conditions used in this experiment, p185 Bcr-Abl exhibited a propensity to drive the expansion of cytokine-independent lymphoid progenitors, whereas p210 and p230 Bcr-Abl generated cytokine-independent cells of the myeloid/monocyte lineage.
Effect of expression of distinct Bcr-Abl proteins on the differentiation, proliferation, and tumorigenicity of primary mouse bone marrow cultures expanded in the presence of exogenous cytokines without stroma.
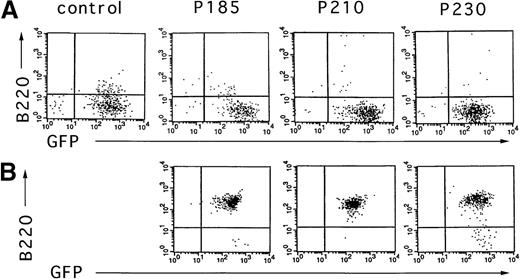
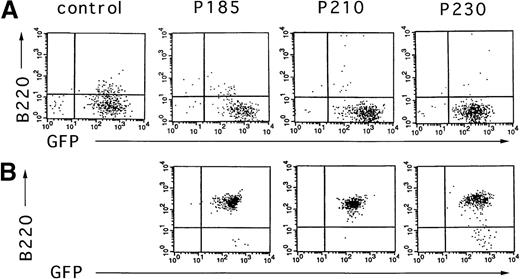
To test the effect of the different Bcr-Abl chimeric proteins on the differentiation of primary mouse bone marrow cultures in the absence of autologous stroma, primary bone marrow cells were transduced with the Bcr-Abl– and GFP-coexpressing retroviruses, selected for GFP expression, and expanded in the presence of cytokines without stroma. After 10 days of growth in the presence of cytokines without autologous stroma, primary bone marrow cultures transduced with Bcr-Abl retroviruses encoding p185, p210, and p230 were either deprived of cytokines or injected subcutaneously into SCID mice. As shown in Figure6, outgrowths of cytokine-independent cells immunophenotypically consistent with pre-B cells were obtained with all 3 Bcr-Abl isoforms from cultures that were initially predominantly B220 negative. The outgrowth of pre-B cells differed markedly among the various Bcr-Abl–expressing cultures. p185 and p210 Bcr-Abl–expressing mouse bone marrow cultures expanded with cytokines alone rapidly evolved into pre-B cells after cytokine withdrawal, whereas pre–B-cell development of p230 Bcr-Abl–expressing cells was delayed (data not shown).
Cytokine withdrawal from Bcr-Abl–expressing primary mouse bone marrow cells after 10 days of cytokine support in the absence of autologous stroma leads to the outgrowth of pre-B cells.
Two days after retroviral-mediated gene transfer, Bcr-Abl and control cells were selected for expression of GFP by FACS. These cells were expanded in the presence of cytokines without autologous stroma for 10 days and analyzed for the expression of GFP and B220 (A). Six days after cytokine withdrawal from Bcr-Abl–expressing cultures (16 days after selection for GFP-expressing cells), cells were again analyzed by flow cytometry for expression of GFP and B220 (B). Vector-expressing cells were not viable after cytokine withdrawal.
Cytokine withdrawal from Bcr-Abl–expressing primary mouse bone marrow cells after 10 days of cytokine support in the absence of autologous stroma leads to the outgrowth of pre-B cells.
Two days after retroviral-mediated gene transfer, Bcr-Abl and control cells were selected for expression of GFP by FACS. These cells were expanded in the presence of cytokines without autologous stroma for 10 days and analyzed for the expression of GFP and B220 (A). Six days after cytokine withdrawal from Bcr-Abl–expressing cultures (16 days after selection for GFP-expressing cells), cells were again analyzed by flow cytometry for expression of GFP and B220 (B). Vector-expressing cells were not viable after cytokine withdrawal.
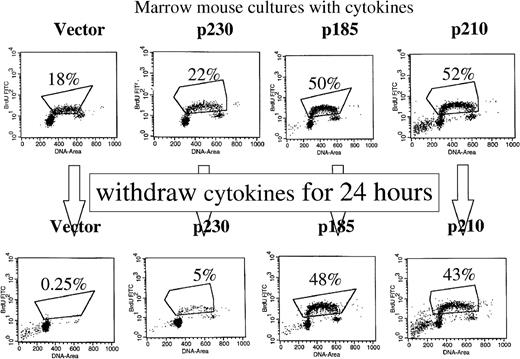
The effect of expression of the various Bcr-Abl tyrosine kinases on the growth of primary mouse bone marrow cells expanded with cytokines and without stroma was assessed by S-phase labeling with bromodeoxyuridine. In the presence of exogenous cytokines and without stroma, a larger percentage of p185 and p210 Bcr-Abl–expressing primary mouse bone marrow cells were in S-phase compared with p230 Bcr-Abl–expressing and control primary mouse bone marrow cells (Figure7, upper panel). Twenty-four hours after cytokine withdrawal, there was a significant reduction in the percentage of p230 Bcr-Abl–expressing and control cells in S-phase (Figure 7, lower panel). In contrast, the percentage of cells in S-phase in the p210 and p185 Bcr-Abl–expressing primary mouse bone marrow cultures decreased only slightly after cytokine withdrawal (Figure 7, lower panel). Thus, in primary mouse bone marrow cultures expanded in the absence of autologous stroma, p230 Bcr-Abl–expressing cells require cytokines for optimal growth, whereas p185 and p210 Bcr-Abl–expressing cells do not.
p230-expressing primary mouse bone marrow cells require cytokines for optimal growth.
GFP-selected, control or Bcr-Abl–expressing primary mouse bone marrow cells were expanded for 10 days with cytokines in the absence of autologous stroma then split into duplicate cultures with (upper panels) and without (lower panels) cytokines. Twenty-four hours later, cultures were incubated in bromodeoxyuridine and analyzed for S-phase fraction.
p230-expressing primary mouse bone marrow cells require cytokines for optimal growth.
GFP-selected, control or Bcr-Abl–expressing primary mouse bone marrow cells were expanded for 10 days with cytokines in the absence of autologous stroma then split into duplicate cultures with (upper panels) and without (lower panels) cytokines. Twenty-four hours later, cultures were incubated in bromodeoxyuridine and analyzed for S-phase fraction.
Next, we tested the tumorigenic potential of primary mouse bone marrow cultures expressing the 3 Bcr-Abl kinases expanded for 10 days in the presence of cytokines without stroma. At this time point, all cultures expressing the 3 Bcr-Abl proteins were negative for B220 (Figure 6A). p185 Bcr-Abl–expressing murine hematopoietic cells expanded with cytokines alone were able to rapidly form tumors when injected subcutaneously into SCID mice (Table 2). These p185 Bcr-Abl–expressing tumors coexpressed B220 and CD24 and were negative for sIgM, again consistent with a pre–B-cell phenotype (data not shown). p210 and p230 Bcr-Abl–expressing murine hematopoietic cells expanded with cytokines were unable to form tumors in SCID mice; however, 2 of 17 mice injected subcutaneously with p230-expressing bone marrow cells developed elevated white blood counts, splenomegaly, and polycythemia with circulating GFP-positive myeloid cells, consistent with the development of a CML-like syndrome. The decreased tumorigenic properties of p210 and p230 Bcr-Abl in this experiment were not due to lack of biologic activity. Transduction of bone marrow cells from 5-fluorouracil–treated mice with normalized retroviral supernatants expressing p230, p210, and p185 Bcr-Abl, revealed that, after injection of the transduced cells into the tail veins of lethally irradiated syngeneic recipient mice,27all mice developed a myeloproliferative disease (J. P. Miller and W. S. Pear, unpublished data). However, p230 Bcr-Abl caused a myeloproliferative disease with a significantly longer latency than that elicited by p185 and p210 Bcr-Abl (up to 27 weeks for p230 compared with 4 to 5 weeks for p185 and p210). Taken together, these data indicate that the proliferative and transforming properties of p230 are weaker than those of p185.
p230 Bcr-Abl elicits a weaker proliferative response than p210 Bcr-Abl in the absence of cytokines and stroma.
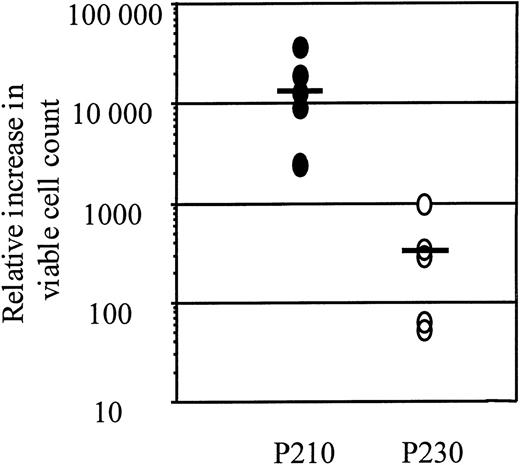
To compare more directly the proliferative responses of p210 and p230 Bcr-Abl, we followed the growth of primary mouse bone marrow cultures expressing p210 or p230 2 days after retroviral-mediated gene transfer and selection for GFP expression in the absence of cytokines and autologous stroma. Ten days after GFP selection, p210 Bcr-Abl–expressing primary mouse bone marrow cells had expanded 13 000-fold, whereas p230 Bcr-Abl–expressing primary mouse bone marrow cells had barely expanded 300-fold, a 40-fold difference that was statistically significant (P < .05, Student ttest) (Figure 8). After 10 days of growth without cytokines or stroma, p185, p210, and p230 Bcr-Abl–expressing cultures were all B220 positive. p185 Bcr-Abl cells grew at the same rate as p210 Bcr-Abl–expressing cells under these conditions, whereas murine bone marrow MNCs infected with the GFP-expressing retrovirus alone did not proliferate under these conditions (data not shown). Thus, although all leukemia-related Bcr-Abl isoforms can cause cytokine independence of primary murine bone marrow cells under a variety of conditions, they are markedly different in their abilities to drive the proliferation of these cells.
Relative increase in viable cell count of p210- and p230-expressing primary mouse bone marrow cultures expanded in the absence of either cytokines or autologous stroma.
Two days after gene transfer, p210 or p230 primary mouse bone marrow MNCs were selected for GFP expression by FACS. Data points represent the increase in total viable cell number relative to the number of GFP-positive cells recovered after FACS. Bars represent the mean of 6 independently derived p210 Bcr-Abl–expressing bone marrow cultures and 5 independently derived p230 Bcr-Abl–expressing bone marrow cultures.
Relative increase in viable cell count of p210- and p230-expressing primary mouse bone marrow cultures expanded in the absence of either cytokines or autologous stroma.
Two days after gene transfer, p210 or p230 primary mouse bone marrow MNCs were selected for GFP expression by FACS. Data points represent the increase in total viable cell number relative to the number of GFP-positive cells recovered after FACS. Bars represent the mean of 6 independently derived p210 Bcr-Abl–expressing bone marrow cultures and 5 independently derived p230 Bcr-Abl–expressing bone marrow cultures.
Discussion
The deregulation of normal hematopoiesis by chimeric oncogenes may result from dual effects on promoting cell growth and inhibiting cell differentiation, as well as enhancing cell survival. CML, considered the prototypical disease of the hematopoietic stem cell, is caused by the protein product of the p210 Bcr-Abl chimeric oncogene. The closely related p185 Bcr-Abl oncogene is the causative agent of a subset of ALL, whereas p230 Bcr-Abl has been associated with CNL, which has a clinical presentation that resembles the more typical p210 Bcr-Abl disease, but may have a more benign natural history.18 Whether patients with p230 Bcr-Abl–related leukemia represent a bona fide stem cell disorder versus a myeloid lineage restricted disease remains to be established.39,40Furthermore, there is overlap between the disease spectrum associated with the different Bcr/Abl fusion proteins.1 For example, p210 Bcr-Abl is associated with 40% of ALL, p185 Bcr-Abl with approximately 2% to 3% of CML,39,40 and p230 Bcr-Abl has been described in patients with typical CML, some of whom have progressed to blast crisis.20-22 It is not clear whether intrinsic differences in the activities of the 3 Bcr-Abl proteins account for their association with different disease phenotypes or whether the expression of each of the 3 Bcr-Abl forms is restricted to a distinct hematopoietic lineage, thus explaining their association with different leukemias. Data exist supporting the 2 possibilities.
It was recently shown by Li and colleagues41that p185 has greater in vitro tyrosine kinase activity than either p210 or p230 and that the proliferation rate of p185-expressing Ba/F3 lymphoid cells was greater than that of p210- and p230-expressing Ba/F3 cells in the absence of cytokines. In addition, mice reconstituted with p185 Bcr-Abl–expressing bone marrow cells that were not pretreated with 5-fluorouracil succumbed to lymphoid as well as other leukemias faster (4 weeks) than mice reconstituted with p210 and p230 Bcr-Abl–expressing marrow, which died by 10 and 11 weeks, respectively.41 These findings are in agreement with our results that show that in primary mouse bone marrow cultures, p230 was markedly impaired in its ability to generate cytokine-independent pre-B cells compared with p185. Indeed, p185 Bcr-Abl was very potent at driving lymphoid expansion, even under conditions that favored myeloid growth. In contrast, p210 and p230 generated cells of the myeloid/monocyte lineage under the same conditions. Thus, the association of p185 with primarily lymphoid diseases and p210 and p230 with primarily myeloid diseases may reflect the inherent ability of the various leukemia-associated Bcr-Abl proteins to promote lymphoid versus myeloid differentiation.
An alternative model that explains the association of the various Bcr-Abl proteins with leukemias of distinct phenotypes proposes thatBcr intron 1 breakpoints are much more common in lymphoid precursors, whereas breakpoints downstream in the Bcr gene occur more often in myeloid precursors. Indirect support for this model was provided by the finding by Li and colleagues41 that all 3 forms of Bcr-Abl (p185, p210, and p230) induce an identical CML-like syndrome in mice receiving transduced bone marrow from 5-fluorouracil pretreated donors.41 In contrast, under similar experimental conditions, we have found that p230 induced a myeloproliferative disease with a much longer latency compared with that induced by p185 and p210 Bcr-Abl (J. P. Miller and W. S. Pear, unpublished data). Future work is needed to explain the basis for the different results. In humans, there is a predominant association of each form of Bcr-Abl with a distinct spectrum of leukemias. However, the murine bone marrow transduction/transplantation model may not reveal marked differences in the disease phenotypes elicited by the 3 Bcr-Abl variants. Thus, alternative models may be required to dissect the molecular basis of the distinct pathologies linked to the expression of the various Bcr-Abl proteins. It has been proposed that the rarity of p190 (p185) Bcr-Abl CML is due to the restriction of m-Bcr breakpoints to progenitor cells already committed to lymphoid development.41 However, this hypothesis fails to explain the frequent occurrence of Ph1+ granulocytic and erythroid colonies in the marrow, and of Ph1+ granulocytes in the blood of patients with Philadelphia chromosome positive ALL.42-44 Our experiments do not specifically address the nature of the cell of origin of Bcr-Abl leukemias and future work will be necessary to directly address this issue.
Hematopoietic tissue culture cells have been useful in the elucidation of signal transduction pathways involved in Bcr-Abl leukemogenesis. These cell lines are typically restricted to either the myeloid or lymphoid lineage and may carry additional mutations as a consequence of adaptation to tissue culture. Using myeloid lineage-restricted hematopoietic cell lines, we have not found any differences in the biologic effects of p185, p210, and p230 Bcr-Abl. However, using lineage-unrestricted primary mouse bone marrow cells, we have shown that p230 Bcr-Abl is limited in its ability to drive the expansion of lymphoid progenitors, but drives the expansion of myeloid lineage cells.
The various Bcr-Abl oncoproteins share the same Abl tyrosine kinase sequences. Thus, the ability of Bcr-Abl to cause myeloid transformation of murine primary bone marrow stem cells may be modulated by the additional Bcr sequences found in the p210 and p230 forms of Bcr-Abl but not in p185 Bcr-Abl. Specifically, the Dbl-like and pleckstrin homology domains of p210, and the CalB and GAPrac domains of p230 may directly influence the ability of these proteins to transform various hematopoietic precursors by inhibition of lymphoid development, promotion of myeloid development, or both. The activity of the small GTP-binding protein Rac was found to be required for p210 Bcr-Abl–mediated transformation of 32D cells.45 It is possible that the additional Bcr sequences included within p230 Bcr-Abl, specifically the GAPrac domain, may function to partially abrogate the properties of activated p21 Rac in p230-expressing hematopoietic cells. Although only the first third of the Bcr GAPrac domain is included in p230 Bcr-Abl, it may function cooperatively with the CalB domain because these 2 domains are often found together.46 Recently, additional support for a role of Bcr sequences in the induction of a myeloproliferative disease by Bcr-Abl in vivo has been provided through direct comparison of the leukemogenic activity of p210 Bcr-Abl with that of an activated Abl kinase that lacks Bcr sequences.47 The latter induced only lymphoid malignancies in mice and did not stimulate the growth of myeloid colonies in vitro. In contrast, p210 Bcr-Abl efficiently induced a CML-like disease under the same conditions.47
The ability of Bcr-Abl to cause malignant transformation is a function of multiple activities such as inhibition of apoptosis and altered cell adhesion and motility. We propose that the ability of Bcr-Abl proteins to direct distinct hematopoietic differentiation pathways is determined by the specific Bcr sequences that are included in the chimeric oncoproteins. The cell culture conditions described here should provide an opportunity to dissect the role of specific Bcr sequences in generating the diverse spectrum of leukemias associated with expression of the various Bcr-Abl tyrosine kinases.
Acknowledgment
We thank Dr Michael Cook for the expert assistance with flow cytometry and cell sorting.
R.C.Q. and G.W.R. contributed equally to this work.
Supported by National Institutes of Health grants CA61033 and CA77570 to A.M.P. and W.S.P., respectively. R.C.Q. was supported by the 4 Schools Physician-Scientist Program, sponsored by the Lucille P. Markey Charitable Trust. G.W.R. was supported by an Environmental Protection Agency Fellowship. K.D.C. was supported by the Medical Scientist Training Program and the Department of Defense Breast Cancer Research Program. A.M.P. and W.S.P. are Scholars of the Leukemia Society of America.
Reprints:Ann Marie Pendergast, Department of Pharmacology & Cancer Biology, Box 3813, Duke University Medical Center, Durham, NC 27710; e-mail: pende014@mc.duke.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.