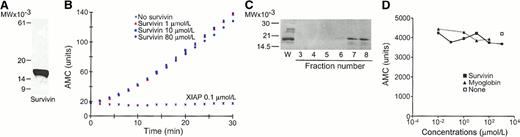
Conway et al recently reported that alternatively spliced forms of mouse survivin exhibit different antiapoptotic properties.1 This was inferred from inhibition of recombinant caspase-3 catalytic activity using a standard chromogenic assay in the presence of increasing concentrations of mammalian-expressed murine survivin isoforms of 140, 121, and 40 amino acids, respectively.1 If confirmed, the ability of survivin to inhibit caspase-3 activity would have a major impact on targeting this ubiquitous cytoprotection pathway in cancer2,3 and during cell cycle progression.4But the data of survivin inhibition of caspase-3 activity presented by Conway et al raise serious concerns of specificity.1First, the experiments contained no controls with genuine inhibitors of caspase-3, including DEVD-CHO or another IAP family protein with well-documented anti–caspase-3 activity (ie, XIAP). Second, from the data presented it was impossible to derive an inhibition constant (Ki) of the cleavage reaction, which is indispensable to quantitatively characterize potential caspase inhibitors.5We have now reinvestigated the data presented by Conway et al1 and attempted to reproduce their caspase-3 inhibition experiments using mouse or human survivin proteins. Mouse recombinant survivin was expressed, purified to homogeneity (Figure1A), and properly folded by 1D-NMR analysis. Concentrations of recombinant mouse survivin up to 80 μmol/L failed to decrease purified, recombinant caspase-3 activity using a peptide substrate cleavage assay similar to that reported by Conway et al1 (Figure 1B). In contrast, 0.1 μmol/L XIAP completely inhibited caspase-3 activity, in agreement with previous data.5 In an attempt to reproduce exactly the mammalian cell expression approach used by Conway et al1, we immunoaffinity-purified native survivin from Jurkat T cells. Eluted fractions contained a single 16.5-kd survivin band by immunoblotting with an antibody to survivin (Figure 1C). But increasing concentrations of native, immunoaffinity-purified survivin did not affect caspase-3 catalytic activity, as determined by substrate peptide cleavage (Figure 1D).
Survivin does not inhibit caspase-3 activity.
(A) SDS gel electrophoresis of purified mouse recombinant survivin. (B) Effect of murine survivin or human XIAP on caspase-3 activity by hydrolysis of the fluorogenic substrate, DEVD-AMC. (C) Western blotting of immunoaffinity-purified human survivin isolated from Jurkat T cells by chromatography on α-survivin-sepharose. (D) Effect of native human survivin on caspase-3 activity by hydrolysis of the fluorogenic substrate, DEVD-AMC. W, whole cell extracts.
Survivin does not inhibit caspase-3 activity.
(A) SDS gel electrophoresis of purified mouse recombinant survivin. (B) Effect of murine survivin or human XIAP on caspase-3 activity by hydrolysis of the fluorogenic substrate, DEVD-AMC. (C) Western blotting of immunoaffinity-purified human survivin isolated from Jurkat T cells by chromatography on α-survivin-sepharose. (D) Effect of native human survivin on caspase-3 activity by hydrolysis of the fluorogenic substrate, DEVD-AMC. W, whole cell extracts.
In conclusion, our data strongly argue against any role of survivin in directly inhibiting caspase-3 activity, at variance with the preliminary work of Conway et al.1 These discrepancies cannot be ascribed to species specificity, protein purification, or differences in experimental protocol. Moreover, available structural data demonstrate that a linker region upstream of the second baculovirus IAP repeat is required for docking IAP proteins (ie, XIAP) to active caspase-3.6 This linker region is absent in survivin, thus further weakening the hypothesis proposed by Conway et al.1 Therefore, the general conclusion of Conway et al1 that alternatively spliced isoforms of survivin have different antiapoptotic functions is not experimentally substantiated. The means by which survivin participates in the apoptosis balance in cancer and during cell cycle progression require further investigation.
Evidence that survivin inhibits caspase-3 activity
Earlier this year, we provided a definitive report indicating that there exist at least 3 murine survivin mRNA variants, each encoding a distinct protein.1-1 We demonstrated, with appropriate controls, that both survivin140 and survivin121 are able to inhibit caspase-3 activity, while survivin40 does not.
In contrast to the claims of Altieri's group in their letter, several reports support our finding that survivin interferes with caspase-3 activity. These include the following: Reed's group reported that “survivin was able to substantially reduce caspase activity, as measured by cleavage of a tetrapeptide substrate, AspGluValAsp-aminofluorocoumarin. Similar results were obtained in intact cells when Survivin was overexpressed.”2(p5315)Altieri's group noted in their Nature paper that “like other IAP proteins, survivin inhibits the terminal effectors caspase-3 and caspase-7.”3(p583) Kobayashi et al demonstrated that overexpression of murine survivin in Rat1 cells inhibited caspase-induced cell death and also that a purified GST fusion protein encoding murine survivin could bind directly to caspase-3.1-4 This group further transfected Jurkat cells with epitope-tagged survivin and showed by immunoprecipitation with anti–caspase-3 antibodies that survivin “can bind efficiently to processed caspase 3.”4(p1460) In theirNature Cell Biology paper, Altieri's group once again noted that survivin regulates apoptosis via caspase-3: “Expression of survivin (C84A) or survivin antisense cDNA also resulted in increased activity of the apoptosis effector caspase-3, as judged by hydrolysis of the fluorogenic caspase-3 substrate.”5(p461)Altieri's group also performed studies on cultured endothelial cells and reported that “[r]ecombinant expression of green fluorescent protein survivin in endothelial cells reduced caspase-3 activity and counteracted apoptosis induced by tumor necrosis factor.”6(p393) And finally, a recent report confirms that the down-regulation of survivin mRNA levels by an antisense approach results in increased caspase-3 activity.1-7
We note that many of the data indicating that survivin interferes with caspase-3 activity predate our own and, in several cases, have actually been provided by Altieri's group. We are not in a position to assess fully the experiments described by Altieri's group and would point out only that they are not identical to our own, in contrast to what that group claims. In addition, we would add that we have observed that the activity of recombinant survivin is highly dependent on the method chosen for its synthesis and purification. Based on our experiments and the current published data, we believe that specific forms of survivin do inhibit caspase-3 activity. Nevertheless, we agree that the means by which survivin participates in the apoptosis balance requires further investigation, and we hypothesize that the alternatively spliced isoforms of survivin may modulate apoptosis differentially.
References
Supported in part by National Institutes of Health grant CA78810.