Abstract
Current techniques for identifying fetal hemoglobin (HbF) inducers are complex and time consuming. We developed a rapid and efficient method for detecting HbF inducers. Our system uses a recombinant DNA construct in which the coding sequences of 2 different luciferase reporter genes, firefly and renilla, are substituted for those of human γ and β globin genes, respectively. The activity of these genes can be distinguished by a simple, highly sensitive enzymatic assay in cell lysates. GM979 cells stably transfected with the construct are cultured in the presence of compounds, and their effects are determined by measuring the changes in activity of the 2 luciferase genes. Specific γ globin gene inducers are recognized by their ability to increase γ-firefly luciferase (γF) gene activity significantly more than β-renilla luciferase (βR) gene activity, identified by an increased ratio of γ-firefly luciferase activity over total luciferase activity. These results suggest that the use of the 2 luciferase reporter genes provides a simple, highly sensitive, and reproducible system for the detection of compounds that increase γ-globin gene expression. It can therefore be used for the screening of chemical agents that may have γ-globin gene inducibility.
Available in vitro techniques for the identification of fetal hemoglobin inducers are based on the use of primary human erythroid cell cultures [BFUe cultures or a 2-phase liquid culture assay].1-5 Peripheral blood mononuclear cells are grown in the presence of the compounds under investigation, and the effects on γ-globin gene expression are evaluated by measuring, in the cultured cells, γ/γ+β mRNA ratios by RNase protection assay2,3 or γ/γ+β protein ratios by globin biosynthesis.1 High-performance liquid chromatography analysis of the synthesized human globins and staining of erythroblasts with globin-specific fluorescent antibodies are also used.1-5 These techniques have led to the discovery of several fetal globin gene inducers, but they are complex and time consuming. The lack of simplicity and efficiency prevents their use as screening methods for large number of compounds.
In this study we describe a simple system for the assessment of globin gene expression that can be used as a screening in vitro assay for potential fetal hemoglobin (HbF) inducers. This is accomplished by using recombinant constructs containing luciferase reporter genes whose activity can be measured by a simple enzymatic assay. In this context, luciferase activity represents human globin gene expression; thus, it can be used for the evaluation of globin gene expression in cells carrying such recombinant DNA constructs. Our results suggest that this system represents a powerful assay that can detect compounds that increase γ-globin gene expression and differentiate their effects from those of general globin gene inducers or agents that do not affect globin gene expression.
Materials and methods
Constructs
To constructμLCRβprRlucAγprFlucplasmids, pRL-null and pGL2-basic, which contain the renilla and firefly luciferase cDNA, respectively, were purchased from Promega (Madison, WI). A 315-bp human β-globin gene promoter sequence (SnaBI–NcoI [blunted]) was inserted upstream of a 1.2-kb renilla luciferase coding sequence fragment (SmaI–XbaI [blunted] from pRL-null) and a 130-bp SV40 polyadenylation signal (HpaI–BamHI fragment from pGL2-basic) was added downstream to generate thePβprRlucconstruct. For the production of thepAγprFluc, a 1.4 kb of human Aγ-globin gene promoter sequence fragment (HindIII–AhaII) was inserted upstream of the 2.7-kb firefly luciferase coding sequence and SV40 polyadenylation signal (HindIII–BamHI from pGL2-basic). For the generation of theμLCRβprRlucAγprFlucconstruct, a 3.1-kb μLCR cassette,6pβprRlucandpAγprFluc were cloned in multiple steps to the pRL-null backbone. BamHI sites partially filled with dGTP and dATP, and XhoI sites partially filled with dTTP and dCTP were ligated to facilitate directional cloning.
Transfection
GM979 cells were cultured in the RPMI-1640 medium (HyCloneLogan, UT) supplemented with 10% fetal calf serum. Stable cell transfection was performed by lipofection. Briefly, the cells were harvested in the logarithmic phase of growth, washed in phosphate-buffered saline, and resuspended in Opti-MEM medium (GIBCO-BRL, Gaithersburg, MD). Eight hundred microliters of a suspension containing 2 × 106 cells were mixed with DNA and lipofectin (GIBCO BRL) and incubated at 37°C for 6 hours, and then 2 mL RPMI-1640 medium with 20% fetal calf serum was added. Cotransfection was accomplished using 2 μg linearizedμLCRβprRlucAγprFlucplasmid and 0.4 μg of PGK-neo plasmid. Lipofectin was used in a 3:1 molar ratio to DNA. After incubation for 24 hours at 37°C, the growth medium was replaced by selection medium containing 900 μg/mL G418. Vigorously growing clumps of cells were observed after 7 to 10 days in culture. Several aliquots of cells were frozen in liquid nitrogen. To test luciferase expression, an aliquot of cells was thawed. Each time cells were maintained in culture in medium containing 400 μg/mL G418, and new aliquots were frozen as soon as possible. To keep our population stable, the cells were not kept in culture for more than 1 to 2 months.
Luciferase assays
Transfected cells were cultured in the presence of increasing concentrations of the various compounds tested (Sigma, St Louis, MO) for 4 days. Firefly and renilla luciferase gene activities were measured sequentially in cell lysates using a commercially available enzymatic assay (Dual Luciferase Reporter Assay System; Promega, Madison, WI) according to the manufacturer's protocol. Measurements were performed in a Lumat LB 9507 luminometer (EG&G Berthold, Germany).
Results
Experimental approach
The goal of this study was to develop a simple and efficient system for the quantitative assessment of globin gene expression. For this purpose we used 2 different luciferase reporter genes, firefly (Photinus pyralis)7 and renilla (Renilla reniformis).8 The activity of each of these genes can be measured sequentially in cell lysates using a simple, commercially available enzymatic assay. The use of the firefly and renilla luciferase genes provides the advantage of extreme sensitivity for both reporter genes (approximately 10−20 and approximately 3 × 10−19 moles of firefly and renilla luciferase, respectively, can be detected) and rapidity of detection. It must be taken into account, however, that in this system, firefly luciferase activity (γF) has approximately 50% greater luminescence than renilla luciferase activity (βR) on a per mole basis. Therefore, adjustments for this difference should be made during analysis of the results. Multiplying the measurements for renilla luciferase activity by a factor of 2 (2βR) adjusts for the difference in measured activity.
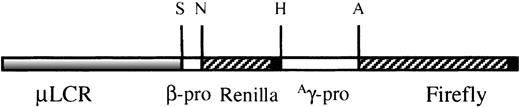
To test the specificity and reproducibility of this approach, the construct shown in Figure 1 was produced. It contains a μLCR cassette linked to a minimal β-globin gene promoter driving the renilla luciferase cDNA and anAγ-globin gene promoter driving the firefly luciferase cDNA. In this context, renilla luciferase gene expression represents β-globin gene expression, whereas firefly luciferase gene expression represents Aγ-globin gene expression.
TheμLCRβprRlucγprFlucconstruct.
It contains a 3.1-kb μLCR cassette linked to a 315-bp human β-globin gene promoter driving the renilla luciferase gene and a 1.4-kb Aγ promoter driving the firefly luciferase gene. S, SnaBI (Genebank, HUMHBB 61869); N, NcoI (Genebank, HUMHBB 62185); H, HindIII (Genebank, HUMBB 388057); A,AhaII (Genebank, HUMHBB 39461). ▪ represent the 130-bp SV40-polyadenylation site.
TheμLCRβprRlucγprFlucconstruct.
It contains a 3.1-kb μLCR cassette linked to a 315-bp human β-globin gene promoter driving the renilla luciferase gene and a 1.4-kb Aγ promoter driving the firefly luciferase gene. S, SnaBI (Genebank, HUMHBB 61869); N, NcoI (Genebank, HUMHBB 62185); H, HindIII (Genebank, HUMBB 388057); A,AhaII (Genebank, HUMHBB 39461). ▪ represent the 130-bp SV40-polyadenylation site.
The μLCR cassette used for the production of theμLCRβprRlucAγprFlucconstruct is a 3.1-kb fragment that contains the DNase I hypersensitive core sequences of 5′HS1, 5′HS2, 5′HS3, and 5′HS4. Using 2 reporter genes in the same construct provided the advantage of assessment of β and γ-globin gene expression and allowed us to differentiate between general globin gene induction and specific γ-globin gene induction. For this purpose, in each experiment the luciferase gene activity in cell lysates was measured for each reporter gene using the enzymatic assay (βR and γF), and the adjusted ratio γF/γF+2βR was calculated. In addition, using this approach of ratio calculations, normalization for protein concentration in the samples was not required. The renilla luciferase cDNA is driven by the native 315-bp β-globin gene promoter, whereas the firefly luciferase cDNA is driven by a 1.4-kbAγ-globin gene promoter that has been shown in previous studies to contain putative butyrate response elements.9Previous studies have also shown that proximity to the LCR increases gene expression, perhaps by influencing the frequency of interaction between the LCR and globin gene promoters10 11; thus, the β promoter was placed between the LCR and the γ promoter. This resulted in lower basal levels of γ gene promoter activity, permitting the detection of a greater range of effects (increase or decrease) on γ-globin gene expression (increased sensitivity of detection).
Specific γ-globin gene inducers can be detected using the 2-luciferase system
TheμLCRβprRlucAγprFlucconstruct shown in Figure 1 was used to stably transfect murine erythroleukemia cells. Murine erythroleukemia cell clone GM979 that expresses embryonic and adult murine globin genes was cotransfected by lipofection with the experimental plasmid and a PGK-neo plasmid conferring resistance to G418. G418-resistant cells were isolated and cultured in increasing concentrations of various compounds for 4 days. Cell lysates were subsequently prepared and analyzed for luciferase activity using the enzymatic assay, and the γF/γF+2βR ratios were calculated.
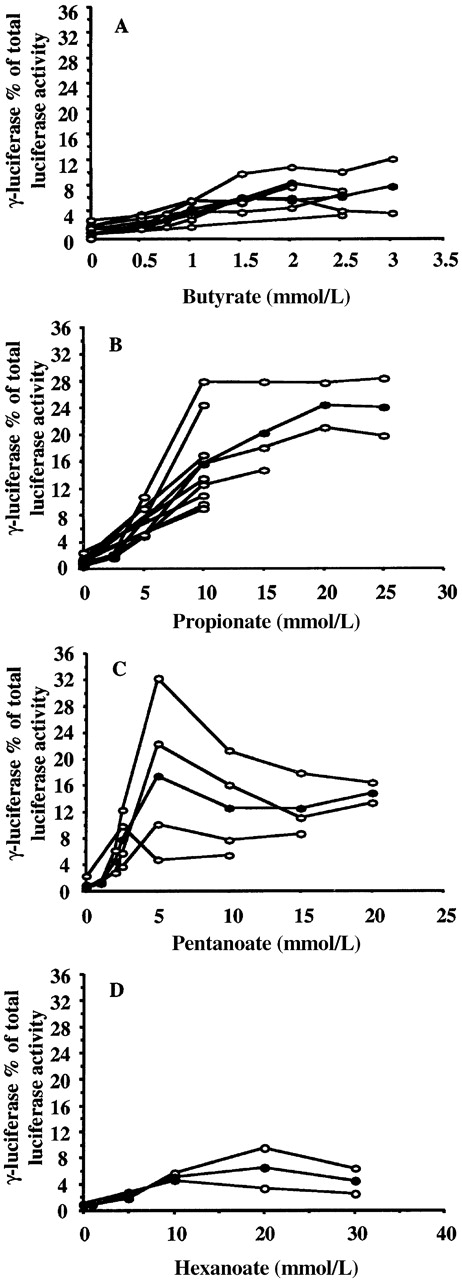
Previously performed studies using BFUe cultures and primates (baboons) have shown that short-chain fatty acids with 2 to 6 carbon atoms have the property of γ-globin gene inducibility.2,3 Therefore, we used 4 compounds to test our experimental system. In Figure 2, dose-response curves for 4 short-chain fatty acids, butyric (panel A), propionic (panel B), pentanoic (panel C), and hexanoic (panel D) are shown. In this figure, γF luciferase activity as a percentage of the adjusted total luciferase activity (γF+2βR) is shown for increasing concentrations of the compound tested. Results from several experiments and the mean values for each short-chain fatty acid are plotted. All 4 compounds increase the γF luciferase activity reaching a plateau phase. Further increases in concentration resulted in the inhibition of cell growth or cell death. However, the effect of each compound is observed at a different concentration range. Notice that butyric acid exerts its effect at low concentrations (1.5-2 mmol/L). Propionic acid induces greater increases in the γFluciferase activity but at concentrations 10 times higher than those for butyric acid; thus, the average γF luciferase activity was 24.32% of total (γF+2βR) luciferase activity at 20 mmol/L. Pentanoic acid is an intermediate inducer that is also active at higher concentrations (average γFluciferase activity 17.35% of total [γF+2βR] luciferase activity at 5 mmol/L) than butyric acid. Finally, hexanoate is the weakest inducer of all 4 compounds (average γF luciferase activity, 6.44% of total (γF+2βR) luciferase activity at 20 mmol/L). These results are in accordance with those obtained in BFUe cultures,3 suggesting that this system is able to identify chemical agents that specifically affect γ-globin gene expression.
Dose-response relationships for 4 short-chain fatty acids.
Butyric (A), propionic (B), pentanoic (C), and hexanoic (D) acid are shown. ○ indicates values from experiments performed on different days. • indicates mean values. All 4 short-chain fatty acids increase the γF/γF+2βR ratio. Higher levels of γF/γF+2βR ratios were observed in propionate- and pentanoate-treated cells, though at higher concentrations than butyrate. Hexanoate was the weakest γ-globin gene inducer among the short-chain fatty acids tested.
Dose-response relationships for 4 short-chain fatty acids.
Butyric (A), propionic (B), pentanoic (C), and hexanoic (D) acid are shown. ○ indicates values from experiments performed on different days. • indicates mean values. All 4 short-chain fatty acids increase the γF/γF+2βR ratio. Higher levels of γF/γF+2βR ratios were observed in propionate- and pentanoate-treated cells, though at higher concentrations than butyrate. Hexanoate was the weakest γ-globin gene inducer among the short-chain fatty acids tested.
Determination of the assay specificity
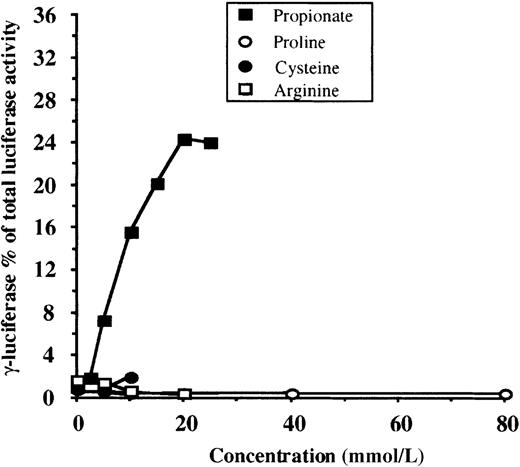
To be useful as a screening method for HbF inducer detection, an assay should differentiate between specific γ-globin gene inducers, general globin gene inducers, and compounds that do not affect globin gene expression. In the latter 2 cases, no change in the adjusted γF/γF+2βR ratio should be observed when transfected cells are exposed to increasing concentrations of the compound. To test the specificity of our system, we studied compounds that are known general globin gene inducers in MEL (dimethyl sulfoxide [DMSO]) or K562 cells (hemin) and “neutral” compounds not expected to have any effect (proline, cysteine, arginine). Firefly (γF) and renilla (βR) luciferase activities, in response to increasing concentrations of DMSO and hemin, are shown in Tables1 and 2, respectively. As expected, neither DMSO nor hemin increased the adjusted γF/γF+2βR ratio. In Figure 3, the results for proline, cysteine, and arginine are shown. Each experiment was performed in triplicate, and means are shown for each data point. The mean dose-response curve for propionate (from Figure 2) was also plotted as reference. Concentrations of cysteine and arginine higher than 10 mmol/L and 20 mmol/L, respectively, were toxic to the cells. Notice that, as expected, no change in the adjusted γF/γF+2βR ratio is observed for any of the compounds. Collectively these results indicate that our system has the required specificity to be used as a screening assay for the detection of potential γ-globin gene inducers.
Dose-response relationships for compounds not anticipated to affect globin gene expression (proline, cysteine, arginine).
For each compound, experiments were performed in triplicate at each concentration shown, and mean values are plotted. Concentrations of cysteine and arginine greater than 10 mmol/L and 20 mmol/L, respectively, were toxic to the cells. ▪ indicates mean values for propionic acid (from Figure 2). Notice that none of the tested compounds increased γ-firefly luciferase activity.
Dose-response relationships for compounds not anticipated to affect globin gene expression (proline, cysteine, arginine).
For each compound, experiments were performed in triplicate at each concentration shown, and mean values are plotted. Concentrations of cysteine and arginine greater than 10 mmol/L and 20 mmol/L, respectively, were toxic to the cells. ▪ indicates mean values for propionic acid (from Figure 2). Notice that none of the tested compounds increased γ-firefly luciferase activity.
Hydroxyl substitutions of butyrate
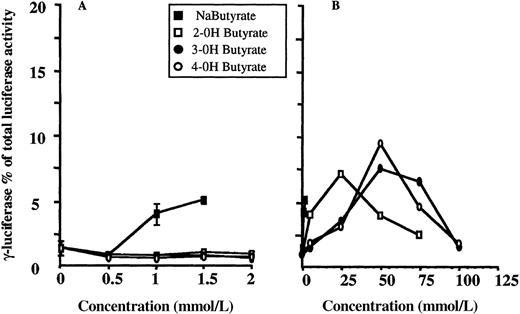
Structural analogues or derivatives of short-chain fatty acids may have the property of γ-globin gene inducibility. We examined whether hydroxyl substitutions of butyrate, the 4-carbon atom short-chain fatty acid, could increase γ-globin gene expression in our system. Results are shown in Figure 4 for 2-, 3-, and 4-hydroxyl-butyrate. Each experiment was performed in triplicate, and means are shown for each data point. Results for control cells cultured in the presence of concentrations of butyrate known to increase γ-globin gene expression in this system are also plotted. As shown in panel A, all 3 compounds failed to induce at the concentrations at which butyrate is active. Interestingly, a significant increase in the adjusted γF/γF+2βR ratio is observed at 10-fold higher concentrations but seems to disappear after a certain concentration is reached (50 mmol/L, 75 mmol/L, and 75 mmol/L for 2-, 3-, and 4-hydroxyl-butyrate, respectively). At the same concentration a significant effect on cell growth is also observed.
Dose-response relationships for 2-, 3-, and 4-hydroxylbutyrate.
(A, B) Results from experiments with low and high concentrations of the compounds, respectively, are shown. For each compound, experiments were performed in triplicate for each concentration shown, and means are plotted. ▪ indicates dose-response relationship for control experiment with sodium butyrate. Experiments were performed in triplicate at each concentration, and means and standard deviations are plotted. Concentrations greater than 2 mmol/L were toxic to the cells.
Dose-response relationships for 2-, 3-, and 4-hydroxylbutyrate.
(A, B) Results from experiments with low and high concentrations of the compounds, respectively, are shown. For each compound, experiments were performed in triplicate for each concentration shown, and means are plotted. ▪ indicates dose-response relationship for control experiment with sodium butyrate. Experiments were performed in triplicate at each concentration, and means and standard deviations are plotted. Concentrations greater than 2 mmol/L were toxic to the cells.
Discussion
A large body of evidence indicates that patients with β-chain hemoglobinopathies who have a genetic propensity for persistent fetal hemoglobin production have milder clinical phenotypes.12,13Elevated levels of fetal hemoglobin improve the severity of sickle cell disease through the inhibition of intracellular polymerization of sickle hemoglobin.12 In β-thalassemia increased synthesis of γ-globin chains is anticipated to reduce the existing globin chain imbalance, thus ameliorating anemia in affected persons.13In the last 2 decades several agents have been explored as potential therapeutic modalities for pharmacologic up-regulation of fetal hemoglobin in vitro and in vivo.
Initial attempts to induce the synthesis of fetal hemoglobin used 5-azacytidine,14,15 a compound known to induce DNA demethylation, under the hypothesis that demethylation results in reactivation of the silent γ genes in the adult. γ gene inducibility by 5-azacytidine was subsequently attributed to the fast erythroid regeneration that follows the 5-azacytidine–caused cytoreduction, under the hypothesis that rapid erythroid regeneration induces the recruitment of progenitor cells with a fetal globin program.16 This was shown in experiments in baboons treated with arabinosylcytosine (AraC), a compound that produces a post-cytoreduction regeneration without affecting DNA methylation.17 Other cytotoxic compounds, such as hydroxyurea18 and vinblastin,19 were also shown to induce fetal hemoglobin in primates. Hydroxyurea was subsequently shown to be an effective inducer of fetal hemoglobin in several patients with sickle cell disease,20,21 and this was confirmed by extensive clinical trials.22-25
The next class of compounds investigated as potential γ-globin gene inducers was butyric acid and its analogues. Co-administration of 5-azacytidine and sodium butyrate was shown to induce embryonic globin expression in the chicken,26 whereas studies in sheep demonstrated that continuous infusion of butyrate to sheep fetuses inhibits the perinatal γ to β switch.27 The administration of butyrate or α-aminobutyric acid induced fetal hemoglobin synthesis in juvenile baboons and in cultures of human erythroid progenitors.28,29 In subsequent phase 1 clinical trials in patients with severe β-thalassemia, intravenous infusions of arginine butyrate were initially considered to result in the consistent induction of fetal hemoglobin and significant hematologic improvement.30 However, the effectiveness of this treatment was subsequently disputed.31 The reasons for the discrepant results remain unclear. Phenylbutyrate was also shown to induce low levels of fetal hemoglobin in humans.32 Induction of fetal hemoglobin has also been reported in the presence of increased 3-hydroxybutyric acid associated with β-ketothiolase deficiency33 and in young women with starvation ketosis.34 Experiments in erythroid cell cultures and in primates have shown that, in addition to butyrate, short-chain fatty acids containing 2 to 9 carbon atoms stimulate fetal hemoglobin synthesis.2,3 In vivo induction of fetal hemoglobin by propionic acid (a 3-carbon fatty acid) has also been observed in patients with propionic acidemia.35 In addition, valproic acid, a pentanoic acid derivative used as an anticonvulsant, induces fetal hemoglobin in one third of treated patients.36
Collectively, these data suggest that structural analogues of short-chain fatty acids and derivatives already approved for human use may have the property of γ-globin gene inducibility and may be useful for the treatment of β-hemoglobinopathies; these compounds are awaiting testing. Efforts have been impeded by the lack of a simple, efficient assay for the detection of potential hemoglobin inducers among large numbers of pharmacologic compounds.
The assay described in this study represents a simple system that can be used for quantitative assessment of globin gene expression. In this system, luciferase reporter genes substitute for the coding sequences of globin genes, and their activity, measured by a simple enzymatic assay, represents globin gene expression. Measurement of luciferase activity for both reporter genes is highly sensitive, simple, and fast. Cells transfected with the experimental recombinant DNA construct (μLCRβprRlucAγprFluc) are cultured in the presence of increasing concentrations of the compound under investigation, and lysates from the cells are prepared and assayed for luciferase activity. Several compounds and multiple concentrations of each compound can be tested simultaneously. The system can differentiate the effects of specific γ-globin gene inducers (eg, butyric, propionic, and pentanoic acid) from general globin gene inducers (eg, DMSO) or compounds than do not affect globin gene expression (eg, proline, cysteine, arginine). Therefore, it can be used as a screening assay for compounds that may be able to induce fetal hemoglobin. Many derivatives of propionic, butyric, and pentanoic acid can be screened using our system, and the structural features of short-chain fatty acids underlying their ability to induce fetal hemoglobin can be delineated.
To examine whether polar substitutions affect the ability of sodium butyrate to induce hemoglobin F production, we tested the effect of 2-, 3-, and 4-hydroxyl butyrate on luciferase expression in this system. Our data suggest that hydroxyl substitutions considerably attenuate the ability of butyric acid to induce γ-globin gene expression. Significantly higher concentrations are needed for an effect to be observed. Previously published studies34 35 suggest that 3-hydroxybutyric acid is associated with increased HbF production. However, in accordance with our results, sustained high concentrations of the compound were necessary for the effect to occur.
It is likely that the mechanism of action of HbF inducers involves either the activation of γ gene transcription or the prevention of γ gene silencing. Whatever the mechanism is, these agents should act by ultimately affecting cis elements of the γ-globin gene or the LCR. Our system may not be able to detect compounds that involvecis elements absent from this construct. Nevertheless, the rapidity of the assay, which allows for the initial screening of large libraries of potential HbF inducers, may compensate for this. A β-locus Yeast Artificial Chromosome (YAC) in which the firefly and renilla luciferase genes substitute for theAγ and β globin gene sequences could be used to overcome this limitation because the effect of compounds on globin gene expression would be assessed in the context of the intact β-locus. Results from a published study37 from our group however, indicated that the LCR of a 155-kb β-YAC cannot function properly when the locus is directly transferred to an erythroid cell environment, precluding the use of YACs in our system.
Our strategy can be applied, though, for the investigation of mechanisms that influence γ-globin gene expression on induction by pharmacologic compounds. For the identification of the involvedcis elements, constructs carrying the truncatedAγ-globin gene promoter driving the luciferase gene and/or various LCR cassettes can be used to stably transfect GM979 cells. If a specific sequence is required for the induction of γ gene expression by a compound, cells that carry constructs in which the sequence is mutated will fail to increase the adjusted γF/γF+2βR ratio in response to the compound.
A limitation of our system is the significant degree of variability in induction observed among experiments performed on different days. This is particularly obvious for sodium butyrate and may be attributed to its effect on cell growth. However, when experiments were performed in triplicate on the same day, such variability was not observed. To overcome this problem, multiple validation assays should be performed to determine the effect of a potential inducer.
In conclusion, our data show that the use of luciferase reporter genes provides a useful system for the assessment of globin gene expression. Compared to the currently used methods for the assessment of human globin gene expression, this system is efficient and less time consuming. By using the in vitro assay, many compounds can effectively be tested. Subsequently, the in vivo effects of compounds that specifically induce γ-globin gene expression in the in vitro system can be evaluated, and valuable information regarding their potential use as therapeutic agents for patients with hemoglobinopathies can be obtained. In addition, our method can be applied for the identification of regulatory elements involved in the pharmacologic induction of HbF, providing useful insight into the possible mechanism of action of these compounds.
Supported by National Institutes of Health grants HL20899 and DK45365, a Cooley's Anemia Foundation Award (E.S.), and an International Fogarty Fellowship Award (G.V.).
Reprints:George Stamatoyannopoulos, Division of Medical Genetics, University of Washington, Box 357720, Seattle, WA 98195; e-mail: gstam@u.washington.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.